

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135604.8 + phase: 0 /pseudo
(1050 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
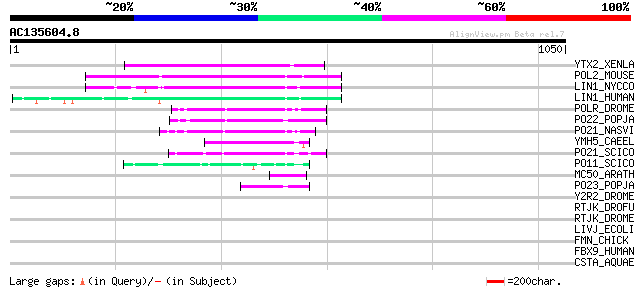
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 137 1e-31
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 128 9e-29
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 127 2e-28
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 117 2e-25
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 82 1e-14
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 81 2e-14
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 75 9e-13
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 66 4e-10
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 64 2e-09
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 62 1e-08
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 57 3e-07
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 51 1e-05
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 45 0.001
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 40 0.025
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 40 0.033
LIVJ_ECOLI (P02917) Leu/Ile/Val-binding protein precursor (LIV-BP) 32 8.9
FMN_CHICK (Q05858) Formin (Limb deformity protein) 32 8.9
FBX9_HUMAN (Q9UK97) F-box only protein 9 32 8.9
CSTA_AQUAE (O67304) Carbon starvation protein A homolog 32 8.9
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 137 bits (346), Expect = 1e-31
Identities = 105/381 (27%), Positives = 179/381 (46%), Gaps = 10/381 (2%)
Query: 218 QSRLNWLRDGDANSKFFHSVLAARRRLNSLSSILV-DGVVVEGVQPFRQAVFTHFKNHFL 276
+SR+ L D D S+FF+++ + ++ + DG +E + R + ++N F
Sbjct: 359 RSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARSFYQNLFS 418
Query: 277 DPRLERPGVGHLQ--FRKLSYLERGGLTKPFSEDEIKAAVWDCDSFKSPGPDGINLGFFK 334
+ L +S + L P + DE+ A+ KSPG DG+ + FF+
Sbjct: 419 PDPISPDACEELWDGLPVVSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEFFQ 478
Query: 335 SFWPEIKTDLVRFVTEFHRNGKLAKGINSTFIALIPKVASPQSLNEFRPISLVGSMYKIL 394
FW + D R +TE + G+L ++L+PK + + +RP+SL+ + YKI+
Sbjct: 479 FFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYKIV 538
Query: 395 AKILANRLRQVIGSVVSEAQSAFVKNRQILDGILIAN*VVDEARKLKKYLLLFKVDFAKA 454
AK ++ RL+ V+ V+ QS V R I D + + ++ AR+ L +D KA
Sbjct: 539 AKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARRTGLSLAFLSLDQEKA 598
Query: 455 YDSVDWGYLDEVMGSMSFPVVWRKWIKECVGTATASVLVNGSPTDEFHLERGLRQGDPLS 514
+D VD YL + + SF + ++K +A V +N S T RG+RQG PLS
Sbjct: 599 FDRVDHQYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINWSLTAPLAFGRGVRQGCPLS 658
Query: 515 PFLFLLAA*GLNVLMKALVDAGMFTGYRVGRADSVVVSHLQFADDTLLIGNKSWANVRAL 574
L+ LA L++ TG + D VV +ADD +L+ + ++
Sbjct: 659 GQLYSLAIEPFLCLLRK-----RLTGLVLKEPDMRVVLS-AYADDVILVA-QDLVDLERA 711
