

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135566.8 - phase: 0
(269 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
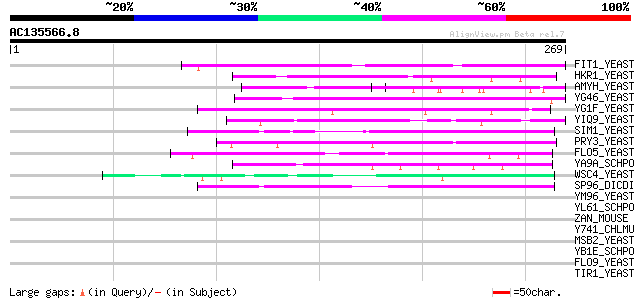
Score E
Sequences producing significant alignments: (bits) Value
FIT1_YEAST (Q04433) Facilitator of iron transport 1 precursor 50 6e-06
HKR1_YEAST (P41809) Hansenula MRAKII killer toxin-resistant prot... 45 2e-04
AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3) (G... 45 2e-04
YG46_YEAST (P53301) Hypothetical 52.8 kDa protein in BUB1-HIP1 i... 45 2e-04
YG1F_YEAST (P53214) Hypothetical 57.5 kDa protein in VMA7-RPS25A... 45 2e-04
YIQ9_YEAST (P40442) Hypothetical 99.7 kDa protein in SDL1 5'regi... 44 4e-04
SIM1_YEAST (P40472) SIM1 protein precursor 44 4e-04
PRY3_YEAST (P47033) PRY3 protein (Pathogen related in Sc 3) 44 5e-04
FLO5_YEAST (P38894) Flocculation protein FLO5 precursor (Floccul... 44 5e-04
YA9A_SCHPO (Q09788) Hypothetical serine-rich protein C13G6.10c i... 43 7e-04
WSC4_YEAST (P38739) Cell wall integrity and stress response comp... 43 7e-04
SP96_DICDI (P14328) Spore coat protein SP96 43 7e-04
YM96_YEAST (Q04893) Hypothetical 113.1 kDa protein in PRE5-FET4 ... 42 0.001
YL61_SCHPO (Q8TFG9) Hypothetical serine/threonine-rich protein P... 42 0.002
ZAN_MOUSE (O88799) Zonadhesin precursor 41 0.004
Y741_CHLMU (Q9PJT6) Hypothetical protein TC0741 precursor 41 0.004
MSB2_YEAST (P32334) MSB2 protein (Multicopy suppressor of bud em... 41 0.004
YB1E_SCHPO (P87179) Serine-rich protein C30B4.01c precursor 40 0.006
FLO9_YEAST (P39712) Flocculation protein FLO9 precursor 40 0.008
TIR1_YEAST (P10863) Cold shock induced protein TIR1 precursor (S... 39 0.010
>FIT1_YEAST (Q04433) Facilitator of iron transport 1 precursor
Length = 528
Score = 50.1 bits (118), Expect = 6e-06
Identities = 48/189 (25%), Positives = 81/189 (42%), Gaps = 13/189 (6%)
Query: 84 WTGYVEK---DTAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIA 140
WTG E+ T ++I ++E S + + +A +S A+ ++A+ A+ + +
Sbjct: 260 WTGEGERAPASTVATSSISAIEIPSTTEASIVEASSAVETSSAAETSSAVETSSAVETSS 319
Query: 141 AASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESD 200
A + ++ VE S E + T A S+ + + +S
Sbjct: 320 AVETSSAAETSSAAETSSAVETSSAV------EISSAVETSAVETSSSSSTIETTSVKSL 373
Query: 201 VPTQTELSSSVQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQT 260
PTQT LSSSVQ+ S E SS+ +T SV S + SS ++ + V S +
Sbjct: 374 SPTQTSLSSSVQASS----PIETSSAAKTSSVVPTFSSTTTENSSNSKSTSAVVASTTTS 429
Query: 261 ETSSVDIVS 269
SS IV+
Sbjct: 430 SESSATIVT 438
>HKR1_YEAST (P41809) Hansenula MRAKII killer toxin-resistant protein
1 precursor
Length = 1802
Score = 45.1 bits (105), Expect = 2e-04
Identities = 47/170 (27%), Positives = 76/170 (44%), Gaps = 20/170 (11%)
Query: 109 ESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLS 168
ES S T+ SS G+ N A+ S + +++ Q G++ + + +G S
Sbjct: 37 ESTPSYSTSAISSTGSSNKEN-----AITSSSETTTMAGQYGESGSTTIMDEQETGTSSQ 91
Query: 169 YYINEFKTQEITQAPSVSTETELSSSVPTESDVPT---------QTELSSSVQSESIAPA 219
Y TQ SV TE+++ P+ S VPT Q + S +S+A +
Sbjct: 92 YISVTTTTQTSDTMSSVKKSTEIAT--PSSSIVPTPLQSYSDESQISQTLSHNPKSVAES 149
Query: 220 KTELSSSIQTESV--ATDASISVPPESSP--AEVSQVQVESQIQTETSSV 265
++ +SS + SV +T S +VP E SP SQ+ E +TSS+
Sbjct: 150 DSDTTSSESSSSVIISTSDSSAVPREISPIITTDSQISKEEGTLAQTSSI 199
Score = 30.8 bits (68), Expect = 3.6
Identities = 27/96 (28%), Positives = 44/96 (45%), Gaps = 10/96 (10%)
Query: 184 SVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTES-------VATDA 236
S ST+T+ S++V E+ V + +S S SES + + T S T + +
Sbjct: 229 SESTQTDFSNTVSFENSVEEEYAMSKSQLSESYSSSSTVYSGGESTADKTSSSPITSFSS 288
Query: 237 SISVPPESSPAEVSQVQV---ESQIQTETSSVDIVS 269
S S + +E S+V V T+T+S+D S
Sbjct: 289 SYSQTTSTETSESSRVAVGVSRPSSITQTTSIDSFS 324
>AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3)
