

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135848.10 - phase: 0
(346 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
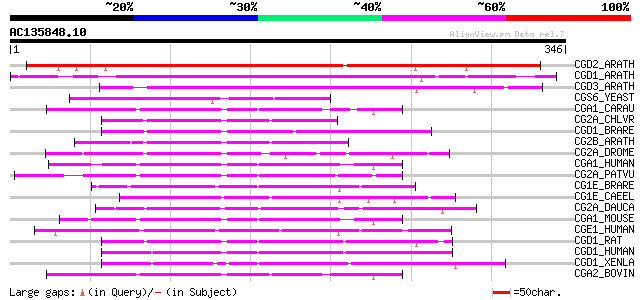
Score E
Sequences producing significant alignments: (bits) Value
CGD2_ARATH (P42752) Cyclin delta-2 269 7e-72
CGD1_ARATH (P42751) Cyclin delta-1 176 1e-43
CGD3_ARATH (P42753) Cyclin delta-3 153 7e-37
CGS6_YEAST (P32943) S-phase entry cyclin 6 73 1e-12
CGA1_CARAU (Q92161) Cyclin A1 (Cyclin A) 70 6e-12
CG2A_CHLVR (P51986) G2/mitotic-specific cyclin A (Fragment) 69 2e-11
CGD1_BRARE (Q90459) G1/S-specific cyclin D1 67 5e-11
CG2B_ARATH (P30183) G2/mitotic-specific cyclin (B-like cyclin) 67 7e-11
CG2A_DROME (P14785) G2/mitotic-specific cyclin A 66 1e-10
CGA1_HUMAN (P78396) Cyclin A1 66 1e-10
CG2A_PATVU (P24861) G2/mitotic-specific cyclin A 65 2e-10
CG1E_BRARE (P47794) G1/S-specific cyclin E 64 6e-10
CG1E_CAEEL (O01501) G1/S-specific cyclin E 64 7e-10
CG2A_DAUCA (P25010) G2/mitotic-specific cyclin C13-1 (A-like cyc... 63 1e-09
CGA1_MOUSE (Q61456) Cyclin A1 62 2e-09
CGE1_HUMAN (P24864) G1/S-specific cyclin E1 62 3e-09
CGD1_RAT (P39948) G1/S-specific cyclin D1 62 3e-09
CGD1_HUMAN (P24385) G1/S-specific cyclin D1 (PRAD1 oncogene) (BC... 62 3e-09
CGD1_XENLA (P50755) G1/S-specific cyclin D1 61 4e-09
CGA2_BOVIN (P30274) Cyclin A2 (Cyclin A) (Fragment) 61 5e-09
>CGD2_ARATH (P42752) Cyclin delta-2
Length = 361
Score = 269 bits (688), Expect = 7e-72
Identities = 156/345 (45%), Positives = 216/345 (62%), Gaps = 26/345 (7%)
Query: 11 NLLCSENNSTCFDDVVVDD----SGISPSWDHTN---VNLDNVGSDSFLCFVAQS----- 58
NL C E + + D DD G + D+ + DN G + + + S
Sbjct: 4 NLACGETSESWIIDNDDDDINYGGGFTNEIDYNHQLFAKDDNFGGNGSIPMMGSSSSSLS 63
Query: 59 EEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVN 118
E+ +K M+ +E + P DY+ RL GDLDLSVR +ALDWI K A+Y FG L +CLS+N
Sbjct: 64 EDRIKEMLVREIEFCPGTDYVKRLLSGDLDLSVRNQALDWILKVCAHYHFGHLCICLSMN 123
Query: 119 YLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQR 178
YLDRFL+ ++ P+ W QLLAV+C SLA+KMEE VP VDLQV +PKFVF+AKTI+R
Sbjct: 124 YLDRFLTSYELPKDKDWAAQLLAVSCLSLASKMEETDVPHIVDLQVEDPKFVFEAKTIKR 183
Query: 179 MELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFLEFR 238
MEL+++++L W+++ALTP SFIDYF+ KIS + ++LI RS + ILN K I+FL+FR
Sbjct: 184 MELLVVTTLNWRLQALTPFSFIDYFVDKIS--GHVSENLIYRSSRFILNTTKAIEFLDFR 241
Query: 239 SSEIAAAVAISLK---ELPTQEVDKAITDFFIVDKERVLKCVELIRDL---------SLI 286
SEIAAA A+S+ E + +KA++ V +ERV +C+ L+R L SL
Sbjct: 242 PSEIAAAAAVSVSISGETECIDEEKALSSLIYVKQERVKRCLNLMRSLTGEENVRGTSLS 301
Query: 287 KVGGNNFASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSHSSPN 331
+ VP SP+GVL+A C+S++S+E T SC NSS SSP+
Sbjct: 302 QEQARVAVRAVPASPVGVLEATCLSYRSEERTVESCTNSSQSSPD 346