Query: 575 RAGLVMFEAMSGLKVNFHKSS 595
+ ++ A S ++N+ KSS
Sbjct: 712 QECQEVYAAASSARINWSKSS 732
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 128 bits (321), Expect = 9e-29
Identities = 126/491 (25%), Positives = 222/491 (44%), Gaps = 19/491 (3%)
Query: 144 LKEKLKLMKVALKDWHAANTHNLPSKIAELKI*QASLDSKGEEEQLSEEELGELHGISTD 203
L+ KL + + K A+T +L + + L+ +A+ + +++ + GE++ + T
Sbjct: 321 LRGKLIALSASKKKRETAHTSSLTTHLKALEKKEANSPKRSRRQEIIKLR-GEINQVETR 379
Query: 204 IHSLSRLITSICWQQSRLNWLRDGDAN-SKFFHSVLAARRRLNSLSSILVDGVVVEG-VQ 261
++ R+ + W ++N + A +K + + N I D ++ ++
Sbjct: 380 -RTIQRINQTRSWFFEKINKIDKPLARLTKGHRDKILINKIRNEKGDITTDPEEIQNTIR 438
Query: 262 PFRQAVF-THFKN-HFLDPRLERPGVGHLQFRKLSYLERGGLTKPFSEDEIKAAVWDCDS 319
F + ++ T +N +D L+R Q KL+ + L P S EI+A + +
Sbjct: 439 SFYKRLYSTKLENLDEMDKFLDR-----YQVPKLNQDQVDHLNSPISPKEIEAVINSLPT 493
Query: 320 FKSPGPDGINLGFFKSFWPEIKTDLVRFVTEFHRNGKLAKGINSTFIALIPKVAS-PQSL 378
KSPGPDG + F+++F ++ L + + G L I LIPK P +
Sbjct: 494 KKSPGPDGFSAEFYQTFKEDLIPILHKLFHKIEVEGTLPNSFYEATITLIPKPQKDPTKI 553
Query: 379 NEFRPISLVGSMYKILAKILANRLRQVIGSVVSEAQSAFVKNRQILDGILIAN*VVDEAR 438
FRPISL+ KIL KILANR+++ I +++ Q F+ Q I + V+
Sbjct: 554 ENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQVGFIPGMQGWFNIRKSINVIHYIN 613
Query: 439 KLK-KYLLLFKVDFAKAYDSVDWGYLDEVMGSMSFPVVWRKWIKECVGTATASVLVNGSP 497
KLK K ++ +D KA+D + ++ +V+ + IK A++ VNG
Sbjct: 614 KLKDKNHMIISLDAEKAFDKIQHPFMIKVLERSGIQGPYLNMIKAIYSKPVANIKVNGEK 673
Query: 498 TDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTGYRVGRADSVVVSHLQFA 557
+ L+ G RQG PLSP+LF + L VL +A+ G ++G+ + V +S L A
Sbjct: 674 LEAIPLKSGTRQGCPLSPYLFNIV---LEVLARAIRQQKEIKGIQIGK-EEVKISLL--A 727
Query: 558 DDTLLIGNKSWANVRALRAGLVMFEAMSGLKVNFHKSSLVGVNINDSWLSEAASVLGCKV 617
DD ++ + + R L + F + G K+N +KS N E +
Sbjct: 728 DDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKSMAFLYTKNKQAEKEIRETTPFSI 787
Query: 618 VKIPFLYLGLS 628
V YLG++
Sbjct: 788 VTNNIKYLGVT 798
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 127 bits (319), Expect = 2e-28
Identities = 126/500 (25%), Positives = 215/500 (42%), Gaps = 37/500 (7%)
Query: 143 VLKEKLKLMKVALKDWHAANTHNLPSKIAELKI*QASLDSKGEEEQLSE--EELGELHGI 200
VL+ K ++ LK +NL + +L+ + S +++++ EL E+
Sbjct: 293 VLRGKFIALQAFLKKTEREEVNNLMGHLKQLEKEEHSNPKPSRRKEITKIRAELNEIE-- 350
Query: 201 STDIHSLSRLITSICWQQSRLNWLRDGDANSKFFHSVLAARRRLNSLSSILVDG------ 254
+ + ++ S W ++N + AN L ++R+ SL S + +G
Sbjct: 351 --NKRIIQQINKSKSWFFEKINKIDKPLAN-------LTRKKRVKSLISSIRNGNDEITT 401
Query: 255 ---VVVEGVQPFRQAVFTHFKNHF--LDPRLERPGVGHLQFRKLSYLERGGLTKPFSEDE 309
+ + + + + +++H + +D LE HL +LS E L +P S E
Sbjct: 402 DPSEIQKILNEYYKKLYSHKYENLKEIDQYLE---ACHLP--RLSQKEVEMLNRPISSSE 456
Query: 310 IKAAVWDCDSFKSPGPDGINLGFFKSFWPEIKTDLVRFVTEFHRNGKLAKGINSTFIALI 369
I + + + KSPGPDG F+++F E+ L+ + G L I LI