(Glucan 1,4-alpha-glucosidase) (1,4-alpha-D-glucan
glucohydrolase)
Length = 1367
Score = 45.1 bits (105), Expect = 2e-04
Identities = 36/96 (37%), Positives = 46/96 (47%), Gaps = 9/96 (9%)
Query: 183 PSVSTETELSSSVPTESDVPTQTEL------SSSVQSESIAPAKTELSSSIQTESVATDA 236
PS ST S+ VPT S T++ + SSS S AP+ T SSS ++ SV
Sbjct: 782 PSSSTTESSSAPVPTPSSSTTESSVAPVPTPSSSSNITSSAPSSTPFSSSTESSSVPVPT 841
Query: 237 SISVPPESSPAEVSQVQVESQ---IQTETSSVDIVS 269
S ESS A VS ES + T +SS +I S
Sbjct: 842 PSSSTTESSSAPVSSSTTESSVAPVPTPSSSSNITS 877
Score = 44.3 bits (103), Expect = 3e-04
Identities = 40/102 (39%), Positives = 53/102 (51%), Gaps = 9/102 (8%)
Query: 176 TQEITQAPSVSTETELSSS-VPTESDVPTQTE---LSSSVQSESIAP---AKTELSSS-I 227
T E + AP S+ TE SS+ VPT S T++ ++SS S AP + TE SS+ +
Sbjct: 336 TTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPV 395
Query: 228 QTESVATDASISVPPESSPAEVSQVQVESQIQTETSSVDIVS 269
T S +T S S P SS E S V S TE+SS + S
Sbjct: 396 PTPSSSTTESSSAPVTSSTTESSSAPVTSS-TTESSSAPVTS 436
Score = 44.3 bits (103), Expect = 3e-04
Identities = 51/170 (30%), Positives = 70/170 (41%), Gaps = 16/170 (9%)
Query: 113 SSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSYYIN 172
SS TSS ++A + + S A +S ++S V S
Sbjct: 405 SSSAPVTSSTTESSSAPVTSSTTESSSAPVTS---STTESSSAPVTSSTTESSSAPVPTP 461
Query: 173 EFKTQEITQAPSVSTETELSSS-VPTESDVPTQTE---LSSSVQSESIAPAKTELSSSIQ 228
T E + AP S+ TE SS+ VPT S T++ ++SS S AP T SS+ +
Sbjct: 462 SSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTE 521
Query: 229 -------TESVATDASISVPPESSPAEVSQ--VQVESQIQTETSSVDIVS 269
T S +T S S P SS E S V S TE+SS + S
Sbjct: 522 SSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSTPVTS 571
Score = 42.4 bits (98), Expect = 0.001
Identities = 49/152 (32%), Positives = 63/152 (41%), Gaps = 17/152 (11%)
Query: 113 SSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSYYIN 172
SS T TSS ++A + + S +SS + +S + PS S
Sbjct: 564 SSSTPVTSSTTESSSAPVPTPSS--STTESSSAPVPTPSSSTTESSSAPAPTPSSS---- 617
Query: 173 EFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESV 232
T E + AP S+ TE SSS P VPT SSS S AP T SS+ ++ S
Sbjct: 618 ---TTESSSAPVTSSTTE-SSSAP----VPTP---SSSTTESSSAPVPTPSSSTTESSSA 666
Query: 233 ATDASISVPPESSPAEVSQVQVESQIQTETSS 264
S ESS A V+ ES TSS
Sbjct: 667 PVPTPSSSTTESSSAPVTSSTTESSSAPVTSS 698
Score = 42.4 bits (98), Expect = 0.001
Identities = 36/104 (34%), Positives = 47/104 (44%), Gaps = 9/104 (8%)
Query: 175 KTQEITQAPSVSTETELSSSVPTESDVPTQTE---LSSSVQSESIAPAKTELSSSIQTES 231
KT PS ST S+ VPT S T++ ++SS S AP T SS+ ++ S
Sbjct: 309 KTTTPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSS 368
Query: 232 V----ATDASISVPPESSPAEVSQ--VQVESQIQTETSSVDIVS 269
+T S S P SS E S V S TE+SS + S
Sbjct: 369 APVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTS 412
Score = 42.4 bits (98), Expect = 0.001
Identities = 56/187 (29%), Positives = 76/187 (39%), Gaps = 17/187 (9%)
Query: 89 EKDTAGQTNIYSVEPAVYVAESAISSGTAG-TSSDGAQNTAAIAAGLALISIAAASSILL 147
E +A T+ + + V S S +A TSS ++A + + S A +
Sbjct: 404 ESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSS 463
Query: 148 QVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTEL 207
++S V S T E + AP S+ TE SSS P VPT
Sbjct: 464 STTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTE-SSSAP----VPTP--- 515
Query: 208 SSSVQSESIAPAKTELSSSIQTESV----ATDASISVP---PESSPAEVSQVQVESQIQT 260
SSS S APA T SS+ ++ S +T S S P P SS E S V S T
Sbjct: 516 SSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSTPVTSS-TT 574
Query: 261 ETSSVDI 267
E+SS +
Sbjct: 575 ESSSAPV 581
Score = 42.0 bits (97), Expect = 0.002
Identities = 49/187 (26%), Positives = 71/187 (37%), Gaps = 13/187 (6%)
Query: 89 EKDTAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQ 148
E +A T+ + + V S S +A + + T + +A + S + S