>CGD1_ARATH (P42751) Cyclin delta-1
Length = 335
Score = 176 bits (445), Expect = 1e-43
Identities = 122/349 (34%), Positives = 181/349 (50%), Gaps = 43/349 (12%)
Query: 1 MAESFDSAESNLLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQSEE 60
M+ SF S + +L C E++ + VD S + +D+ DS CF+
Sbjct: 12 MSVSF-SNDMDLFCGEDSGVFSGESTVDFS---------SSEVDSWPGDSIACFI----- 56
Query: 61 IVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYL 120
E E+ +P DYL R + LD S R +++ WI K AYY F PL+ L+VNY+
Sbjct: 57 ------EDERHFVPGHDYLSRFQTRSLDASAREDSVAWILKVQAYYNFQPLTAYLAVNYM 110
Query: 121 DRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRME 180
DRFL + P W +QLLAVAC SLAAKMEE+ VP D QV K++F+AKTI+RME
Sbjct: 111 DRFLYARRLPETSGWPMQLLAVACLSLAAKMEEILVPSLFDFQVAGVKYLFEAKTIKRME 170
Query: 181 LMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFLEFRSS 240
L++LS L W++R++TP FI +F KI I+ + ++IL+ IK FLE+ S
Sbjct: 171 LLVLSVLDWRLRSVTPFDFISFFAYKIDPSGTFLGFFISHATEIILSNIKEASFLEYWPS 230
Query: 241 EIAAAVAISL-KELPT-------QEVDKAITDFFIVDKERVLKCVELIRDLSLIKVGGNN 292
IAAA + + ELP+ E + D + KE++++C L++ ++ I+ N
Sbjct: 231 SIAAAAILCVANELPSLSSVVNPHESPETWCDG--LSKEKIVRCYRLMKAMA-IENNRLN 287
Query: 293 FASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSHSSPNAKRMKFDGPS 341
+ + + V + ++ SDE SS KR K G S
Sbjct: 288 TPKVIAKLRVSVRASSTLTRPSDE-----------SSSPCKRRKLSGYS 325
>CGD3_ARATH (P42753) Cyclin delta-3
Length = 376
Score = 153 bits (386), Expect = 7e-37
Identities = 98/292 (33%), Positives = 159/292 (53%), Gaps = 26/292 (8%)
Query: 57 QSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSV-RREALDWIWKAHAYYGFGPLSLCL 115
+ E++V + ++E+ L D D+ LS R+EA+ WI + +A+YGF L+ L
Sbjct: 59 EDEDLVTLFSKEEEQGLSCLD--------DVYLSTDRKEAVGWILRVNAHYGFSTLAAVL 110
Query: 116 SVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKT 175
++ YLD+F+ + R W +QL++VAC SLAAK+EE +VP +D QV E K+VF+AKT
Sbjct: 111 AITYLDKFICSYSLQRDKPWMLQLVSVACLSLAAKVEETQVPLLLDFQVEETKYVFEAKT 170
Query: 176 IQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFL 235
IQRMEL+ILS+L WKM +TP SF+D+ + ++ + + + +L+L++I F+
Sbjct: 171 IQRMELLILSTLEWKMHLITPISFVDHIIRRLGLKNNAHWDFLNKCHRLLLSVISDSRFV 230
Query: 236 EFRSSEIAAAVAISLKE----LPTQEVDKAITDFFIVDKERVLKCVELIRDLSLIKV--- 288
+ S +AAA + + E + + KE+V C +LI L + ++
Sbjct: 231 GYLPSVVAAATMMRIIEQVDPFDPLSYQTNLLGVLNLTKEKVKTCYDLILQLPVDRICLQ 290
Query: 289 --------GGNNFASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSHSSPNA 332
++ +S SP V+DA F SDE +N S SS + P +
Sbjct: 291 IQIQSSKKRKSHDSSSSLNSPSCVIDAN--PFNSDESSNDSWSASSCNPPTS 340
>CGS6_YEAST (P32943) S-phase entry cyclin 6
Length = 380
Score = 72.8 bits (177), Expect = 1e-12
Identities = 52/166 (31%), Positives = 83/166 (49%), Gaps = 9/166 (5%)
