

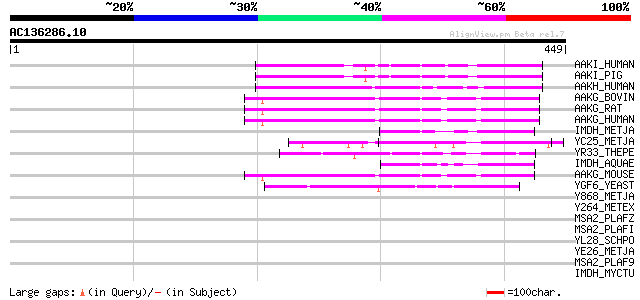
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.10 + phase: 0
(449 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AAKI_HUMAN (Q9UGI9) 5'-AMP-activated protein kinase, gamma-3 sub... 65 4e-10
AAKI_PIG (Q9MYP4) 5'-AMP-activated protein kinase, gamma-3 subun... 64 6e-10
AAKH_HUMAN (Q9UGJ0) 5'-AMP-activated protein kinase, gamma-2 sub... 62 3e-09
AAKG_BOVIN (P58108) 5'-AMP-activated protein kinase, gamma-1 sub... 50 9e-06
AAKG_RAT (P80385) 5'-AMP-activated protein kinase, gamma-1 subun... 50 1e-05
AAKG_HUMAN (P54619) 5'-AMP-activated protein kinase, gamma-1 sub... 50 1e-05
IMDH_METJA (Q59011) Inosine-5'-monophosphate dehydrogenase (EC 1... 49 3e-05
YC25_METJA (Q58622) Hypothetical protein MJ1225 49 3e-05
YR33_THEPE (P15889) Hypothetical 33.4 kDa protein in ribosomal R... 48 5e-05
IMDH_AQUAE (O67820) Inosine-5'-monophosphate dehydrogenase (EC 1... 45 3e-04
AAKG_MOUSE (O54950) 5'-AMP-activated protein kinase, gamma-1 sub... 45 3e-04
YGF6_YEAST (P53172) Hypothetical 58.1 kDa protein in UBC2-OLE1 i... 44 8e-04
Y868_METJA (Q58278) Hypothetical protein MJ0868 43 0.001
Y264_METEX (Q8GEK8) Hypothetical UPF0264 protein ORF22 43 0.001
MSA2_PLAFZ (Q03645) Merozoite surface antigen 2 precursor (MSA-2) 43 0.002
MSA2_PLAFI (Q03644) Merozoite surface antigen 2 precursor (MSA-2) 43 0.002
YL28_SCHPO (Q10343) Hypothetical protein C1556.08c in chromosome I 42 0.004
YE26_METJA (Q58821) Hypothetical protein MJ1426 42 0.004
MSA2_PLAF9 (Q03994) Merozoite surface antigen 2 precursor (MSA-2) 41 0.006
IMDH_MYCTU (P65167) Inosine-5'-monophosphate dehydrogenase (EC 1... 40 0.009
>AAKI_HUMAN (Q9UGI9) 5'-AMP-activated protein kinase, gamma-3
subunit (AMPK gamma-3 chain) (AMPK gamma3)
Length = 464
Score = 65.1 bits (157), Expect = 4e-10
Identities = 54/238 (22%), Positives = 111/238 (45%), Gaps = 23/238 (9%)
Query: 200 ILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVN 259
I + E K T R I + P V ++ N ++ + L K R+ +PV++P ++++
Sbjct: 236 IYEIEQHKIETWREIYLQGCFKPLVSISPNDSLFEAVYTLIKNRIHRLPVLDPVSGNVLH 295
Query: 260 FITQSAVIQGLEGCRGRDWFDCIAARP------MADLGLPFMSADKVISIQSNELILEAF 313
+T +++ L F + RP + DLG+ D + +++ IL A
Sbjct: 296 ILTHKRLLKFLH------IFGSLLPRPSFLYRTIQDLGIGTFR-DLAVVLETAP-ILTAL 347
Query: 314 KIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASY 373
I D ++ LPVV + +VG S D+ +L + N +++V + +++
Sbjct: 348 DIFVDRRVSALPVVNECGQ-VVGLYSRFDVIHLAAQQTY--NHLDMSVGEALRQRTLC-- 402
Query: 374 ESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEP 431
+ ++C+P +L VI +A + +HR+ V+ ++GV++L D++ + P
Sbjct: 403 ----LEGVLSCQPHESLGEVIDRIAREQVHRLVLVDETQHLLGVVSLSDILQALVLSP 456
>AAKI_PIG (Q9MYP4) 5'-AMP-activated protein kinase, gamma-3 subunit
(AMPK gamma-3 chain) (AMPK gamma3)
Length = 514
Score = 64.3 bits (155), Expect = 6e-10
Identities = 54/238 (22%), Positives = 108/238 (44%), Gaps = 23/238 (9%)
