

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.11 - phase: 0
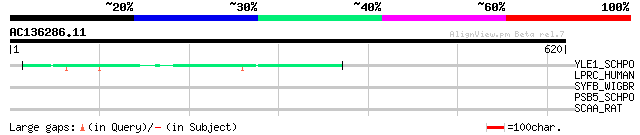
(620 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YLE1_SCHPO (Q10451) Hypothetical protein C1093.01 in chromosome I 51 8e-06
LPRC_HUMAN (P42704) 130 kDa leucine-rich protein (LRP 130) (GP13... 40 0.014
SYFB_WIGBR (Q8D3B5) Phenylalanyl-tRNA synthetase beta chain (EC ... 33 1.7
PSB5_SCHPO (P30655) Proteasome subunit beta type 5 precursor (EC... 32 3.8
SCAA_RAT (P37089) Amiloride-sensitive sodium channel alpha-subun... 32 6.5
>YLE1_SCHPO (Q10451) Hypothetical protein C1093.01 in chromosome I
Length = 1261
Score = 51.2 bits (121), Expect = 8e-06
Identities = 79/380 (20%), Positives = 154/380 (39%), Gaps = 54/380 (14%)
Query: 15 LYKEALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLF-----KTGFHSH 69
+++ + +LY + + S + ++L A LSS Q ++LF G+
Sbjct: 829 VFEHSKHLYRKISTKSLEKANWFMALILDAMI-LSSSFARQFKSSNLFCDNMKMLGYIPR 887
Query: 70 PHTSTALI--ASYAANTRSFHYALELFDEMPQ----PTITAFNAVLSGLSRNGPRGQAVW 123
T LI ++ +T AL +F+E + P++ +NAVLS L R +
Sbjct: 888 ASTFAHLINNSTRRGDTDDATTALNIFEETKRHNVKPSVFLYNAVLSKLGRARRTTECWK 947
Query: 124 LFRQIGFWNIRPNSVTIVSLLSARDVKNQSHVQQVHCLACKLGVEYDVYVSTSLVTAYSK 183
LF+++ + P SVT ++++A AC++G E S
Sbjct: 948 LFQEMKESGLLPTSVTYGTVINA---------------ACRIGDE-------------SL 979
Query: 184 CGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVS 243
L + + N + + V YN + +Q F+R + + + +P+ T
Sbjct: 980 AEKLFAEMENQPNYQPR-VAPYNTMIQFEVQTMFNREKALFYYNRLCATDIEPSSHTYKL 1038
Query: 244 VVSACATLSNIRLGK-----------QVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSA 292
++ A TL + +G V LSM A H++ + D+ + C+ +A
Sbjct: 1039 LMDAYGTLKPVNVGSVKAVLELMERTDVPILSMHYAAYIHILG-NVVSDVQAATSCYMNA 1097
Query: 293 FDVFSRSE-KRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQ 351
E + + + S I ++ N +++ M + ++ N+LI GF +
Sbjct: 1098 LAKHDAGEIQLDANLFQSQIESLIANDRIVEGIQIVSDMKRYNVSLNAYIVNALIKGFTK 1157
Query: 352 KGVCVEAFKYFSKMQCAGVA 371
G+ +A YF ++C G++
Sbjct: 1158 VGMISKARYYFDLLECEGMS 1177
Score = 34.3 bits (77), Expect = 1.00
Identities = 22/90 (24%), Positives = 39/90 (42%)
Query: 453 GTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFRMIRKYGLDPKP 512
G D +A +F E V+P+ + +VLS + + + F+ +++ GL P
Sbjct: 902 GDTDDATTALNIFEETKRHNVKPSVFLYNAVLSKLGRARRTTECWKLFQEMKESGLLPTS 961
Query: 513 EHFGCVVDLLGRAGQLGEARDLVQELAEPP 542
+G V++ R G A L E+ P
Sbjct: 962 VTYGTVINAACRIGDESLAEKLFAEMENQP 991
>LPRC_HUMAN (P42704) 130 kDa leucine-rich protein (LRP 130) (GP130)
(Leucine-rich PPR-motif containing protein)
Length = 1273
Score = 40.4 bits (93), Expect = 0.014
Identities = 26/116 (22%), Positives = 52/116 (44%), Gaps = 2/116 (1%)
Query: 415 LVDTYMKCGCVSFARFVFDQFDVK--PDDPAFWNAMIGGYGTNGDYESAFEVFYEMLDEM 472
L+ +Y G + A + K P A ++A++ G+ GD E+A + M D
Sbjct: 82 LIASYCNVGDIEGASKILGFMKTKDLPVTEAVFSALVTGHARAGDMENAENILTVMRDAG 141
Query: 473 VQPNSATFVSVLSACSHSGQIERGLRFFRMIRKYGLDPKPEHFGCVVDLLGRAGQL 528
++P T++++L+A + G I+ + + K+ L ++ +AG L
Sbjct: 142 IEPGPDTYLALLNAYAEKGDIDHVKQTLEKVEKFELHLMDRDLLQIIFSFSKAGYL 197