Query: 38 HTNVNLDNVGSDSFLCFVAQSEEIVKVMVEKEKDHLPREDYLIRLRGG-DLDLSVRREAL 96
H NLD++ D ++ I + EKE LP +YL+ + L S+R +
Sbjct: 101 HQWKNLDSIEMDDPFMVAEYTDSIFSHLYEKEIQMLPTHNYLMDTQSPYHLKSSMRALLI 160
Query: 97 DWIWKAHAYYGFGPLSLCLSVNYLDRFLS--VFQFPRGVTWTVQLLAVACFSLAAKMEEV 154
DW+ + H + P +L L++N LDRFLS V + + +QLL + C +A K EEV
Sbjct: 161 DWLVEVHEKFHCLPETLFLAINLLDRFLSQNVVKLNK-----LQLLCITCLFIACKFEEV 215
Query: 155 KVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFI 200
K+P+ + + I++ EL +LSSLG+ + P +FI
Sbjct: 216 KLPKITNFAY-VTDGAATVEGIRKAELFVLSSLGYNISLPNPLNFI 260
>CGA1_CARAU (Q92161) Cyclin A1 (Cyclin A)
Length = 391
Score = 70.5 bits (171), Expect = 6e-12
Identities = 69/228 (30%), Positives = 108/228 (47%), Gaps = 20/228 (8%)
Query: 24 DVVVDDSGISPSWDHTNVNL-DNVGSDSFLCFVAQSEEIVKVMVEKEKDHLPREDYLIRL 82
D VV D G D + +L + + LC +E+I + + E E + P+ Y+
Sbjct: 103 DDVVQDLGSGSCMDSSMQSLPEEAAYEDILCVPEYAEDIHRYLRECEVKYRPKPGYM--R 160
Query: 83 RGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAV 142
+ D+ +R +DW+ + Y +L L+VNYLDRFLS RG +QL+
Sbjct: 161 KQPDITNCMRVILVDWLVEVGEEYKLCSETLFLAVNYLDRFLSCMSVLRG---KLQLVGT 217
Query: 143 ACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDY 202
A LAAK EEV P VD V + K + RME +L L + M A T +
Sbjct: 218 AAVLLAAKYEEV-YPPEVDEFVYITDDTYTKKQLLRMEQHLLRVLAFDMTAPT----VHQ 272
Query: 203 FLAKISCEKYPDKSLIARSVQLI-----LNIIKGIDFLEFRSSEIAAA 245
FL + + E + + AR+V L L++++ F+++ S+ AAA
Sbjct: 273 FLMQYTLEGH----ICARTVNLALYLSELSLLEVDPFVQYLPSKTAAA 316
>CG2A_CHLVR (P51986) G2/mitotic-specific cyclin A (Fragment)
Length = 420
Score = 68.6 bits (166), Expect = 2e-11
Identities = 45/147 (30%), Positives = 78/147 (52%), Gaps = 6/147 (4%)
Query: 58 SEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSV 117
+++I + + E + P+ +Y+ + D++ S+R +DW+ + Y P +L LSV
Sbjct: 166 AQDIHNYLKKSEAKYRPKSNYMRKQT--DINSSMRAILIDWLVEVSEEYKLIPQTLYLSV 223
Query: 118 NYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQ 177
+Y+DRFLS RG +QL+ AC +AAK EE+ P+ + V + AK +
Sbjct: 224 SYIDRFLSHMSVLRG---KLQLVGAACMLVAAKFEEIYPPEVAEF-VYITDDTYTAKQVL 279
Query: 178 RMELMILSSLGWKMRALTPCSFIDYFL 204
RME +IL +L + + T F+ +L
Sbjct: 280 RMEHLILKTLAFDLSVPTCRDFLSRYL 306
>CGD1_BRARE (Q90459) G1/S-specific cyclin D1
Length = 291
Score = 67.4 bits (163), Expect = 5e-11
Identities = 53/207 (25%), Positives = 104/207 (49%), Gaps = 6/207 (2%)
Query: 58 SEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSV 117
++ +++ M++ E+++LP +Y ++ ++ +R+ W+ + L++
Sbjct: 24 NDRVLQTMLKAEENYLPSPNYFKCVQK-EIVPKMRKIVATWMLEVCEEQKCEEEVFPLAM 82
Query: 118 NYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQ 177
NYLDRFLSV + +QLL C LA+KM+E + L + V + +Q