Sbjct: 677 ESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAP 736
Query: 149 VGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTEL- 207
V P E S ++ E + + PS ST S+ VPT S T++
Sbjct: 737 V---PTPSSSTTESSSAPVTSSTTESSSAPVP-TPSSSTTESSSAPVPTPSSSTTESSSA 792
Query: 208 -----SSSVQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTET 262
SSS S+AP T SSS T S + S ESS V S TE+
Sbjct: 793 PVPTPSSSTTESSVAPVPTPSSSSNITSSAPSSTPFSSSTESSSV---PVPTPSSSTTES 849
Query: 263 SSVDIVS 269
SS + S
Sbjct: 850 SSAPVSS 856
Score = 40.4 bits (93), Expect = 0.005
Identities = 44/170 (25%), Positives = 69/170 (39%), Gaps = 10/170 (5%)
Query: 100 SVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQK 159
S P + S S +A + + T + +A + S + S + V +P
Sbjct: 760 SSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSVAPVP--TPSSSSN 817
Query: 160 VEYSGPSLSYYINEFKTQEITQAPSVSTETELS----SSVPTESDVPTQTELSSSVQSES 215
+ S PS + + + ++ + S+ TE S SS TES V SSS S
Sbjct: 818 ITSSAPSSTPFSSSTESSSVPVPTPSSSTTESSSAPVSSSTTESSVAPVPTPSSSSNITS 877
Query: 216 IAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSV 265
AP+ SS+ ++ S T +V P SS SQ + TET+ V
Sbjct: 878 SAPSSIPFSSTTESFSTGT----TVTPSSSKYPGSQTETSVSSTTETTIV 923
Score = 39.3 bits (90), Expect = 0.010
Identities = 34/98 (34%), Positives = 43/98 (43%), Gaps = 8/98 (8%)
Query: 176 TQEITQAPSVSTETELSSSVPT------ESDVPTQTELSSSVQSESIAPAKTELSSSIQT 229
T E + AP S+ TE SSS P S P T SS+ +S S + SS
Sbjct: 363 TTESSSAPVTSSTTE-SSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAP 421
Query: 230 ESVATDASISVPPESSPAEVSQVQVESQIQTETSSVDI 267
+ +T S S P SS E S V S TE+SS +
Sbjct: 422 VTSSTTESSSAPVTSSTTESSSAPVTSS-TTESSSAPV 458
Score = 38.9 bits (89), Expect = 0.013
Identities = 32/93 (34%), Positives = 43/93 (45%), Gaps = 7/93 (7%)
Query: 183 PSVSTETELSSSVPTESDV--PTQTELSSSVQSESIAPAKTELSSSIQTESV----ATDA 236
P+ S+ T SSS P S + ++SS S AP T SS+ ++ S +T
Sbjct: 357 PTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTE 416
Query: 237 SISVPPESSPAEVSQVQVESQIQTETSSVDIVS 269
S S P SS E S V S TE+SS + S
Sbjct: 417 SSSAPVTSSTTESSSAPVTSS-TTESSSAPVTS 448
Score = 37.7 bits (86), Expect = 0.030
Identities = 45/171 (26%), Positives = 68/171 (39%), Gaps = 19/171 (11%)
Query: 103 PAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEY 162
P+ ES+ + +SS ++A + + S +SS + +S +
Sbjct: 599 PSSSTTESSSAPAPTPSSSTTESSSAPVTS-----STTESSSAPVPTPSSSTTESSSAPV 653
Query: 163 SGPSLSYYINEFKTQEITQAP--SVSTETELSSSVPTESDVPTQTE--LSSSVQSESIAP 218
PS S T E + AP + S+ T SSS P S + ++SS S AP
Sbjct: 654 PTPSSS-------TTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAP 706
Query: 219 AKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSVDIVS 269
T SS+ ++ S S ESS A V S TE+SS + S
Sbjct: 707 VPTPSSSTTESSSAPVPTPSSSTTESSSA---PVPTPSSSTTESSSAPVTS 754
Score = 31.2 bits (69), Expect = 2.8
Identities = 23/99 (23%), Positives = 39/99 (39%), Gaps = 1/99 (1%)
Query: 172 NEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTES 231
+E T T AP+ T T + PT + T+ + P + +++ +T +
Sbjct: 248 SESSTSSSTTAPATPTTTSCTKEKPTPPTTTSCTKEKPTPPHHDTTPCTKKKTTTSKTCT 307
Query: 232 VATDASISVPPESSPAEVS-QVQVESQIQTETSSVDIVS 269
T + P S+ S V S TE+SS + S
Sbjct: 308 KKTTTPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTS 346
>YG46_YEAST (P53301) Hypothetical 52.8 kDa protein in BUB1-HIP1
intergenic region
Length = 507
Score = 44.7 bits (104), Expect = 2e-04
Identities = 40/164 (24%), Positives = 79/164 (47%), Gaps = 9/164 (5%)
Query: 110 SAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSY 169
+ +++G + +SS + +T + +A S +SS+ V ++ V S S S
Sbjct: 294 AVLANGGSISSSSTSSSTVSSSA-----SSTVSSSVSSTVSSSASSTVSSSVSSTVSSSS 348
Query: 170 YINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQT 229
++ + + + + S++T SSSV T S + + + SSS +++A + T S T
Sbjct: 349 SVSSSSSTSPSSSTATSSKTLASSSVTTSSSISSFEKQSSSSSKKTVASSSTSESIISST 408