Query: 200 ILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVN 259
I + E K T R I + P V ++ N ++ + L K R+ +PV++P +++
Sbjct: 286 IYEIEEHKIETWREIYLQGCFKPLVSISPNDSLFEAVYALIKNRIHRLPVLDPVSGAVLH 345
Query: 260 FITQSAVIQGLEGCRGRDWFDCIAARP------MADLGLPFMSADKVISIQSNELILEAF 313
+T +++ L F + RP + DLG+ D + +++ IL A
Sbjct: 346 ILTHKRLLKFLH------IFGTLLPRPSFLYRTIQDLGIGTFR-DLAVVLETAP-ILTAL 397
Query: 314 KIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASY 373
I D ++ LPVV + +VG S D+ +L + N ++ V + +++
Sbjct: 398 DIFVDRRVSALPVVNETGQ-VVGLYSRFDVIHLAAQQTY--NHLDMNVGEALRQRTLC-- 452
Query: 374 ESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEP 431
+ ++C+P TL VI + + +HR+ V+ ++GV++L D++ + P
Sbjct: 453 ----LEGVLSCQPHETLGEVIDRIVREQVHRLVLVDETQHLLGVVSLSDILQALVLSP 506
>AAKH_HUMAN (Q9UGJ0) 5'-AMP-activated protein kinase, gamma-2
subunit (AMPK gamma-2 chain) (AMPK gamma2) (H91620p)
Length = 569
Score = 62.0 bits (149), Expect = 3e-09
Identities = 52/232 (22%), Positives = 105/232 (44%), Gaps = 11/232 (4%)
Query: 200 ILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVN 259
I + E K T R + + P V ++ ++++ + L K ++ +PVI+P + +
Sbjct: 338 IYELEEHKIETWRELYLQETFKPLVNISPDASLFDAVYSLIKNKIHRLPVIDPISGNALY 397
Query: 260 FITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDN 319
+T +++ L+ + + +LG+ + + I + I++A I +
Sbjct: 398 ILTHKRILKFLQLFMSDMPKPAFMKQNLDELGIG--TYHNIAFIHPDTPIIKALNIFVER 455
Query: 320 QIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVT 379
+I LPVV+ K +V S D+ + L E N ++TV ++ + Y G V
Sbjct: 456 RISALPVVDESGK-VVDIYSKFDV--INLAAEKTYNNLDITVTQALQH--RSQYFEGVVK 510
Query: 380 RPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEP 431
C L++++ + +HR+ VN D +VG+I+L D++ I P
Sbjct: 511 ----CNKLEILETIVDRIVRAEVHRLVVVNEADSIVGIISLSDILQALILTP 558
>AAKG_BOVIN (P58108) 5'-AMP-activated protein kinase, gamma-1
subunit (AMPK gamma-1 chain) (AMPKg)
Length = 330
Score = 50.4 bits (119), Expect = 9e-06
Identities = 47/242 (19%), Positives = 105/242 (42%), Gaps = 15/242 (6%)
Query: 191 NLGEDFYKVILQE----EPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRN 246
N+ +YK L + E K T R + + P V ++ N+++ + L + ++
Sbjct: 93 NILHRYYKSALVQIYELEEHKIETWREVFLQDSFKPLVCISPNASLFDAVSSLIRNKIHR 152
Query: 247 VPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSN 306
+PVI+P + + +T +++ L+ ++ + +L + + + +++
Sbjct: 153 LPVIDPESGNTLYILTHKRILKFLKLFITEFPKPEFMSKSLEELQIGTYA--NIAMVRTT 210
Query: 307 ELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMK 366
+ A I +++ LPVV+ + +V S D+ L + + NL V
Sbjct: 211 TPVYVALGIFVQHRVSALPVVDEKGR-VVDIYSKFDVINLAAE----KTYNNLDVSVTKA 265
Query: 367 KIVSASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISC 426
+ Y G + C TL+++I+ L +HR+ V+ D V G+++L D++
Sbjct: 266 LQHRSHYFEGV----LKCYLHETLETIINRLVEAEVHRLVVVDENDVVKGIVSLSDILQA 321
Query: 427 FI 428
+
Sbjct: 322 LV 323