Sbjct: 83 NYLDRFLSVEPTKKT---RLQLLGATCMFLASKMKETVPLTAEKLCIYTDNSVRPGELLQ 139
Query: 178 RMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFLEF 237
MEL+ L+ L W + ++TP FI++FLAK+ + + L + + ++F+
Sbjct: 140 -MELLALNKLKWDLASVTPHDFIEHFLAKLPIHQSSKQILRKHAQTFVALCATDVNFIAS 198
Query: 238 RSSEIAA-AVAISLKELPTQEVDKAIT 263
S IAA +VA +++ L + D ++
Sbjct: 199 PPSMIAAGSVAAAVQGLYLKSTDSCLS 225
>CG2B_ARATH (P30183) G2/mitotic-specific cyclin (B-like cyclin)
Length = 428
Score = 67.0 bits (162), Expect = 7e-11
Identities = 55/172 (31%), Positives = 85/172 (48%), Gaps = 8/172 (4%)
Query: 41 VNLDNVGSDSFLCFVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIW 100
V++D+ ++ L V E+I E + PR DY+ D++ +R ++W+
Sbjct: 149 VDIDSADVENDLAAVEYVEDIYSFYKSVESEWRPR-DYMASQP--DINEKMRLILVEWLI 205
Query: 101 KAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSV 160
H + P + L+VN LDRFLSV PR +QL+ ++ ++AK EE+ PQ
Sbjct: 206 DVHVRFELNPETFYLTVNILDRFLSVKPVPRK---ELQLVGLSALLMSAKYEEIWPPQVE 262
Query: 161 DLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFL-AKISCEK 211
DL V + K I ME ILS+L W + T F+ F+ A I+ EK
Sbjct: 263 DL-VDIADHAYSHKQILVMEKTILSTLEWYLTVPTHYVFLARFIKASIADEK 313
>CG2A_DROME (P14785) G2/mitotic-specific cyclin A
Length = 491
Score = 66.2 bits (160), Expect = 1e-10
Identities = 71/261 (27%), Positives = 123/261 (46%), Gaps = 24/261 (9%)
Query: 23 DDVVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQSEEIVKVMVEKEKDHLPREDYLIRL 82
D V ++G+SP+ + N FL V +I++ E EK H P+ Y+ R
Sbjct: 170 DISVGTETGVSPTGRVKELPPRN-DRQRFLEVVQYQMDILEYFRESEKKHRPKPLYMRRQ 228
Query: 83 RGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAV 142
+ D+ ++R +DW+ + Y +L LSV YLDRFLS R +QL+
Sbjct: 229 K--DISHNMRSILIDWLVEVSEEYKLDTETLYLSVFYLDRFLSQMAVVRS---KLQLVGT 283
Query: 143 ACFSLAAKMEEVKVPQSVDLQVGEPKFV----FQAKTIQRMELMILSSLGWKMRALTPCS 198
A +AAK EE+ P +VGE F+ + + RME +IL L + + TP +
Sbjct: 284 AAMYIAAKYEEIYPP-----EVGEFVFLTDDSYTKAQVLRMEQVILKILSFDL--CTPTA 336
Query: 199 FIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFLEF-----RSSEIAAAVAISLKEL 253
++ + C+ P+K L++++G +L++ S+ +A A I E+
Sbjct: 337 YVFINTYAVLCD-MPEKLKYMTLYISELSLMEGETYLQYLPSLMSSASVALARHILGMEM 395
Query: 254 PTQEVDKAITDFFIVDKERVL 274
T +++ IT + + D + V+
Sbjct: 396 WTPRLEE-ITTYKLEDLKTVV 415
>CGA1_HUMAN (P78396) Cyclin A1
Length = 465
Score = 65.9 bits (159), Expect = 1e-10
Identities = 61/226 (26%), Positives = 107/226 (46%), Gaps = 25/226 (11%)
Query: 25 VVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQSEEIVKVMVEKEKDHLPREDYLIRLRG 84
++VD S +S S D +++ D + + +EEI + + E E H P+ Y+ +
Sbjct: 185 MLVDSSLLSQSEDISSLGTDVINVTEY------AEEIYQYLREAEIRHRPKAHYM--KKQ 236
Query: 85 GDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVAC 144