Sbjct: 457 IASTIQNLPKKKSPGPDGFTSEFYQTFKEELVPILLNLFQNIEKEGILPNTFYEANITLI 516
Query: 370 PKVAS-PQSLNEFRPISLVGSMYKILAKILANRLRQVIGSVVSEAQSAFVKNRQILDGIL 428
PK P +RPISL+ KIL KIL NR++Q I ++ Q F+ Q I
Sbjct: 517 PKPGKDPTRKENYRPISLMNIDAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQGWFNIR 576
Query: 429 IAN*VVDEARKLK-KYLLLFKVDFAKAYDSVDWGYLDEVMGSMSFPVVWRKWIKECVGTA 487
+ V+ KLK K ++ +D KA+D++ ++ + + + K I+
Sbjct: 577 KSINVIQHINKLKNKDHMILSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKLIEAIYSKP 636
Query: 488 TASVLVNGSPTDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTGYRVGRAD 547
TA++++NG F L G RQG PLSP LF + + VL A+ + G +G
Sbjct: 637 TANIILNGVKLKSFPLRSGTRQGCPLSPLLFNIV---MEVLAIAIREEKAIKGIHIG--- 690
Query: 548 SVVVSHLQFADDTLLIGNKSWANVRALRAGLVMFEAMSGLKVNFHKSSLVGVNINDSWLS 607
S + FADD ++ + + L + + +SG K+N HKS N+
Sbjct: 691 SEEIKLSLFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKSVAFIYTNNNQAEK 750
Query: 608 EAASVLGCKVVKIPFLYLGL 627
+ VV YLG+
Sbjct: 751 TVKDSIPFTVVPKKMKYLGV 770
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 117 bits (293), Expect = 2e-25
Identities = 147/650 (22%), Positives = 259/650 (39%), Gaps = 48/650 (7%)
Query: 6 VCGDFNTVRSVEERRSVRGSQVVDDCTPFNDFIEDRVLIDLLLS----GRKFTWFKGDGR 61
+ GDFNT S +R + + ++ D N + LID+ + ++T+F
Sbjct: 141 IMGDFNTPLSTLDRSTRQ--KINKDIQELNSALHQADLIDIYRTLHPKSTEYTFFSAPHH 198
Query: 62 SMRRLDRFLLSVEWCMVWPNCIQVAQLRG-LSDHCPILLSV--------DEENWGPRPVR 112
+ + D L S + C + + LSDH I L + W +
Sbjct: 199 TYSKTDHILGSKT---LLSKCKRTEIITNCLSDHSAIKLELRIKKLTQNHSTTWKLNNLL 255
Query: 113 LLKCW------QDMPGYIDFIREKWVFFQVSGW--GGHVLKEKLKLMKVALKDWHAANTH 164
L W ++ + + K +Q + W V + K + + +
Sbjct: 256 LNDYWVHNEMKAEIKKFFETNENKDTTYQ-NLWDTAKAVCRGKFIALNAHKRKQERSKID 314
Query: 165 NLPSKIAELKI*QASLDSKGEEEQLSEEELGELHGISTDIHSLSRLITSICWQQSRLNWL 224
L S++ EL+ Q +SK Q + EL I T +L ++ S W ++N +
Sbjct: 315 TLISQLKELEK-QEQTNSKASRRQEIIKIRAELKEIETQ-KTLQKINESRSWFFEKINKI 372
Query: 225 RDGDANSKFFHSVLAARRRLNSLSSILVDGVVVEGVQPFRQAVFTHFKNHFLDPRLER-- 282
+ ++ +R N + +I D + Q + H +LE
Sbjct: 373 ------DRPLARLIKKKREKNQIDTIKNDRGDITTDPTEIQTTIREYYKHLYANKLENLE 426
Query: 283 ---PGVGHLQFRKLSYLERGGLTKPFSEDEIKAAVWDCDSFKSPGPDGINLGFFKSFWPE 339
+ +L+ E L +P + EI+A + + KSPGP+G F++ + E
Sbjct: 427 EMDKFLDTYTLPRLNQEEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQRYKEE 486
Query: 340 IKTDLVRFVTEFHRNGKLAKGINSTFIALIPKVASPQSLNE-FRPISLVGSMYKILAKIL 398
+ L++ + G L I LIPK + E FRPISL+ KIL KIL
Sbjct: 487 LVPFLLKLFQSIEKEGILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNKIL 546
Query: 399 ANRLRQVIGSVVSEAQSAFVKNRQILDGILIAN*VVDEARKLK-KYLLLFKVDFAKAYDS 457
AN+++Q I ++ Q F+ Q I + ++ + K ++ +D KA+D
Sbjct: 547 ANQIQQHIKKLIHHDQVGFIPAMQGWFNIRKSINIIQHINRTKDTNHMIISIDAEKAFDK 606
Query: 458 VDWGYLDEVMGSMSFPVVWRKWIKECVGTATASVLVNGSPTDEFHLERGLRQGDPLSPFL 517