>AAKG_RAT (P80385) 5'-AMP-activated protein kinase, gamma-1 subunit
(AMPK gamma-1 chain) (AMPKg)
Length = 330
Score = 50.1 bits (118), Expect = 1e-05
Identities = 47/242 (19%), Positives = 105/242 (42%), Gaps = 15/242 (6%)
Query: 191 NLGEDFYKVILQE----EPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRN 246
N+ +YK L + E K T R + + P V ++ N+++ + L + ++
Sbjct: 92 NILHRYYKSALVQIYELEEHKIETWREVYLQDSFKPLVCISPNASLFDAVSSLIRNKIHR 151
Query: 247 VPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSN 306
+PVI+P + + +T +++ L+ ++ + +L + + + +++
Sbjct: 152 LPVIDPESGNTLYILTHKRILKFLKLFITEFPKPEFMSKSLEELQIGTYA--NIAMVRTT 209
Query: 307 ELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMK 366
+ A I +++ LPVV+ + +V S D+ L + + NL V
Sbjct: 210 TPVYVALGIFVQHRVSALPVVDEKGR-VVDIYSKFDVINLAAE----KTYNNLDVSVTKA 264
Query: 367 KIVSASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISC 426
+ Y G + C TL+++I+ L +HR+ V+ D V G+++L D++
Sbjct: 265 LQHRSHYFEGV----LKCYLHETLEAIINRLVEAEVHRLVVVDEHDVVKGIVSLSDILQA 320
Query: 427 FI 428
+
Sbjct: 321 LV 322
>AAKG_HUMAN (P54619) 5'-AMP-activated protein kinase, gamma-1
subunit (AMPK gamma-1 chain) (AMPKg)
Length = 331
Score = 50.1 bits (118), Expect = 1e-05
Identities = 47/242 (19%), Positives = 105/242 (42%), Gaps = 15/242 (6%)
Query: 191 NLGEDFYKVILQE----EPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRN 246
N+ +YK L + E K T R + + P V ++ N+++ + L + ++
Sbjct: 93 NILHRYYKSALVQIYELEEHKIETWREVYLQDSFKPLVCISPNASLFDAVSSLIRNKIHR 152
Query: 247 VPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSN 306
+PVI+P + + +T +++ L+ ++ + +L + + + +++
Sbjct: 153 LPVIDPESGNTLYILTHKRILKFLKLFITEFPKPEFMSKSLEELQIGTYA--NIAMVRTT 210
Query: 307 ELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMK 366
+ A I +++ LPVV+ + +V S D+ L + + NL V
Sbjct: 211 TPVYVALGIFVQHRVSALPVVDEKGR-VVDIYSKFDVINLAAE----KTYNNLDVSVTKA 265
Query: 367 KIVSASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISC 426
+ Y G + C TL+++I+ L +HR+ V+ D V G+++L D++
Sbjct: 266 LQHRSHYFEGV----LKCYLHETLETIINRLVEAEVHRLVVVDENDVVKGIVSLSDILQA 321
Query: 427 FI 428
+
Sbjct: 322 LV 323
>IMDH_METJA (Q59011) Inosine-5'-monophosphate dehydrogenase (EC
1.1.1.205) (IMP dehydrogenase) (IMPDH) (IMPD)
Length = 496
Score = 48.9 bits (115), Expect = 3e-05
Identities = 31/126 (24%), Positives = 63/126 (49%), Gaps = 23/126 (18%)
Query: 300 VISIQSNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNL 359
VI++ ++ + EA +M I GLPVV+ K +VG ++ RD++
Sbjct: 100 VITVSPDDTVGEAINVMETYSISGLPVVDNEDK-LVGIITHRDVK--------------- 143
Query: 360 TVMDFMKKIVSASYESGKVTRPITC-KPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVI 418
+ D KK+ +T+ + C K D + + + + + R+ V+ +++++G+I
Sbjct: 144 AIEDKTKKVKDV------MTKDVVCAKEDVEEEEALELMYANRVERLPIVDDENRLIGII 197
Query: 419 TLRDVI 424
TLRD++
Sbjct: 198 TLRDIL 203
Score = 37.0 bits (84), Expect = 0.10