D+ +R +DW+ + Y +L L+VN+LDRFLS RG +QL+ A
Sbjct: 237 PDITEGMRTILVDWLVEVGEEYKLRAETLYLAVNFLDRFLSCMSVLRG---KLQLVGTAA 293
Query: 145 FSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFL 204
LA+K EE+ P VD V + + + +ME ++L L + + T F+ +L
Sbjct: 294 MLLASKYEEI-YPPEVDEFVYITDDTYTKRQLLKMEHLLLKVLAFDLTVPTTNQFLLQYL 352
Query: 205 AKISCEKYPDKSLIARSVQLI-----LNIIKGIDFLEFRSSEIAAA 245
+ + + R+ L L++++ FL++ S IAAA
Sbjct: 353 RR--------QGVCVRTENLAKYVAELSLLEADPFLKYLPSLIAAA 390
>CG2A_PATVU (P24861) G2/mitotic-specific cyclin A
Length = 426
Score = 65.5 bits (158), Expect = 2e-10
Identities = 67/243 (27%), Positives = 112/243 (45%), Gaps = 21/243 (8%)
Query: 4 SFDSAESNLLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQ-SEEIV 62
S + E + +EN S DV ++D+ P + ++ + V + +E+I
Sbjct: 128 SLQALEDIQVDNENGSPMVLDVTIEDAEKKP-----------IDREAIILSVPEYAEDIY 176
Query: 63 KVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDR 122
K + E E H + Y+ + D+ S+R +DW+ + Y +L L++NY+DR
Sbjct: 177 KHLREAESRHRSKPGYM--KKQPDITNSMRSILVDWMVEVSEEYKLHRETLFLAINYIDR 234
Query: 123 FLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELM 182
FLS RG +QL+ A +A+K EE+ P V V ++ K + RME +
Sbjct: 235 FLSQMSVLRG---KLQLVGAASMFIASKYEEI-YPPEVSEFVYITDDTYEQKQVLRMEHL 290
Query: 183 ILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFLEFRSSEI 242
IL L + + T F D + AK++ KSL +L L + +L++ S I
Sbjct: 291 ILKVLSFDVAQPTINWFTDTY-AKMADTDETTKSLSMYLSELTL--VDADPYLKYLPSTI 347
Query: 243 AAA 245
AAA
Sbjct: 348 AAA 350
>CG1E_BRARE (P47794) G1/S-specific cyclin E
Length = 410
Score = 63.9 bits (154), Expect = 6e-10
Identities = 53/210 (25%), Positives = 98/210 (46%), Gaps = 15/210 (7%)
Query: 52 LCFVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPL 111
LC+ A +E+ ++ K+K +L D + R +L +R LDW+ + Y
Sbjct: 110 LCW-ASKDEVWNNLLGKDKLYL--RDTRVMERHPNLQPKMRAILLDWLMEVCEVYKLHRE 166
Query: 112 SLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVF 171
+ L +Y DRF++ + + T+QL+ ++C +AAKMEE+ P V
Sbjct: 167 TFYLGQDYFDRFMATQE--NVLKTTLQLIGISCLFIAAKMEEI-YPPKVHQFAYVTDGAC 223
Query: 172 QAKTIQRMELMILSSLGWKMRALTPCSFIDYFL--------AKISCEKYPDKSLIARSVQ 223
I ME++I+ L W + LTP ++++ ++ A++ +YP + + + +
Sbjct: 224 TEDDILSMEIIIMKELNWSLSPLTPVAWLNIYMQMAYLKETAEVLTAQYPQATFV-QIAE 282
Query: 224 LILNIIKGIDFLEFRSSEIAAAVAISLKEL 253
L+ I + LEF S +AA+ L
Sbjct: 283 LLDLCILDVRSLEFSYSLLAASALFHFSSL 312
>CG1E_CAEEL (O01501) G1/S-specific cyclin E
Length = 524
Score = 63.5 bits (153), Expect = 7e-10
Identities = 57/224 (25%), Positives = 99/224 (43%), Gaps = 17/224 (7%)
Query: 69 EKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQ 128
++D +PR + D+D RR +DW+ + + L+V+Y+DR+L
Sbjct: 239 KRDEIPRATRFLLGNHPDMDDEKRRILIDWMMEVCESEKLHRETFHLAVDYVDRYLESSN 298