+ ++ + + + + K I+ TA++++NG + L+ G RQG PLSP L
Sbjct: 607 IQQPFMLKPLNKLGIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGCPLSPLL 666
Query: 518 FLLAA*GLNVLMKALVDAGMFTGYRVGRADSVVVSHLQFADDTLLIGNKSWANVRALRAG 577
+ L VL +A+ G ++G+ + V FADD ++ + + L
Sbjct: 667 PNIV---LEVLARAIRQEKEIKGIQLGKEE---VKLSLFADDMIVYLENPIVSAQNLLKL 720
Query: 578 LVMFEAMSGLKVNFHKSSLVGVNINDSWLSEAASVLGCKVVKIPFLYLGL 627
+ F +SG K+N KS N S+ S L + YLG+
Sbjct: 721 ISNFSKVSGYKINVQKSQAFLYTNNRQTESQIMSELPFTIASKRIKYLGI 770
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 81.6 bits (200), Expect = 1e-14
Identities = 79/294 (26%), Positives = 134/294 (44%), Gaps = 17/294 (5%)
Query: 306 SEDEIKAAVWDCDSFKSPGPDGINLGFFKSFWPEIKTDLVRFVTEFHRNGKLAKGINSTF 365
+E +++A+ S SPGPDGI KS ++R + G L I
Sbjct: 345 TEQDLRASRVSLSS--SPGPDGITP---KSAREVPSGIMLRIMNLILWCGNLPHSIRLAR 399
Query: 366 IALIPKVASPQSLNEFRPISLVGSMYKILAKILANRLRQVIGSVVSEAQSAFVKNRQILD 425
IPK + + +FRPIS+ + + L ILA RL I Q F+ D
Sbjct: 400 TVFIPKTVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSINW--DPRQRGFLPTDGCAD 457
Query: 426 GILIAN*VVDEARKLKKYLLLFKVDFAKAYDSVDWGYLDEVMGSMSFPVVWRKWIKECVG 485
I + V+ + K + + +D +KA+DS+ + + + + P + +++
Sbjct: 458 NATIVDLVLRHSHKHFRSCYIANLDVSKAFDSLSHASIYDTLRAYGAPKGFVDYVQNTYE 517
Query: 486 TATASVLVNGSPTDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTGYRVGR 545
S+ +G ++EF RG++QGDPLSP LF L ++ L++ L G +VG
Sbjct: 518 GGGTSLNGDGWSSEEFVPARGVKQGDPLSPILFNLV---MDRLLRTLPSE---IGAKVGN 571
Query: 546 ADSVVVSHLQFADDTLLIGNKSWANVRALRAGLVMFEAMSGLKVNFHKSSLVGV 599
A + + FADD +L ++ ++ L + F ++ GLK+N K VG+
Sbjct: 572 A---ITNAAAFADDLVLFA-ETRMGLQVLLDKTLDFLSIVGLKLNADKCFTVGI 621
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 80.9 bits (198), Expect = 2e-14
Identities = 74/299 (24%), Positives = 134/299 (44%), Gaps = 19/299 (6%)
Query: 303 KPFSEDEIKAAVWDCDSFKSPGPDGINLGFFKSFWPEIKTDLVRFVTEFHR-NGKLAKGI 361
+P + +EI+ A+ +PG DG+ + +T L R + H G +
Sbjct: 4 RPIAREEIQCAIKGWKP-SAPGSDGLTVQAIT------RTRLPRNFVQLHLLRGHVPTPW 56
Query: 362 NSTFIALIPKVASPQSLNEFRPISLVGSMYKILAKILANRLRQVIGSVVSEAQSAFVKNR 421
+ LIPK ++ + +RPI++ ++ ++L +ILA RL + + AQ + +
Sbjct: 57 TAMRTTLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVE--LHPAQKGYARID 114
Query: 422 QILDGILIAN*VVDEARKLKKYLLLFKVDFAKAYDSVDWGYLDEVMGSMSFPVVWRKWIK 481
L L+ + + R+ +K + +D KA+D+V + + + +I
Sbjct: 115 GTLVNSLLLDTYISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYIT 174
Query: 482 ECVGTATASVLVN-GSPTDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTG 540
+ +T ++ V GS T + + RG++QGDPLSPFLF N ++ L+ + T
Sbjct: 175 GSLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLF-------NAVLDELLCSLQSTP 227
Query: 541 YRVGRADSVVVSHLQFADDTLLIGNKSWANVRALRAGLVMFEAMSGLKVNFHKSSLVGV 599
G + L FADD LL+ + L A + F + G+ +N KS + V
Sbjct: 228 GIGGTIGEEKIPVLAFADDLLLLEDNDVLLPTTL-ATVANFFRLRGMSLNAKKSVSISV 285