Query: 230 ESVATDASISVPPESSPAEVSQVQVESQIQTE----TSSVDIVS 269
++ AT +S + + + S V +S +Q + TSS D+ S
Sbjct: 409 KTPATVSSTTRSTVAPTTQQSSVSSDSPVQDKGGVATSSNDVTS 452
Score = 34.7 bits (78), Expect = 0.25
Identities = 46/171 (26%), Positives = 74/171 (42%), Gaps = 17/171 (9%)
Query: 108 AESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQK-VEYSGPS 166
A S +SS + T S A +T + + + S ++ SS +S K + S +
Sbjct: 316 ASSTVSSSVSSTVSSSASSTVSSSVSSTVSSSSSVSSSSSTSPSSSTATSSKTLASSSVT 375
Query: 167 LSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELS-SSVQSESIAPAKTELSS 225
S I+ F+ Q + S S +T SSS T+T + SS ++AP T S
Sbjct: 376 TSSSISSFEKQ----SSSSSKKTVASSSTSESIISSTKTPATVSSTTRSTVAP--TTQQS 429
Query: 226 SIQTESVATD----ASISVPPESSPAEVSQ-----VQVESQIQTETSSVDI 267
S+ ++S D A+ S SS ++S +Q S + T+SV I
Sbjct: 430 SVSSDSPVQDKGGVATSSNDVTSSTTQISSKYTSTIQSSSSEASSTNSVQI 480
>YG1F_YEAST (P53214) Hypothetical 57.5 kDa protein in VMA7-RPS25A
intergenic region
Length = 551
Score = 44.7 bits (104), Expect = 2e-04
Identities = 41/178 (23%), Positives = 84/178 (47%), Gaps = 8/178 (4%)
Query: 92 TAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGK 151
T T+ + + S + S ++ T + +++++++ + S++ +S +
Sbjct: 151 TVASTSSTVASSTLSTSSSLVISTSSSTFTFSSESSSSLISSSISTSVSTSSVYVPSSST 210
Query: 152 NSPP----QVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESD--VPTQT 205
+SPP ++ YS S S + + + + + S S+ + SSS + S T
Sbjct: 211 SSPPSSSSELTSSSYSSSSSSSTLFSYSSSFSSSSSSSSSSSSSSSSSSSSSSSYFTLST 270
Query: 206 ELSSSVQSESIAPAKTELSSSIQTESV-ATDASISVPPESSPAEVSQVQVESQIQTET 262
SSS+ S S P+ + SSS T S+ +T AS S+ P S + +++ + S I+ +T
Sbjct: 271 SSSSSIYSSSSYPSFSSSSSSNPTSSITSTSASSSITPASEYSNLAKT-ITSIIEGQT 327
Score = 37.7 bits (86), Expect = 0.030
Identities = 51/182 (28%), Positives = 82/182 (45%), Gaps = 23/182 (12%)
Query: 110 SAISSGTAGT--SSDGAQNTAAIAAGLALISIAAASSIL-LQVGKNSPPQVQKVEYSGPS 166
SA+ S T+ T +S + +T +IA+ ++ S +SS+L L +S ++ S S
Sbjct: 56 SAVESFTSSTDATSSASLSTPSIAS-VSFTSFPQSSSLLTLSSTLSSELSSSSMQVSSSS 114
Query: 167 LSYYINEF----KTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSV----------Q 212
S +E + I+ + S ST SSS+PT + T + ++SS
Sbjct: 115 TSSSSSEVTSSSSSSSISPSSSSSTIISSSSSLPTFTVASTSSTVASSTLSTSSSLVIST 174
Query: 213 SESIAPAKTELSSSIQTESVATDASIS---VPPE--SSPAEVSQVQVESQIQTETSSVDI 267
S S +E SSS+ + S++T S S VP SSP S S + +SS +
Sbjct: 175 SSSTFTFSSESSSSLISSSISTSVSTSSVYVPSSSTSSPPSSSSELTSSSYSSSSSSSTL 234
Query: 268 VS 269
S
Sbjct: 235 FS 236
Score = 33.9 bits (76), Expect = 0.43
Identities = 40/172 (23%), Positives = 68/172 (39%), Gaps = 6/172 (3%)
Query: 92 TAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGK 151
++ + + S + ++ S+ SS +SS T A + S + SS L+ +
Sbjct: 116 SSSSSEVTSSSSSSSISPSSSSSTIISSSSSLPTFTVASTSSTVASSTLSTSSSLV-IST 174
Query: 152 NSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSV 211
+S E S +S I+ + PS ST S P+ S T + SSS
Sbjct: 175 SSSTFTFSSESSSSLISSSISTSVSTSSVYVPSSSTS-----SPPSSSSELTSSSYSSSS 229
Query: 212 QSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETS 263
S ++ + SSS + S ++ +S S SS S I + +S
Sbjct: 230 SSSTLFSYSSSFSSSSSSSSSSSSSSSSSSSSSSSYFTLSTSSSSSIYSSSS 281
Score = 30.4 bits (67), Expect = 4.7
Identities = 34/150 (22%), Positives = 63/150 (41%), Gaps = 4/150 (2%)
Query: 117 AGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFKT 176
A TSS A +T + ++ L + + ++S+ +S + S + S Y+ T
Sbjct: 153 ASTSSTVASSTLSTSSSLVIST--SSSTFTFSSESSSSLISSSISTSVSTSSVYVPSSST 210
Query: 177 QEITQAPSVSTETELSSSVPTES--DVPTQTELSSSVQSESIAPAKTELSSSIQTESVAT 234
+ S T + SSS + + + SSS S S + + + SSS +++T
Sbjct: 211 SSPPSSSSELTSSSYSSSSSSSTLFSYSSSFSSSSSSSSSSSSSSSSSSSSSSSYFTLST 270
Query: 235 DASISVPPESSPAEVSQVQVESQIQTETSS 264
+S S+ SS S + + TS+
Sbjct: 271 SSSSSIYSSSSYPSFSSSSSSNPTSSITST 300
>YIQ9_YEAST (P40442) Hypothetical 99.7 kDa protein in SDL1 5'region