Query: 129 FPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLG 188
T QL+ A +AAK EE+ P+ +D F I+ ME++I+ +G
Sbjct: 299 VECS-TDNFQLVGTAALFIAAKYEEIYPPKCIDF-AHLTDSAFTCDNIRTMEVLIVKYIG 356
Query: 189 WKMRALTPCSFIDYFL------AKISCEKYPDKSLIARSV--QLILNIIKGIDFLEFR-- 238
W + +T ++ +L K + Y ++++ + L + K +DFL F
Sbjct: 357 WSLGPITSIQWLSTYLQLLGTGKKNKSDHYEEQNMYVPELLRSEYLEMCKILDFLLFEID 416
Query: 239 ----SSEIAAAVAISLKELPTQEVDKAITDFFIVDKERVLKCVE 278
S AA + + PT V+KA T F E+V++ VE
Sbjct: 417 SFTFSYRTIAAAVLFVNYEPTCAVEKA-TGFMQAQLEKVIEYVE 459
>CG2A_DAUCA (P25010) G2/mitotic-specific cyclin C13-1 (A-like
cyclin) (Fragment)
Length = 341
Score = 62.8 bits (151), Expect = 1e-09
Identities = 63/246 (25%), Positives = 123/246 (49%), Gaps = 19/246 (7%)
Query: 54 FVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSL 113
+V+ E +K M E E P +Y+ +++ D+ ++R +DW+ + Y P +L
Sbjct: 74 YVSDVYEYLKQM-EMETKRRPMMNYIEQVQK-DVTSNMRGVLVDWLVEVSLEYKLLPETL 131
Query: 114 CLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQA 173
L+++Y+DR+LSV R +QLL V+ F +A+K EE+K P++V V +
Sbjct: 132 YLAISYVDRYLSVNVLNR---QKLQLLGVSSFLIASKYEEIK-PKNVADFVDITDNTYSQ 187
Query: 174 KTIQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGID 233
+ + +ME +L +L ++M + T +F+ + A ++ PD + + + N + +
Sbjct: 188 QEVVKMEADLLKTLKFEMGSPTVKTFLGFIRA---VQENPDVPKL--KFEFLANYLAELS 242
Query: 234 FLEFRSSE-IAAAVAISLKELPTQEVDKAITDFFIV------DKERVLK-CVELIRDLSL 285
L++ E + + +A S+ L + + + I K + LK CV L+ DL +
Sbjct: 243 LLDYGCLEFVPSLIAASVTFLARFTIRPNVNPWSIALQKCSGYKSKDLKECVLLLHDLQM 302
Query: 286 IKVGGN 291
+ GG+
Sbjct: 303 GRRGGS 308
>CGA1_MOUSE (Q61456) Cyclin A1
Length = 421
Score = 62.4 bits (150), Expect = 2e-09
Identities = 58/219 (26%), Positives = 101/219 (45%), Gaps = 20/219 (9%)
Query: 32 ISPSWDHTNVNLDNVGSDSFLCFVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSV 91
+ P+ + + GSD + +EEI + + E E H P+ Y+ + D+ +
Sbjct: 143 VDPTTHAQSEEATDFGSD-VINVTEYAEEIHRYLPEAEVRHRPKAHYM--RKQPDITEGM 199
Query: 92 RREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKM 151
R +DW+ + Y +L L+VN+LDRFLS RG +QL+ A LA+K
Sbjct: 200 RAILVDWLVEVGEEYKLRTETLYLAVNFLDRFLSCMSVLRG---KLQLVGTAAILLASKY 256
Query: 152 EEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEK 211
EE+ P VD V + + + RME ++L L + + T F+ +L +
Sbjct: 257 EEI-YPPDVDEFVYITDDTYTKRQLLRMEHLLLKVLAFDLTVPTTNQFLLQYLRR----- 310
Query: 212 YPDKSLIARSVQLI-----LNIIKGIDFLEFRSSEIAAA 245
+ + R+ L L++++ FL++ S +AAA
Sbjct: 311 ---QGVCIRTENLAKYVAELSLLEADPFLKYLPSLVAAA 346
>CGE1_HUMAN (P24864) G1/S-specific cyclin E1
Length = 410
Score = 61.6 bits (148), Expect = 3e-09
Identities = 65/276 (23%), Positives = 125/276 (44%), Gaps = 25/276 (9%)
Query: 16 ENNSTCFDDVVV-------DDSGISP-SWDHTNVNLDNVGSDSFLCFVAQSEEIVKVMVE 67