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 75.1 bits (183), Expect = 9e-13
Identities = 75/299 (25%), Positives = 131/299 (43%), Gaps = 22/299 (7%)
Query: 283 PGVGHLQFRKLSYLERGGLTKPFSEDEIKAAVWDCDSFKSPGPDGINLGFFKSFWPEIKT 342
P HL+ R ++ L +P S DEIK + + GPDG+ + W I
Sbjct: 299 PTTRHLRSRMQGEIKN--LWRPISNDEIKEV--EACKRTAAGPDGMTT----TAWNSIDE 350
Query: 343 DLVRFVTEFHRNGKLAKGINSTFIALIPKVASPQSLNEFRPISLVGSMYKILAKILANRL 402
+ +G+ + + LIPK FRP+S+ + +ILANR+
Sbjct: 351 CIKSLFNMIMYHGQCPRRYLDSRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANRI 410
Query: 403 RQVIGSVVSEAQSAFVKNRQILDGILIAN*VVDEARKLKKYLLLFKVDFAKAYDSVDWGY 462
+ ++ Q AF+ + + + + ++ EAR K L + +D KA+DSV+
Sbjct: 411 GE--HGLLDTRQRAFIVADGVAENTSLLSAMIKEARMKIKGLYIAILDVKKAFDSVEHRS 468
Query: 463 LDEVMGSMSFPVVWRKWIKECVGTATASVLVNGSPTDEFHLERGLRQGDPLSPFLFLLAA 522
+ + + P+ R +I + + V + RG+RQGDPLSP LF
Sbjct: 469 ILDALRRKKLPLEMRNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLFNCV- 527
Query: 523 *GLNVLMKALVDAGMFTGYRVGRADSVVVSHLQFADDTLLIGNKS---WANVRALRAGL 578
++ +++ L + TG+ +G + + L FADD +L+ A++ + AGL
Sbjct: 528 --MDAVLRRLPEN---TGFLMG---AEKIGALVFADDLVLLAETREGLQASLSRIEAGL 578
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 66.2 bits (160), Expect = 4e-10
Identities = 53/204 (25%), Positives = 93/204 (44%), Gaps = 13/204 (6%)
Query: 369 IPKVASPQSLNEFRPISLVGSMYKILAKILANRLRQVIGSVVSEAQSAFVKNRQILDGIL 428
IPK +P S + +RPISL +I+ +I+ +R+R ++S Q F+ R ++
Sbjct: 673 IPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGFLNFRSCPSSLV 732
Query: 429 IAN*VVDEARKLKKYLLLFKVDFAKAYDSVDWGYLDEVMGSMSFPVVWRKWIKECVGTAT 488
+ + K +K L + DFAKA+D V L + + + W KE + T
Sbjct: 733 RSISLYHSILKNEKSLDILFFDFAKAFDKVSHPILLKKLALFGLDKLTCSWFKEFLHLRT 792
Query: 489 ASVLVNG-SPTDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTGYRVGRAD 547
SV +N ++ + + G+ QG P LF+L L + ++ + F AD
Sbjct: 793 FSVKINKFVSSNAYPISSGVPQGSVSGPLLFILFINDLLIDLEPNIHVSCF-------AD 845
Query: 548 SVVVSH-----LQFADDTLLIGNK 566
+ + H LQ + DT++ +K
Sbjct: 846 DIKIFHHNPSTLQNSIDTIVKWSK 869
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 64.3 bits (155), Expect = 2e-09
Identities = 79/302 (26%), Positives = 125/302 (41%), Gaps = 24/302 (7%)
Query: 301 LTKPFSEDEIKAAVWDCDSFKSPGPDGINLGFFKSFWPEIKTDLVRFVTEFHRNGKLAKG 360
L P S EIK+A + K GPDG+ + + K L + R KG
Sbjct: 155 LWDPVSLIEIKSA--RASNEKGAGPDGVTPRSWNALDDRYKRLLYNIFVFYGRVPSPIKG 212
Query: 361 INSTFIALIPKVASPQSLNEFRPISLVGSMYKILAKILANRLRQVIGSVVSEAQSAFVKN 420
+ F PK+ FRP+S+ + + KILA R V E Q+A++
Sbjct: 213 SRTVFT---PKIEGGPDPGVFRPLSICSVILREFNKILARRF--VSCYTYDERQTAYLPI 267
Query: 421 RQILDGILIAN*VVDEARKLKKYLLLFKVDFAKAYDSVDWGYLDEVMGSMSFPVVWRKWI 480
+ + + ++ EA++L+K L + +D KA++SV L + + P +I
Sbjct: 268 DGVCINVSMLTAIIAEAKRLRKELHIAILDLVKAFNSVYHSALIDAITEAGCPPGVVDYI 327