precursor
Length = 995
Score = 43.9 bits (102), Expect = 4e-04
Identities = 46/166 (27%), Positives = 80/166 (47%), Gaps = 17/166 (10%)
Query: 106 YVAESAISSGTAGTS-SDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSG 164
Y ++ISS ++ TS + + +I++ +A S ++A+ IL + +++ G
Sbjct: 26 YSNSTSISSNSSSTSVVSSSSGSVSISSSIAETS-SSATDILSSITQSASSTSGVSSSVG 84
Query: 165 PSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELS 224
PS S ++ +Q + VS+ SSS + +++SSSV A + +++S
Sbjct: 85 PSSSSVVSSSVSQSSSSVSDVSSSVSQSSS--------SASDVSSSVSQS--ASSTSDVS 134
Query: 225 SSI-QTESVATDASISVPPESSPAEVSQVQVESQIQTETSSVDIVS 269
SS+ Q+ S A+D S SV SS A V S + SS VS
Sbjct: 135 SSVSQSSSSASDVSSSVSQSSSSAS----DVSSSVSQSASSASDVS 176
Score = 38.9 bits (89), Expect = 0.013
Identities = 46/160 (28%), Positives = 71/160 (43%), Gaps = 9/160 (5%)
Query: 110 SAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSY 169
S +SS + S + + +A L SI ++S V + P V S S S
Sbjct: 40 SVVSSSSGSVSISSSIAETSSSATDILSSITQSASSTSGVSSSVGPSSSSVVSSSVSQSS 99
Query: 170 YINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQT 229
+ ++Q S S+ +++SSSV + +++SSSV S+S + A SS Q+
Sbjct: 100 SSVSDVSSSVSQ--SSSSASDVSSSV--SQSASSTSDVSSSV-SQSSSSASDVSSSVSQS 154
Query: 230 ESVATDASISVPPESSPAEVSQVQVESQIQTETSSVDIVS 269
S A+D S SV +S A V S + SS VS
Sbjct: 155 SSSASDVSSSVSQSASSAS----DVSSSVSQSASSTSDVS 190
Score = 35.8 bits (81), Expect = 0.11
Identities = 46/160 (28%), Positives = 73/160 (44%), Gaps = 16/160 (10%)
Query: 110 SAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSY 169
S +SS + +SS + +++++ + S + SS + Q ++ V S S S
Sbjct: 103 SDVSSSVSQSSSSASDVSSSVSQSAS--STSDVSSSVSQSSSSASDVSSSVSQSSSSASD 160
Query: 170 YINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQT 229
++ +Q + A VS+ +S S + SDV SSSV S+S + A SS Q+
Sbjct: 161 -VSSSVSQSASSASDVSSS--VSQSASSTSDV------SSSV-SQSSSSASDVSSSVSQS 210
Query: 230 ESVATDASISVPPESSPAEVSQVQVESQIQTETSSVDIVS 269
S A+D S SV +S S V S + SS VS
Sbjct: 211 SSSASDVSSSVSQSAS----STSDVSSSVSQSASSTSGVS 246
Score = 33.1 bits (74), Expect = 0.73
Identities = 29/99 (29%), Positives = 47/99 (47%), Gaps = 4/99 (4%)
Query: 166 SLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSS 225
+L+ Y Q + + S+S+ + +S V + S +SSS+ +E+ + A LSS
Sbjct: 13 ALALYSQSALGQYYSNSTSISSNSSSTSVVSSSSG---SVSISSSI-AETSSSATDILSS 68
Query: 226 SIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSS 264
Q+ S + S SV P SS S V S ++ SS
Sbjct: 69 ITQSASSTSGVSSSVGPSSSSVVSSSVSQSSSSVSDVSS 107
Score = 32.0 bits (71), Expect = 1.6
Identities = 42/165 (25%), Positives = 75/165 (45%), Gaps = 8/165 (4%)
Query: 110 SAISSGTAGTSSDGAQNTAAIAAGLALISIAAA--SSILLQVGKNSPPQVQKVEYSGPSL 167
S+ +S + + S A +T+ +++ ++ S +A+ SS + Q ++ V S S
Sbjct: 113 SSSASDVSSSVSQSASSTSDVSSSVSQSSSSASDVSSSVSQSSSSASDVSSSVSQSASSA 172
Query: 168 SYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSE-SIAPAKTELSSS 226
S ++ +Q + VS+ SSS + SDV + SSS S+ S + +++ S+S
Sbjct: 173 SD-VSSSVSQSASSTSDVSSSVSQSSS--SASDVSSSVSQSSSSASDVSSSVSQSASSTS 229
Query: 227 IQTESVATDASIS--VPPESSPAEVSQVQVESQIQTETSSVDIVS 269
+ SV+ AS + V S + S S TSS S
Sbjct: 230 DVSSSVSQSASSTSGVSSSGSQSVSSASGSSSSFPQSTSSASTAS 274
>SIM1_YEAST (P40472) SIM1 protein precursor
Length = 475
Score = 43.9 bits (102), Expect = 4e-04
Identities = 48/178 (26%), Positives = 75/178 (41%), Gaps = 27/178 (15%)
Query: 87 YVEKDTAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSIL 146
YV + ++ ++V PA +A +S TS T++ AAG+A SIA +S+ L
Sbjct: 41 YVYETVVVDSDGHTVTPAASEVATAATSAIITTSV--LAPTSSAAAGIAA-SIAVSSAAL 97
Query: 147 LQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTE 206
+ NE K + + + ST SSS + S T
Sbjct: 98 AK-----------------------NE-KISDAAASATASTSQGASSSSSSSSATSTLES 133
Query: 207 LSSSVQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSS 264
S S SE AP T +S+S T+S A+ A+ S +SP+ + S + +SS