Identities = 20/68 (29%), Positives = 39/68 (56%), Gaps = 1/68 (1%)
Query: 357 RNLTVMDFMKKIVSASYESGKVTRP-ITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVV 415
RN+++ + + ++ + V + IT PD T+ I+ + + SI + V+ +D++V
Sbjct: 75 RNMSIEEQVHQVQAVKKADEVVIKDVITVSPDDTVGEAINVMETYSISGLPVVDNEDKLV 134
Query: 416 GVITLRDV 423
G+IT RDV
Sbjct: 135 GIITHRDV 142
>YC25_METJA (Q58622) Hypothetical protein MJ1225
Length = 280
Score = 48.5 bits (114), Expect = 3e-05
Identities = 40/153 (26%), Positives = 71/153 (46%), Gaps = 25/153 (16%)
Query: 299 KVISIQSNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDI--------RYLLLKP 350
K++++ I +A M +N+ LPVV +VG ++ DI +Y L++
Sbjct: 13 KIVTVYPTTTIRKALMTMNENKYRRLPVVNAGNNKVVGIITSMDIVDFMGGGSKYNLIRE 72
Query: 351 EIFSNFR---NLTVMDFMKKIVSASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYT 407
+ NF N V + M++ V IT K +A + I T ++++
Sbjct: 73 KHERNFLAAINEPVREIMEENV------------ITLKENADIDEAIETFLTKNVGGAPI 120
Query: 408 VNGQDQVVGVITLRDVISCFITEPDYH--FDDY 438
VN ++Q++ +IT RDVI + + D + DDY
Sbjct: 121 VNDENQLISLITERDVIRALLDKIDENEVIDDY 153
Score = 45.1 bits (105), Expect = 4e-04
Identities = 55/241 (22%), Positives = 105/241 (42%), Gaps = 44/241 (18%)
Query: 226 VAKNSAMLTV---------LLLLSKYRLRNVPVIEPGKADIVNFITQSAVIQGLEG---- 272
+A+N ++TV L+ +++ + R +PV+ G +V IT ++ + G
Sbjct: 8 IAQNKKIVTVYPTTTIRKALMTMNENKYRRLPVVNAGNNKVVGIITSMDIVDFMGGGSKY 67
Query: 273 --CRGRDWFDCIAA--RPMADLGLPFMSADKVISIQSNELILEAFKIMRDNQIGGLPVVE 328
R + + +AA P+ ++ + VI+++ N I EA + +GG P+V
Sbjct: 68 NLIREKHERNFLAAINEPVREI-----MEENVITLKENADIDEAIETFLTKNVGGAPIV- 121
Query: 329 GPAKTIVGNLSIRD-IRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVTRPITCKPD 387
++ ++ RD IR LL K + N + D++ + V I P
Sbjct: 122 NDENQLISLITERDVIRALLDKID-----ENEVIDDYITRDV------------IVATPG 164
Query: 388 ATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEPDYHFDDYYGFAVKEML 447
L+ V T+ R+ V+ + ++VG+IT D I + D+ F+ V+E+
Sbjct: 165 ERLKDVARTMVRNGFRRLPVVS-EGRLVGIITSTDFIK--LLGSDWAFNHMQTGNVREIT 221
Query: 448 N 448
N
Sbjct: 222 N 222
>YR33_THEPE (P15889) Hypothetical 33.4 kDa protein in ribosomal RNA
operon
Length = 300
Score = 48.1 bits (113), Expect = 5e-05
Identities = 49/211 (23%), Positives = 100/211 (47%), Gaps = 25/211 (11%)
Query: 219 RWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVNFITQSAVIQGLEGCRGRD- 277
R+ P V +S +L VL+ + + R+R+VP+++ + + ++ ++ L G R RD
Sbjct: 10 RFPPLAVVPSSSRVLDVLVAMGRNRVRHVPLVDE-RGVLKGMVSARDLVDFLGGRRFRDV 68
Query: 278 ---WFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDNQIGGLPVVEGPAKTI 334
F+ + + G+ F+ D ++L E ++M + IG L VV+ + +
Sbjct: 69 VEARFNGDVYKALEQTGVEFLKYDPPYVYTRSDL-REVIELMVERGIGALAVVDEDLR-V 126
Query: 335 VGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVTRPITCKPDATLQSVI 394
VG +S R + LL E ++ V + M V + P +L +
Sbjct: 127 VGIVSERHVISLLANVE-----THVKVKEIMTSEV------------VYLSPMDSLFEGM 169
Query: 395 HTLASQSIHRIYTVNGQDQVVGVITLRDVIS 425