Query: 481 KECVGTATASVLVNGSPTDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTG 540
+ + G + + G+ QGDPLS LF LA ++AL + G F
Sbjct: 328 ADMYNNVITEMQFEGK-CELASILAGVYQGDPLSGPLFTLA---YEKALRALNNEGRFD- 382
Query: 541 YRVGRADSVVVSHLQFADDTLLIGNKSWANVRALRAGLVMFE---AMSGLKVNFHKSSLV 597
V V+ ++DD LL+ V L+ L F A GL++N KS V
Sbjct: 383 -----IADVRVNASAYSDDGLLLA----MTVIGLQHNLDKFGETLAKIGLRINSRKSKTV 433
Query: 598 GV 599
+
Sbjct: 434 SL 435
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 61.6 bits (148), Expect = 1e-08
Identities = 93/365 (25%), Positives = 147/365 (39%), Gaps = 53/365 (14%)
Query: 215 CWQQSRLNWLRDGDANSKFFHSVLAARRRLNSLSSILVDGVVVEGVQPFRQAVFTHFKNH 274
CWQ+ + + F + RR+ + S+ VDGV + + A+ F
Sbjct: 326 CWQEFVASESNENPWGRVF--KLCRGRRKPVDVCSVKVDGVYTDTWEGSVNAMMNVFFPA 383
Query: 275 FLDPRLERPGVGHLQFRKLSYLERGGLTKPFSEDEIKAAVWDCDSFKSPGPDGINLGFFK 334
+D E +L + R L DE+ +V C KSPGPDGI +
Sbjct: 384 SIDDASE--------IDRLKAIARP-LPPDLEMDEVSDSVRRCKVRKSPGPDGIVGEMVR 434
Query: 335 SFW---PEIKTDLVR---FVTEFHRNGKLAKGINSTFIALIPKVASPQSLNEFRPISLVG 388
+ W PE L + + F + K+A + + L+ ++ S +RPI L+
Sbjct: 435 AVWGAIPEYMFCLYKQCLLESYFPQKWKIASLV--ILLKLLDRIRSDP--GSYRPICLLD 490
Query: 389 SMYKILAKILANRLRQVIGSV-VSEAQSAFVKNRQILDGILIAN*VVDEARKLKKYLLLF 447
++ K+L I+ RL Q + V VS Q AF + D V+ + KY++
Sbjct: 491 NLGKVLEGIMVKRLDQKLMDVEVSPYQFAFTYGKSTEDAWRCVQRHVECSE--MKYVIGL 548
Query: 448 KVDFAKAYDSVDW----GYLDEVMGSMSFPVVWRKWIKECVGTATASVLVNGSPTDEFHL 503
+DF A+D++ W LDE + + W+ G V G + +
Sbjct: 549 NIDFQGAFDNLGWLSMLLKLDEAQSN-----EFGLWMSYFGGRKVYYVGKTGIVRKD--V 601
Query: 504 ERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTGYRVGRADSVVVSHLQFADD-TLL 562
RG QG P ++ L +N L+ ALV AG F + FADD T++
Sbjct: 602 TRGCPQGSKSGPAMWKLV---MNELLLALVAAGFFI--------------VAFADDGTIV 644
Query: 563 IGNKS 567
IG S
Sbjct: 645 IGANS 649
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 57.0 bits (136), Expect = 3e-07
Identities = 31/69 (44%), Positives = 42/69 (59%), Gaps = 1/69 (1%)
Query: 492 LVNGSPTDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTGYRVGRADSVVV 551
++NG+P RGLRQGDPLSP+LF+L L+ L + + G G RV +S +
Sbjct: 13 IINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSN-NSPRI 71
Query: 552 SHLQFADDT 560
+HL FADDT
Sbjct: 72 NHLLFADDT 80
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 51.2 bits (121), Expect = 1e-05
Identities = 35/130 (26%), Positives = 63/130 (47%), Gaps = 8/130 (6%)
Query: 438 RKLK-KYLLLFKVDFAKAYDSVDWGYLDEVMGSMSFPVVWRKWIKECVGTATASVLVNGS 496
R+LK K + +D KA+D+V + M + + +I + A +++V G
Sbjct: 16 RRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMSTITDAYTNIVVGGR 75
Query: 497 PTDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTGYRVGRADSVVVSHLQF 556
T++ ++ G++QGDPLSP LF N+++ LV + ++ L F
Sbjct: 76 TTNKIYIRNGVKQGDPLSPVLF-------NIVLDELVTRLNDEQPGASMTPACKIASLAF 128