Sbjct: 134 SSVSSSSEEAAPTSTVVSTSSATQSSASSATKSSTSSTSPSTSTSTSTSSTSSSSSSS 191
>PRY3_YEAST (P47033) PRY3 protein (Pathogen related in Sc 3)
Length = 881
Score = 43.5 bits (101), Expect = 5e-04
Identities = 47/174 (27%), Positives = 75/174 (43%), Gaps = 10/174 (5%)
Query: 101 VEPAVY-VAESAISSGTAGTSSDGAQNTA-----AIAAGLALISIAAASSILLQVGKNSP 154
VEP + V+ S+ SS + T+SD + A+A G +AASS L+ +P
Sbjct: 158 VEPLISTVSSSSSSSSSTSTTSDTVSTISSSIMPAVAQGYTTTVSSAASSSSLKSTTINP 217
Query: 155 PQVQKVEYSGPSLSYYINEF---KTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSV 211
+ + S ++ E T + SV+T SSS SD + T +SSV
Sbjct: 218 AKTATLTASSSTVITSSTESVGSSTVSSASSSSVTTSYATSSSTVVSSDATSSTTTTSSV 277
Query: 212 QSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSV 265
+ S + ++ +SS S + AS S SS + + S I + +S V
Sbjct: 278 ATSS-STTSSDPTSSTAAASSSDPASSSAAASSSASTENAASSSSAISSSSSMV 330
Score = 40.0 bits (92), Expect = 0.006
Identities = 51/204 (25%), Positives = 82/204 (40%), Gaps = 19/204 (9%)
Query: 66 AEEKTKVVGTYPPRRKQGWTGYVEKDTAGQTNIYSVEPAVY-----VAESAISSGTAGTS 120
AEE ++ T + DT T S+ PAV SA SS + ++
Sbjct: 155 AEEVEPLISTVSSSSSSSSSTSTTSDTVS-TISSSIMPAVAQGYTTTVSSAASSSSLKST 213
Query: 121 SDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEIT 180
+ TA + A + + ++ S+ ++ Y+ S S ++ T T
Sbjct: 214 TINPAKTATLTASSSTVITSSTESVGSSTVSSASSSSVTTSYATSS-STVVSSDATSSTT 272
Query: 181 QAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDASISV 240
SV+T + +SS PT S SSS + S A A SSS TE+ A+ +S
Sbjct: 273 TTSSVATSSSTTSSDPTSSTAAA----SSSDPASSSAAA----SSSASTENAASSSSAI- 323
Query: 241 PPESSPAEVSQVQVESQIQTETSS 264
SS + + + S + T T+S
Sbjct: 324 ---SSSSSMVSAPLSSTLTTSTAS 344
Score = 38.5 bits (88), Expect = 0.017
Identities = 43/177 (24%), Positives = 68/177 (38%), Gaps = 32/177 (18%)
Query: 92 TAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGK 151
T+ T + S + S++++ ++ TSSD +TAA ++ S AAASS
Sbjct: 257 TSSSTVVSSDATSSTTTTSSVATSSSTTSSDPTSSTAAASSSDPASSSAAASS------- 309
Query: 152 NSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSV 211
T+ + S + + S P S + T T S SV
Sbjct: 310 ---------------------SASTENAASSSSAISSSSSMVSAPLSSTLTTSTASSRSV 348
Query: 212 QSESIAPAK---TELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSV 265
S S+ K T + S+ T SV+ S SV + S+ + S + TS V
Sbjct: 349 TSNSVNSVKFANTTVFSAQTTSSVSASLSSSVAADDIQGSTSK-EATSSVSEHTSIV 404
Score = 37.0 bits (84), Expect = 0.051
Identities = 42/190 (22%), Positives = 76/190 (39%), Gaps = 5/190 (2%)
Query: 78 PRRKQGWTGYVEKDTAGQT-NIYSVEPAVYVAESAISSG--TAGTSSDGAQNTAAIAAGL 134
P QG+T V + + ++ PA +A SS T+ T S G+ ++ ++
Sbjct: 191 PAVAQGYTTTVSSAASSSSLKSTTINPAKTATLTASSSTVITSSTESVGSSTVSSASSSS 250
Query: 135 ALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSS 194
S A +SS ++ S + S + T + + S+ SSS
Sbjct: 251 VTTSYATSSSTVVSSDATSSTTTTSSVATSSSTTSSDPTSSTAAASSSDPASSSAAASSS 310
Query: 195 VPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQV 254
TE+ + + +SSS S + T +S+ + SV +++ SV + + V Q
Sbjct: 311 ASTENAASSSSAISSSSSMVSAPLSSTLTTSTASSRSVTSNSVNSV--KFANTTVFSAQT 368
Query: 255 ESQIQTETSS 264
S + SS
Sbjct: 369 TSSVSASLSS 378
>FLO5_YEAST (P38894) Flocculation protein FLO5 precursor (Flocculin
5)
Length = 1075
Score = 43.5 bits (101), Expect = 5e-04
Identities = 48/204 (23%), Positives = 88/204 (42%), Gaps = 26/204 (12%)
Query: 79 RRKQGWTGY-----VEKDTAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAG 133
R + WTG E T TN + V V + ++ ++ SS Q T++I +
Sbjct: 593 RTTEPWTGTFTSTSTEVTTITGTNGQPTDETVIVIRTPTTAISSSLSSSSGQITSSITSS 652
Query: 134 LALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSS 193
+I+ S+ + + + S S + F + + + S +T T + S
Sbjct: 653 RPIITPFYPSNGTSVISSSV------ISSSVTSSLVTSSSFISSSVISS-STTTSTSIFS 705
Query: 194 SVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTES--VATDASISVPPESS------ 245