++ + I R+ V+G +++ G++T++DV+S
Sbjct: 170 RVMSERRIRRLPLVSG-EELRGIVTIKDVLS 199
>IMDH_AQUAE (O67820) Inosine-5'-monophosphate dehydrogenase (EC
1.1.1.205) (IMP dehydrogenase) (IMPDH) (IMPD)
Length = 490
Score = 45.4 bits (106), Expect = 3e-04
Identities = 33/124 (26%), Positives = 61/124 (48%), Gaps = 18/124 (14%)
Query: 301 ISIQSNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLT 360
++++ + + EA IM +I G+PVV+ K ++G L+ RD+R++ KPE +S
Sbjct: 101 VTVKPDTRVKEALDIMAKYKISGVPVVDEERK-LIGILTNRDLRFI--KPEDYSK----P 153
Query: 361 VMDFMKKIVSASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITL 420
V +FM K IT TL I ++ V+ + ++ G+IT+
Sbjct: 154 VSEFMTK-----------ENLITAPEGITLDEAEEIFRKYKIEKLPIVDKEGKIKGLITI 202
Query: 421 RDVI 424
+D++
Sbjct: 203 KDIV 206
Score = 40.0 bits (92), Expect = 0.012
Identities = 19/60 (31%), Positives = 34/60 (56%), Gaps = 2/60 (3%)
Query: 374 ESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEPDY 433
ESG + P+T KPD ++ + +A I + V+ + +++G++T RD+ FI DY
Sbjct: 93 ESGMIINPVTVKPDTRVKEALDIMAKYKISGVPVVDEERKLIGILTNRDL--RFIKPEDY 150
Score = 34.3 bits (77), Expect = 0.67
Identities = 29/120 (24%), Positives = 63/120 (52%), Gaps = 15/120 (12%)
Query: 224 VPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIA 283
V V ++ + L +++KY++ VPV++ + ++ +T + L + D+
Sbjct: 101 VTVKPDTRVKEALDIMAKYKISGVPVVDEERK-LIGILTN----RDLRFIKPEDY----- 150
Query: 284 ARPMADLGLPFMSADKVISIQSNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDI 343
++P+++ FM+ + +I+ + EA +I R +I LP+V+ K I G ++I+DI
Sbjct: 151 SKPVSE----FMTKENLITAPEGITLDEAEEIFRKYKIEKLPIVDKEGK-IKGLITIKDI 205
>AAKG_MOUSE (O54950) 5'-AMP-activated protein kinase, gamma-1
subunit (AMPK gamma-1 chain) (AMPKg)
Length = 330
Score = 45.4 bits (106), Expect = 3e-04
Identities = 46/238 (19%), Positives = 102/238 (42%), Gaps = 15/238 (6%)
Query: 191 NLGEDFYKVILQE----EPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRN 246
N+ +YK L + E K T R + + P V ++ N++ + L + ++
Sbjct: 92 NILHRYYKSALVQIYELEEHKIETWREVYLQDSFKPLVCISPNASSFDAVSSLIRNKIHR 151
Query: 247 VPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSN 306
+PVI+P + + +T +++ L+ ++ + +L + + + +++
Sbjct: 152 LPVIDPESGNTLYILTHKRILKFLKLFIIEFPKPEFMSKSLQELQIGTYA--NIAMVRTT 209
Query: 307 ELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMK 366
+ A I +++ LPVV+ + +V S D+ L + + NL V
Sbjct: 210 TPVYVALGIFVQHRVSALPVVDEKGR-VVDIYSKFDVINLAAE----KTYNNLDVSVTKA 264
Query: 367 KIVSASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVI 424
+ Y G + C TL+++I+ L +HR+ V+ V G+++L D++
Sbjct: 265 LXHRSHYFEGV----LKCYLHETLETIINRLVEAEVHRLVVVDEHXXVKGIVSLSDIL 318
>YGF6_YEAST (P53172) Hypothetical 58.1 kDa protein in UBC2-OLE1
intergenic region
Length = 527
Score = 43.9 bits (102), Expect = 8e-04
Identities = 51/218 (23%), Positives = 96/218 (43%), Gaps = 16/218 (7%)