Score = 33.9 bits (76), Expect = 1.3
Identities = 26/116 (22%), Positives = 51/116 (43%), Gaps = 4/116 (3%)
Query: 441 DPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFF 500
D + +NA++ Y N S + +M + +QPN T+ ++++ + G IE +
Sbjct: 40 DVSHYNALLKVYLQNEYKFSPTDFLAKMEEANIQPNRVTYQRLIASYCNVGDIEGASKIL 99
Query: 501 RMIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQEL----AEPPASVFDSLLGA 552
++ L F +V RAG + A +++ + EP + +LL A
Sbjct: 100 GFMKTKDLPVTEAVFSALVTGHARAGDMENAENILTVMRDAGIEPGPDTYLALLNA 155
>SYFB_WIGBR (Q8D3B5) Phenylalanyl-tRNA synthetase beta chain (EC
6.1.1.20) (Phenylalanine--tRNA ligase beta chain)
(PheRS)
Length = 803
Score = 33.5 bits (75), Expect = 1.7
Identities = 30/135 (22%), Positives = 54/135 (39%), Gaps = 2/135 (1%)
Query: 124 LFRQIGFWNIRPNSVTIVSLLSARD--VKNQSHVQQVHCLACKLGVEYDVYVSTSLVTAY 181
L+ +GF+ + I+ LL + +KN V C + +E D+ S +
Sbjct: 418 LYEILGFFISKKLIENILLLLGYKYEFIKNYWKVSVPSWRMCNVKIEEDLISDISRFYGF 477
Query: 182 SKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTL 241
K + +S K+ N N+ T N L G+ ++ F D + PNK+ +
Sbjct: 478 EKIPIDLSYKKIKYNNCKYNINTLNNAKKLLTNRGYQEIISYTFVDKNIQNIIHPNKIPV 537
Query: 242 VSVVSACATLSNIRL 256
+ +S +RL
Sbjct: 538 PIINPISTEMSVLRL 552
>PSB5_SCHPO (P30655) Proteasome subunit beta type 5 precursor (EC
3.4.25.1) (Proteasome component pts1) (Macropain subunit
pts1) (Multicatalytic endopeptidase complex subunit
pts1)
Length = 272
Score = 32.3 bits (72), Expect = 3.8
Identities = 25/78 (32%), Positives = 37/78 (47%), Gaps = 2/78 (2%)
Query: 189 SSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVF-DVFKDMTMNLEEKPNKVTLVSVVSA 247
S+ + + VK V+ N ++ G L G F + M L + NK L+SV +A
Sbjct: 82 SAGPLIASQTVKKVIEINPYLLGTLAGGAADCQFWETVLGMECRLHQLRNK-ELISVSAA 140
Query: 248 CATLSNIRLGKQVHGLSM 265
LSNI + +GLSM
Sbjct: 141 SKILSNITYSYKGYGLSM 158
>SCAA_RAT (P37089) Amiloride-sensitive sodium channel alpha-subunit
(Epithelial Na+ channel alpha subunit) (Alpha ENaC)
(Nonvoltage-gated sodium channel 1 alpha subunit)
(SCNEA) (Alpha NaCH)
Length = 698
Score = 31.6 bits (70), Expect = 6.5
Identities = 24/108 (22%), Positives = 49/108 (45%), Gaps = 6/108 (5%)
Query: 173 VSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNL 232
++ L YS+ + S + +FE L ++N T N +G+ + FK++
Sbjct: 509 INYKLSAGYSRWPSVKSQDWIFEMLSLQNNYTINNKRNGVAKLNIF------FKELNYKT 562
Query: 233 EEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLV 280
+ VT+VS++S + ++ G V + E ++V+T L+
Sbjct: 563 NSESPSVTMVSLLSNLGSQWSLWFGSSVLSVVEMAELIFDLLVITLLM 610
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,152,804
Number of Sequences: 164201
Number of extensions: 2860767
Number of successful extensions: 6590
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 6574
Number of HSP's gapped (non-prelim): 15
length of query: 620
length of database: 59,974,054
effective HSP length: 116
effective length of query: 504
effective length of database: 40,926,738
effective search space: 20627075952
effective search space used: 20627075952
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC136286.11