T S +PT + S S +S++ + + + SSSI +ES T++S S+PP +S
Sbjct: 706 ESSTSSVIPTSSSTSGSSESKTSSASSSSSSSSISSESPKSPTNSSSSLPPVTSATTGQE 765
Query: 246 ------PAEVSQVQVESQIQTETS 263
PA ++ ++ + T TS
Sbjct: 766 TASSLPPATTTKTSEQTTLVTVTS 789
Score = 30.8 bits (68), Expect = 3.6
Identities = 25/105 (23%), Positives = 53/105 (49%), Gaps = 13/105 (12%)
Query: 166 SLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSV---QSESIAPAKTE 222
S+S +N ++ T + T+T ++SS+ + TQT ++ V S ++ ++T
Sbjct: 953 SVSSKMNSATSETTTNTGAAETKTAVTSSLSRFNHAETQTASATDVIGHSSSVVSVSETG 1012
Query: 223 LSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSVDI 267
+ S+ + ++T +S P S+PA S + + T+S++I
Sbjct: 1013 NTMSLTSSGLST---MSQQPRSTPA-------SSMVGSSTASLEI 1047
>YA9A_SCHPO (Q09788) Hypothetical serine-rich protein C13G6.10c in
chromosome I precursor
Length = 530
Score = 43.1 bits (100), Expect = 7e-04
Identities = 52/185 (28%), Positives = 76/185 (40%), Gaps = 33/185 (17%)
Query: 109 ESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLS 168
ES + + +A +S A + A L ++ ++ V N+ V E S S S
Sbjct: 88 ESVVVTTSAPAASSSATSYPATFVSTPLYTM---DNVTAPVWSNTSVPVSTPETSATSSS 144
Query: 169 YYINEF---KTQEITQAPSVSTE---------TELSSSVPTESDVPTQTE-----LSSSV 211
+ + ++ + P+ STE TE++SS P S+V T T +SSSV
Sbjct: 145 EFFTSYPATSSESSSSYPASSTEVASSYSASSTEVTSSYPASSEVATSTSSYVAPVSSSV 204
Query: 212 QSESIAPAKTEL------SSSIQTESVATDAS-------ISVPPESSPAEVSQVQVESQI 258
S S A + SSSI SV AS +S P SS A S V S +
Sbjct: 205 ASSSEISAGSATSYVPTSSSSIALSSVVASASVSAANKGVSTPAVSSAAASSSAVVSSVV 264
Query: 259 QTETS 263
+ TS
Sbjct: 265 SSATS 269
>WSC4_YEAST (P38739) Cell wall integrity and stress response
component 4 precursor
Length = 605
Score = 43.1 bits (100), Expect = 7e-04
Identities = 56/232 (24%), Positives = 92/232 (39%), Gaps = 54/232 (23%)
Query: 46 CNDEECAPDKEVGKVSMEWLAEEKTKVVGTYPPRRKQGWTGYVEKDT-----AGQTNIYS 100
C+D E + VG S GT P + G +KD GQT + S
Sbjct: 71 CSDSEPSTQTSVGDCS------------GTCPGYGYED-CGNADKDLFGYIYLGQTPLSS 117
Query: 101 V-------EPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNS 153
V E +VYV+ S+I+S +SS +T I+ L + +++ L ++
Sbjct: 118 VQSVETSTESSVYVSSSSITS----SSSTSIVDTTTISPTLT----STSTTPLTTASTST 169
Query: 154 PPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELS-SSVQ 212
P +IT A +T T+LS+S+PT + T T S SS
Sbjct: 170 TPST--------------------DITSALPTTTSTKLSTSIPTSTTSSTSTTTSTSSST 209
Query: 213 SESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSS 264
S +++ + +++ T S S S S+P S + + + TS+
Sbjct: 210 STTVSVTSSTSTTTSTTSSTLISTSTSSSSSSTPTTTSSAPISTSTTSSTST 261
Score = 42.7 bits (99), Expect = 0.001
Identities = 41/170 (24%), Positives = 70/170 (41%), Gaps = 7/170 (4%)
Query: 100 SVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQK 159
S + + + S SS + TS+ + +T + + SS L+ +S
Sbjct: 184 STKLSTSIPTSTTSSTSTTTSTSSSTSTTVSVTSSTSTTTSTTSSTLISTSTSSSSSSTP 243
Query: 160 VEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPA 219
S +S + + + S ST + SSS PT S T T + + S S AP+
Sbjct: 244 TTTSSAPIS------TSTTSSTSTSTSTTSPTSSSAPTSSSNTTPTSTTFTTTSPSTAPS 297
Query: 220 KTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSVDIVS 269
T ++ + T S T SV ++S + S + V S +T+ +I S
Sbjct: 298 STTVTYTSTTASPITSTITSVNLQTS-LKYSVITVTSVHTMDTNISEITS 346
>SP96_DICDI (P14328) Spore coat protein SP96
Length = 600
Score = 43.1 bits (100), Expect = 7e-04
Identities = 40/173 (23%), Positives = 74/173 (42%), Gaps = 19/173 (10%)
Query: 92 TAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGK 151
T+ + + S + SA+SS +G+S+ A ++ + +A + S +AASS
Sbjct: 426 TSDSSALGSTSESSASGSSAVSSSASGSSA--ASSSPSSSAASSSPSSSAASSSPSSSAA 483
Query: 152 NSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSV 211
+S P S PS S ++ + SS + S P+ + SSS
Sbjct: 484 SSSPSSSASSSSSPS-----------------SSASSSSAPSSSASSSSAPSSSASSSSA 526