+NN+ C D + DD + P S + + GS + A EE+ K+M+
Sbjct: 63 DNNAVCADPCSLIPTPDKEDDDRVYPNSTCKPRIIAPSRGSPLPVLSWANREEVWKIMLN 122
Query: 68 KEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVF 127
KEK +L + +L + L +R LDW+ + Y + L+ ++ DR+++
Sbjct: 123 KEKTYLRDQHFLEQ--HPLLQPKMRAILLDWLMEVCEVYKLHRETFYLAQDFFDRYMATQ 180
Query: 128 QFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSL 187
+ V +QL+ ++ +AAK+EE+ P+ I MELMI+ +L
Sbjct: 181 E--NVVKTLLQLIGISSLFIAAKLEEIYPPKLHQFAYVTDG-ACSGDEILTMELMIMKAL 237
Query: 188 GWKMRALTPCSFIDYF--------LAKISCEKYPDKSLIARSVQLILNIIKGIDFLEFRS 239
W++ LT S+++ + L ++ +YP + I + +L+ + +D LEF
Sbjct: 238 KWRLSPLTIVSWLNVYMQVAYLNDLHEVLLPQYPQQIFI-QIAELLDLCVLDVDCLEFPY 296
Query: 240 SEIAAAVAISLKELPTQEVDKAITDFFIVDKERVLK 275
+AA+ +L + E+ + ++ + D E +K
Sbjct: 297 GILAAS---ALYHFSSSELMQKVSGYQWCDIENCVK 329
>CGD1_RAT (P39948) G1/S-specific cyclin D1
Length = 295
Score = 61.6 bits (148), Expect = 3e-09
Identities = 53/225 (23%), Positives = 108/225 (47%), Gaps = 15/225 (6%)
Query: 58 SEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSV 117
++ +++ M++ E+ P Y ++ ++ S+R+ W+ + L++
Sbjct: 24 NDRVLRAMLKTEETCAPSVSYFKCVQR-EIVPSMRKIVATWMLEVCEEQKCEEEVFPLAM 82
Query: 118 NYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQ 177
NYLDRFLS+ + +QLL C +A+KM+E +P + + + + +
Sbjct: 83 NYLDRFLSLEPLKKS---RLQLLGATCMFVASKMKET-IPLTAEKLCIYTDNSIRPEELL 138
Query: 178 RMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNI-IKGIDFLE 236
+MEL++++ L W + A+TP FI++FL+K+ E +K +I + Q + + + F+
Sbjct: 139 QMELLLVNKLKWNLAAMTPHDFIEHFLSKMP-EADENKQIIRKHAQTFVALCATDVKFIS 197
Query: 237 FRSSEIAAAVAISLKE-----LPTQEVDKAITDFFIVDKERVLKC 276
S +AA ++ + P + T F+ RV+KC
Sbjct: 198 NPPSMVAAGSVVAAMQGLNLGSPNNFLSCYRTTHFL---SRVIKC 239
>CGD1_HUMAN (P24385) G1/S-specific cyclin D1 (PRAD1 oncogene) (BCL-1
oncogene)
Length = 295
Score = 61.6 bits (148), Expect = 3e-09
Identities = 53/222 (23%), Positives = 111/222 (49%), Gaps = 9/222 (4%)
Query: 58 SEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSV 117
++ +++ M++ E+ P Y ++ L S+R+ W+ + L++
Sbjct: 24 NDRVLRAMLKAEETCAPSVSYFKCVQKEVLP-SMRKIVATWMLEVCEEQKCEEEVFPLAM 82
Query: 118 NYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQ 177
NYLDRFLS+ + +QLL C +A+KM+E +P + + + + +
Sbjct: 83 NYLDRFLSLEPVKKS---RLQLLGATCMFVASKMKET-IPLTAEKLCIYTDNSIRPEELL 138
Query: 178 RMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNI-IKGIDFLE 236
+MEL++++ L W + A+TP FI++FL+K+ E +K +I + Q + + + F+
Sbjct: 139 QMELLLVNKLKWNLAAMTPHDFIEHFLSKMP-EAEENKQIIRKHAQTFVALCATDVKFIS 197
Query: 237 FRSSEIAA-AVAISLKELPTQEVDKAITDFFIVD-KERVLKC 276
S +AA +V +++ L + + ++ + + RV+KC
Sbjct: 198 NPPSMVAAGSVVAAVQGLNLRSPNNFLSYYRLTRFLSRVIKC 239