Query: 207 KSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVNFITQSAV 266
KS V I+K PF + + + TV+ +L + V + I ++Q +
Sbjct: 175 KSVPVAEIVKLTPKNPFYKLPETENLSTVIGILGS-GVHRVAITNVEMTQIKGILSQRRL 233
Query: 267 IQGL-EGCRGRDWFDCIAARPMADLGLPFMSA---------DKVISIQSNELILEAFKIM 316
I+ L E R + + +L + ++A +VISIQ +E ++ A M
Sbjct: 234 IKYLWENARSFPNLKPLLDSSLEELNIGVLNAARDKPTFKQSRVISIQGDEHLIMALHKM 293
Query: 317 RDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVS-ASYES 375
+I + VV+ P ++GN+S+ D+++ + + + N T F+ I++ E+
Sbjct: 294 YVERISSIAVVD-PQGNLIGNISVTDVKH-VTRTSQYPLLHN-TCRHFVSVILNLRGLET 350
Query: 376 GKVTRPI-TCKPDATLQSVIHTLASQSIHRIYTVNGQD 412
GK + PI P ++L L + HR++ V D
Sbjct: 351 GKDSFPIFHVYPTSSLARTFAKLVATKSHRLWIVQPND 388
>Y868_METJA (Q58278) Hypothetical protein MJ0868
Length = 127
Score = 43.1 bits (100), Expect = 0.001
Identities = 18/50 (36%), Positives = 32/50 (64%), Gaps = 1/50 (2%)
Query: 379 TRPITCKPDATLQSVIHTLASQSIHRIYTVNG-QDQVVGVITLRDVISCF 427
T PIT P+A L+ + +A + IH +Y + +D++VGV++ +D+I F
Sbjct: 73 TNPITVSPEAPLEKAVEIMAEKGIHHLYVKSPCEDKIVGVLSSKDIIKLF 122
>Y264_METEX (Q8GEK8) Hypothetical UPF0264 protein ORF22
Length = 236
Score = 43.1 bits (100), Expect = 0.001
Identities = 50/185 (27%), Positives = 78/185 (42%), Gaps = 21/185 (11%)
Query: 121 AELAAVALSAGTATAAGVGAG--------TVGALGA----IALGATGPAAIAGLTAAAVG 168
A +A V A T+ AG G G T+ A G IA+G AA+A A A G
Sbjct: 55 AIVAGVGGRAVTSAVAGDGTGREIAAAIATIAATGVDFIKIAVGGADDAALAEAAAQAPG 114
Query: 169 AAVVGGVAADKTMAKDAPQAANNLGEDFYKVILQEEPFKSTTVRSILKSYRWAPFVPVAK 228
V+G + A+ +A+D P A F ++ TT+ S++ + + A FV +
Sbjct: 115 -RVIGVLFAEDDVAEDGP--ARLAAAGFVGAMIDTRGKSGTTLTSLMAAPQLAAFVAGCR 171
Query: 229 NSAMLTVLLLLSKYRLRNVPVIEPGKADIVNF----ITQSAVIQGLEGCRGRDWFDCIAA 284
+++ L L ++PV+ D + F S Q L+G R + + A
Sbjct: 172 THGLMSG--LAGSLGLGDIPVLARLDPDYLGFRGGLCRASDRRQALDGARVAQAVEAMRA 229
Query: 285 RPMAD 289
P AD
Sbjct: 230 GPRAD 234
>MSA2_PLAFZ (Q03645) Merozoite surface antigen 2 precursor (MSA-2)
Length = 300
Score = 42.7 bits (99), Expect = 0.002
Identities = 25/66 (37%), Positives = 31/66 (46%)
Query: 130 AGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAKDAPQAA 189
AG+ AG GAG V GA A+ +G A+AG A AV + G VA +A
Sbjct: 54 AGSGAGAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAVAGSGAGNG 113
Query: 190 NNLGED 195
N G D
Sbjct: 114 ANPGAD 119
Score = 33.5 bits (75), Expect = 1.1
Identities = 18/47 (38%), Positives = 22/47 (46%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAA 170
A +G AG GAG V GA A+ +G A+AG A A A
Sbjct: 64 AGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAVAGSGA 110
Score = 32.3 bits (72), Expect = 2.6
Identities = 19/45 (42%), Positives = 23/45 (50%), Gaps = 2/45 (4%)
Query: 132 TATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVA 176
T AG GAG GA+A +G A+AG A AV + G VA
Sbjct: 50 TGAVAGSGAGAGSGAGAVA--GSGAGAVAGSGAGAVAGSGAGAVA 92
Score = 30.4 bits (67), Expect = 9.7