Query: 557 ADDTLLIGNK 566
ADD LL+ ++
Sbjct: 129 ADDLLLLEDR 138
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 45.1 bits (105), Expect = 0.001
Identities = 52/214 (24%), Positives = 85/214 (39%), Gaps = 12/214 (5%)
Query: 309 EIKAAVWDCDSFKSPGPDGINLGFFKSFWPEIKTDLVRFVTEFHRNGKLAKGINSTFIAL 368
E+ V S +SPG DGIN K+ W I L + R G +
Sbjct: 438 EVDTCVARLKSRRSPGLDGINGTICKAVWRAIPEHLASLFSRCIRLGYFPAEWKCPRVVS 497
Query: 369 IPKVASPQSL--NEFRPISLVGSMYKILAKILANRLRQVIGSVVSEAQSAFVKNRQILDG 426
+ K + +R I L+ K+L I+ NR+R+V+ Q F + R + D
Sbjct: 498 LLKGPDKDKCEPSSYRGICLLPVFGKVLEAIMVNRVREVLPE-GCRWQFGFRQGRCVEDA 556
Query: 427 ILIAN*VVDEARKLKKYLLLFKVDFAKAYDSVDWGYLDEVMGSMSFPV--VWRKWIKECV 484
V +Y+L VDF A+D+V+W + + +W+ +
Sbjct: 557 WRHVKSSV--GASAAQYVLGTFVDFKGAFDNVEWSAALSRLADLGCREMGLWQSFF---- 610
Query: 485 GTATASVLVNGSPTDEFHLERGLRQGDPLSPFLF 518
+ +V+ + S T E + RG QG PF++
Sbjct: 611 -SGRRAVIRSSSGTVEVPVTRGCPQGSISGPFIW 643
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 40.4 bits (93), Expect = 0.025
Identities = 51/218 (23%), Positives = 91/218 (41%), Gaps = 4/218 (1%)
Query: 304 PFSEDEIKAAVWDCDSFKSPGPDGINLGFFKSFWPEIKTDLVRFVTEFHRNGKLAKGINS 363
P + +E+K V K+PG D ++ + + LV R G K +
Sbjct: 437 PVTLEEVKELVSKLKPKKAPGEDLLDNRTIRLLPDQALLYLVLIFNSILRVGYFPKARPT 496
Query: 364 TFIALIPKVAS-PQSLNEFRPISLVGSMYKILAKILANRL--RQVIGSVVSEAQSAFVKN 420
I +I K P ++ +RP SL+ S+ K+L +++ NR+ + + + + Q F
Sbjct: 497 ASIIMILKPGKQPLDVDSYRPTSLLPSLGKMLERLILNRILTSEEVTRAIPKFQFGFRLQ 556
Query: 421 RQILDGILIAN*VVDEARKLKKYLLLFKVDFAKAYDSVDWGYLDEVMGSMSFPVVWRKWI 480
+ + EA + K+Y +D +A+D V L S+ P ++ + I
Sbjct: 557 HGTPEQLHRVVNFALEALEKKEYAGSCFLDIQQAFDRVWHPGLLYKAKSLLSPQLF-QLI 615
Query: 481 KECVGTATASVLVNGSPTDEFHLERGLRQGDPLSPFLF 518
K SV +G + +E G+ QG L P L+
Sbjct: 616 KSFWEGRKFSVTADGCRSSVKFIEAGVPQGSVLGPTLY 653
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 40.0 bits (92), Expect = 0.033
Identities = 58/259 (22%), Positives = 101/259 (38%), Gaps = 16/259 (6%)
Query: 308 DEIKAAVWDCDSFKSPGPDGINLGFFKSFWPEIKTDLVRFVTEFHRNGKLAKGINSTFIA 367
+E+K + K+PG D ++ + + L G K S I
Sbjct: 441 EEVKNLIAKLPLKKAPGEDLLDNRTIRLLPDQALQFLALIFNSVLDVGYFPKAWKSASII 500
Query: 368 LIPKVA-SPQSLNEFRPISLVGSMYKILAKILANRLR--QVIGSVVSEAQSAFVKNRQIL 424
+I K +P ++ +RP SL+ S+ KI+ +++ NRL + + + + Q F
Sbjct: 501 MIHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRLLTCKDVTKAIPKFQFGFRLQHGTP 560
Query: 425 DGILIAN*VVDEARKLKKYLLLFKVDFAKAYDSVDWGYLDEVMGSMSFPVVWRKWIKECV 484
+ + EA + K+Y + +D +A+D V W FP +K +
Sbjct: 561 EQLHRVVNFALEAMENKEYAVGAFLDIQQAFDRV-WHPGLLYKAKRLFPPQLYLVVKSFL 619
Query: 485 GTATASVLVNGSPTDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTGYRVG 544
T V V+G + + G+ QG L P L+ + A + M T V
Sbjct: 620 EERTFHVSVDGYKSSIKPIAAGVPQGSVLGPTLYSVFA------------SDMPTHTPVT 667
Query: 545 RADSVVVSHLQFADDTLLI 563