>CGD1_XENLA (P50755) G1/S-specific cyclin D1
Length = 291
Score = 61.2 bits (147), Expect = 4e-09
Identities = 65/266 (24%), Positives = 123/266 (45%), Gaps = 20/266 (7%)
Query: 58 SEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSV 117
++ +++ M++ E+ P Y ++ L ++R+ W+ + L++
Sbjct: 22 TDRVLQTMLKAEETSCPSMSYFKCVQKEILP-NMRKIVATWMLEVCEEQKCEEEVFPLAM 80
Query: 118 NYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQ 177
NYLDRFLSV P +W +QLL C LA+KM+E +P + + + +
Sbjct: 81 NYLDRFLSVE--PLRKSW-LQLLGATCMFLASKMKET-IPLTAEKLCIYTDNSIRPDELL 136
Query: 178 RMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNI-IKGIDFLE 236
MEL +L+ L W + ++TP FI++FL K+ + K +I + Q + + ++F+
Sbjct: 137 IMELRVLNKLKWDLASVTPHDFIEHFLNKMPLTE-DTKQIIRKHAQTFVALCATDVNFIS 195
Query: 237 FRSSEIAA-AVAISLKELPTQEVDKAI-TDFFIVDKERVLKC-----------VELIRDL 283
S IAA +VA +++ L D T + +V+KC +E + +
Sbjct: 196 NPPSMIAAGSVAAAVQGLNLGNADSVFSTQRLTLFLSQVIKCDPDCLRACQEQIESLLES 255
Query: 284 SLIKVGGNNFASFVPQSPIGVLDAGC 309
SL + + AS ++ + +D C
Sbjct: 256 SLRQAQQQHNASSDTKNMVDEVDISC 281
>CGA2_BOVIN (P30274) Cyclin A2 (Cyclin A) (Fragment)
Length = 406
Score = 60.8 bits (146), Expect = 5e-09
Identities = 62/224 (27%), Positives = 101/224 (44%), Gaps = 12/224 (5%)
Query: 24 DVVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQSEEIVKVMVEKEKDHLPREDYLIRLR 83
D +D S SP +V L++ S E+I + E E P+ Y+ +
Sbjct: 119 DYPMDGSFESPHTMEMSVVLEDEKPVSVNEVPDYHEDIHTYLREMEVKCKPKVGYM--KK 176
Query: 84 GGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVA 143
D+ S+R +DW+ + Y +L L+VNY+DRFLS RG +QL+ A
Sbjct: 177 QPDITNSMRAILVDWLVEVGEEYKLQNETLHLAVNYIDRFLSSMSVLRG---KLQLVGTA 233
Query: 144 CFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYF 203
LA+K EE+ P+ + V + K + RME ++L L + + A T I+ F
Sbjct: 234 AMLLASKFEEIYPPEVAEF-VYITDDTYTKKQVLRMEHLVLKVLAFDLAAPT----INQF 288
Query: 204 LAKISCEKYPDKSLIARSVQLI--LNIIKGIDFLEFRSSEIAAA 245
L + + P + + L++I +L++ S IAAA
Sbjct: 289 LTQYFLHQQPANCKVESLAMFLGELSLIDADPYLKYLPSVIAAA 332
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,666,913
Number of Sequences: 164201
Number of extensions: 1621200
Number of successful extensions: 3739
Number of sequences better than 10.0: 122
Number of HSP's better than 10.0 without gapping: 53
Number of HSP's successfully gapped in prelim test: 69
Number of HSP's that attempted gapping in prelim test: 3597
Number of HSP's gapped (non-prelim): 124
length of query: 346
length of database: 59,974,054
effective HSP length: 111
effective length of query: 235
effective length of database: 41,747,743
effective search space: 9810719605
effective search space used: 9810719605
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC135848.10