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 3/55 (5%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAV-GAAVVGGVAA 177
A AG + GAG V GA A+ +G A+AG A AV G+ V G A
Sbjct: 58 AGAGSGAGAVAGS--GAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAVAGSGA 110
>MSA2_PLAFI (Q03644) Merozoite surface antigen 2 precursor (MSA-2)
Length = 300
Score = 42.7 bits (99), Expect = 0.002
Identities = 25/66 (37%), Positives = 31/66 (46%)
Query: 130 AGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAKDAPQAA 189
AG+ AG GAG V GA A+ +G A+AG A AV + G VA +A
Sbjct: 54 AGSGAGAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAVAGSGAGNG 113
Query: 190 NNLGED 195
N G D
Sbjct: 114 ANPGAD 119
Score = 33.9 bits (76), Expect = 0.88
Identities = 22/64 (34%), Positives = 27/64 (41%), Gaps = 1/64 (1%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTA-AAVGAAVVGGVAADKTMA 182
A +G AG GAG V GA A+ +G A+AG A A GA AD +
Sbjct: 64 AGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAVAGSGAGNGANPGADAERS 123
Query: 183 KDAP 186
P
Sbjct: 124 PSTP 127
Score = 32.3 bits (72), Expect = 2.6
Identities = 19/45 (42%), Positives = 23/45 (50%), Gaps = 2/45 (4%)
Query: 132 TATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVA 176
T AG GAG GA+A +G A+AG A AV + G VA
Sbjct: 50 TGAVAGSGAGAGSGAGAVA--GSGAGAVAGSGAGAVAGSGAGAVA 92
Score = 30.4 bits (67), Expect = 9.7
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 3/55 (5%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAV-GAAVVGGVAA 177
A AG + GAG V GA A+ +G A+AG A AV G+ V G A
Sbjct: 58 AGAGSGAGAVAGS--GAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAVAGSGA 110
>YL28_SCHPO (Q10343) Hypothetical protein C1556.08c in chromosome I
Length = 334
Score = 41.6 bits (96), Expect = 0.004
Identities = 46/240 (19%), Positives = 110/240 (45%), Gaps = 22/240 (9%)
Query: 196 FYKVILQEEPFKSTTVRSILKSYRWAP--FVPVAKNSAMLTVLLLLSKYRLRNVPVI--- 250
F + I + + F+ +R + + P + V +++ L +SK R R +P+I
Sbjct: 90 FPEAIAEIDKFRLLGLREVERKIGAIPPETIYVHPMHSLMDACLAMSKSRARRIPLIDVD 149
Query: 251 -EPGKADIVNFITQSAVIQGLE-GCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNEL 308
E G IV+ +TQ +++ + C+ + P+ + + S S+++
Sbjct: 150 GETGSEMIVSVLTQYRILKFISMNCKET----AMLRVPLNQMTIGTWSNLATASMETK-- 203
Query: 309 ILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKI 368
+ + K++ + I +P+V T++ D+ +L+ + +SN +L+V + + K
Sbjct: 204 VYDVIKMLAEKNISAVPIVNSEG-TLLNVYESVDVMHLIQDGD-YSNL-DLSVGEALLKR 260
Query: 369 VSASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFI 428
A+++ TC+ L + + +HR++ V+ ++ G+++L D+++ I
Sbjct: 261 -PANFDGVH-----TCRATDRLDGIFDAIKHSRVHRLFVVDENLKLEGILSLADILNYII 314
>YE26_METJA (Q58821) Hypothetical protein MJ1426
Length = 168
Score = 41.6 bits (96), Expect = 0.004
Identities = 31/146 (21%), Positives = 62/146 (42%), Gaps = 19/146 (13%)
Query: 300 VISIQSNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYL------------- 346