D V +ADDT ++
Sbjct: 668 EVDEEDVLIATYADDTAVL 686
>LIVJ_ECOLI (P02917) Leu/Ile/Val-binding protein precursor (LIV-BP)
Length = 367
Score = 32.0 bits (71), Expect = 8.9
Identities = 31/114 (27%), Positives = 49/114 (42%), Gaps = 15/114 (13%)
Query: 224 LRDGDANSKFFHSVLAARRRLNSLSSIL----VDGVVVEGVQP-----FRQAVFTHFKNH 274
L+ G+AN FF + A + ++L + L +D V G P RQA K
Sbjct: 185 LKKGNANVVFFDGITAGEKDFSTLVARLKKENIDFVYYGGYHPEMGQILRQARAAGLKTQ 244
Query: 275 FLDPRLERPGVGHLQFRKLSYLERGGL--TKPFSEDEIKAAVWDCDSFKSPGPD 326
F+ P GV ++ ++ GL TKP + D++ A D+ K+ D
Sbjct: 245 FMGPE----GVANVSLSNIAGESAEGLLVTKPKNYDQVPANKPIVDAIKAKKQD 294
>FMN_CHICK (Q05858) Formin (Limb deformity protein)
Length = 1213
Score = 32.0 bits (71), Expect = 8.9
Identities = 23/102 (22%), Positives = 46/102 (44%), Gaps = 7/102 (6%)
Query: 144 LKEKLKLMKVALKDWHAANTHNLPSKIAELKI*QASLDSKGEEEQLSEEELGELHGISTD 203
LK + +L ++ HA +T L IA LK LD+K ++G +ST+
Sbjct: 522 LKSQFELRVFHIRGEHAVSTAQLEETIAHLK---NELDNKLNRRNEEARDIG----VSTE 574
Query: 204 IHSLSRLITSICWQQSRLNWLRDGDANSKFFHSVLAARRRLN 245
+L + ++C Q R +++ + ++ + ++LN
Sbjct: 575 DDNLPKTYRNVCIQTDRETFIKPSEEENRAVKNNQIVPKKLN 616
>FBX9_HUMAN (Q9UK97) F-box only protein 9
Length = 447
Score = 32.0 bits (71), Expect = 8.9
Identities = 24/96 (25%), Positives = 37/96 (38%), Gaps = 26/96 (27%)
Query: 102 DEENWGPRPVRLLKCWQDMPGYIDFIREKWVFFQVSGWGGHVLKEKLKLMKVALKDWHAA 161
DEEN P D+ + R +W+F G ++
Sbjct: 28 DEENESPAET-------DLQAQLQMFRAQWMFELAPGVS-------------------SS 61
Query: 162 NTHNLPSKIAELKI*QASLDSKGEEEQLSEEELGEL 197
N N P + A + + S D+KG++EQ EE+ EL
Sbjct: 62 NLENRPCRAARGSLQKTSADTKGKQEQAKEEKAREL 97
>CSTA_AQUAE (O67304) Carbon starvation protein A homolog
Length = 555
Score = 32.0 bits (71), Expect = 8.9
Identities = 32/116 (27%), Positives = 52/116 (44%), Gaps = 15/116 (12%)
Query: 512 PLSPFLFLLAA*GLNVLMKALVDAGMFTGYRVGRADSVVVSHLQFADDTLLIGNKSWANV 571
P+ PFLF+ A G ALV +G + +D+ V +L F ++ L + A +
Sbjct: 289 PIFPFLFITIACGALSGFHALVSSGTSSKQLEKESDARAVGYLGFMGESSL----ALAVI 344
Query: 572 RALRAGLVMFEAMSGLKV-----NFHKSSLVGVNINDSWLSEAASVLGCKVVKIPF 622
A AG+ MF A ++ + SSLV V + A++LG + + F
Sbjct: 345 IATTAGIAMFSAEKFQELYSSWSSIKSSSLVAV------VDGGANLLGAVGIPVDF 394
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.338 0.149 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 118,002,972
Number of Sequences: 164201
Number of extensions: 4878915
Number of successful extensions: 14208
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 14164
Number of HSP's gapped (non-prelim): 29
length of query: 1050
length of database: 59,974,054
effective HSP length: 121
effective length of query: 929
effective length of database: 40,105,733
effective search space: 37258225957
effective search space used: 37258225957
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC135604.8