++ + N+LI + ++ R N+I G PV+ K +VG +S DI
Sbjct: 25 IVVYEDNDLI-DVIRLFRKNKISGAPVLNKDGK-LVGIISESDIVKTIVTHNEDLNLILP 82
Query: 347 ----LLKPEIFSNFRNLTVMDFMKKIVSASYESGKVTRPITCKPDATLQSVIHTLASQSI 402
L++ + + + M+ +K + + I KPD T+ + +I
Sbjct: 83 SPLDLIELPLKTALKIEEFMEDLKNALKTKVRDVMTRKVIVAKPDMTINDAAKLMVKNNI 142
Query: 403 HRIYTVNGQDQVVGVITLRDVISCFI 428
R+ V+ + ++G++T D+I I
Sbjct: 143 KRLPVVDDEGNLIGIVTRGDLIEALI 168
>MSA2_PLAF9 (Q03994) Merozoite surface antigen 2 precursor (MSA-2)
Length = 302
Score = 41.2 bits (95), Expect = 0.006
Identities = 26/64 (40%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Query: 130 AGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAV---GAAVVGGVAADKTMAKDAP 186
AG+ AG GAG V GA A+ +G A+AG A AV GA V G A A
Sbjct: 54 AGSGAGAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAG 113
Query: 187 QAAN 190
AN
Sbjct: 114 NGAN 117
Score = 37.7 bits (86), Expect = 0.061
Identities = 22/63 (34%), Positives = 29/63 (45%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAK 183
A +G AG GAG V GA A+ +G A+AG A AV + G A A+
Sbjct: 64 AGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGNGANPGADAE 123
Query: 184 DAP 186
+P
Sbjct: 124 RSP 126
Score = 32.3 bits (72), Expect = 2.6
Identities = 20/53 (37%), Positives = 25/53 (46%), Gaps = 2/53 (3%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVA 176
A AG + GAG V GA A+ +G A+AG A AV + G VA
Sbjct: 58 AGAGSGAGAVAGS--GAGAVAGSGAGAVAGSGAGAVAGSGAGAVAGSGAGAVA 108
Score = 32.3 bits (72), Expect = 2.6
Identities = 19/45 (42%), Positives = 23/45 (50%), Gaps = 2/45 (4%)
Query: 132 TATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVA 176
T AG GAG GA+A +G A+AG A AV + G VA
Sbjct: 50 TGAVAGSGAGAGSGAGAVA--GSGAGAVAGSGAGAVAGSGAGAVA 92
>IMDH_MYCTU (P65167) Inosine-5'-monophosphate dehydrogenase (EC
1.1.1.205) (IMP dehydrogenase) (IMPDH) (IMPD)
Length = 529
Score = 40.4 bits (93), Expect = 0.009
Identities = 21/50 (42%), Positives = 29/50 (58%)
Query: 374 ESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDV 423
E+G VT P+TC+PD TL V A I + V+ +VG+IT RD+
Sbjct: 126 EAGMVTDPVTCRPDNTLAQVDALCARFRISGLPVVDDDGALVGIITNRDM 175
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.135 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 49,251,730
Number of Sequences: 164201
Number of extensions: 2016571
Number of successful extensions: 9546
Number of sequences better than 10.0: 155
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 102
Number of HSP's that attempted gapping in prelim test: 7999
Number of HSP's gapped (non-prelim): 1079
length of query: 449
length of database: 59,974,054
effective HSP length: 113
effective length of query: 336
effective length of database: 41,419,341
effective search space: 13916898576
effective search space used: 13916898576
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC136286.10