Query: 212 QSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSS 264
S S + A T +++I T + T A+ + ++ A + + I T T++
Sbjct: 527 SSSSASSAATTAATTIATTAATTTATTTATTATTTATTTATTTAATIATTTAA 579
Score = 29.6 bits (65), Expect = 8.1
Identities = 33/167 (19%), Positives = 66/167 (38%), Gaps = 7/167 (4%)
Query: 110 SAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSY 169
++I+S A T S GA + ++A A +++ + + + + + S +
Sbjct: 377 ASIASTIASTGSTGATSPCSVAQCPTGYVCVAQNNVAVSLPRPTTTTGSTSDSSALGSTS 436
Query: 170 YINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSE-------SIAPAKTE 222
+ + ++ + S S+ S S S P+ + SSS S S A + +
Sbjct: 437 ESSASGSSAVSSSASGSSAASSSPSSSAASSSPSSSAASSSPSSSAASSSPSSSASSSSS 496
Query: 223 LSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSVDIVS 269
SSS + S + ++ S SS A S S T++ ++
Sbjct: 497 PSSSASSSSAPSSSASSSSAPSSSASSSSASSSSASSAATTAATTIA 543
>YM96_YEAST (Q04893) Hypothetical 113.1 kDa protein in PRE5-FET4
intergenic region
Length = 1140
Score = 42.4 bits (98), Expect = 0.001
Identities = 48/175 (27%), Positives = 79/175 (44%), Gaps = 18/175 (10%)
Query: 103 PAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAA------SSILLQVGKNSPPQ 156
P A SA SS + +S ++ + A LA S+ ++ SS++ +S
Sbjct: 259 PVSSEAPSATSSSVSSEASSSTSSSVSSEAPLATSSVVSSEAPSSTSSVVSSEAPSSTSS 318
Query: 157 VQKVEYSGPSLSYYINE--FKTQEI--TQAPSVSTETELSSSVP--TESDVPTQTELSSS 210
E S + S +E T + ++APS ST + +SS + T S V ++ L++S
Sbjct: 319 SVSSEISSTTSSSVSSEAPLATSSVVSSEAPS-STSSSVSSEISSTTSSSVSSEAPLATS 377
Query: 211 VQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSV 265
S AP+ T S S + S + + S P S+ + VS S+I + SSV
Sbjct: 378 SVVSSEAPSSTSSSVSSEAPSSTSSSVSSEAPSSTSSSVS-----SEISSTKSSV 427
Score = 38.1 bits (87), Expect = 0.023
Identities = 43/168 (25%), Positives = 71/168 (41%), Gaps = 21/168 (12%)
Query: 110 SAISSGTAGTSSDGAQNTAAIAAGLALISI--AAASSILLQVGKNSPPQVQKVEYSGPSL 167
S+ISS A +S+ N+ +A L + + SSI+ + + + S SL
Sbjct: 580 SSISSNLASSSAPSDNNSTIASASLIVTKTKNSVVSSIVSSITSSETTNESNLATSSTSL 639
Query: 168 SYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSI 227
A S+ST S+ + S+VPT T S S + I P + S+S+
Sbjct: 640 --------LSNKATARSLST-----SNATSASNVPTGTFSSMSSHTSVITPGFSTSSASL 686
Query: 228 QTESVATDASIS----VPPESSPAEVSQVQVE--SQIQTETSSVDIVS 269
S +S++ PESSP + V E S + + T+S ++
Sbjct: 687 AINSTVVSSSLAGYSFSTPESSPTTSTLVTSEAPSTVSSMTTSAPFIN 734
Score = 38.1 bits (87), Expect = 0.023
Identities = 43/177 (24%), Positives = 75/177 (42%), Gaps = 14/177 (7%)
Query: 88 VEKDTAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILL 147
VE T+G +N YS A+SS A SS + + ++ + S +SS
Sbjct: 108 VESSTSGPSNSYS----------ALSSTNAQLSSSTTETDSISSSAIQTSSPQTSSSN-- 155
Query: 148 QVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTEL 207
G +S P + + S + T VS+ ++SSS + V T ++
Sbjct: 156 GGGSSSEPLGKSSVLETTASSSDTTAVTSSTFTTLTDVSSSPKISSSGSAVTSVGTTSDA 215
Query: 208 SSSVQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSS 264
S V S S + + LSS+ + ++ S ++P S+ ++ V S+ + TSS
Sbjct: 216 SKEVFSSSTSDVSSLLSST--SSPASSTISETLPFSSTILSITSSPVSSEAPSATSS 270
Score = 33.9 bits (76), Expect = 0.43
Identities = 44/183 (24%), Positives = 66/183 (36%), Gaps = 24/183 (13%)
Query: 99 YSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILL----------- 147
Y++ V V+ + GT + S G + + I A++
Sbjct: 868 YNITKTVIVSRETTAIGTVTSCSGGCTKNRKSTTLITITDIDASTVTTCPEKEVTSTTSG 927
Query: 148 -QVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTE 206
+ + ++ E S S S+ + KT + T+ +ET SSS E PT
Sbjct: 928 DEAEHTTSTKISNFETSTFSESF--KDMKTSQETKKAKPGSETVRSSSSFVEKTSPTTKA 985
Query: 207 LSSSVQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSVD 266
S+ SES A T SVAT+AS S P S VE T + +
Sbjct: 986 SPSTSPSESKAAGNT---------SVATNASPSTSPSESQG-TGSTSVEGAKSKSTKNSE 1035
Query: 267 IVS 269
VS
Sbjct: 1036 GVS 1038
Score = 32.0 bits (71), Expect = 1.6
Identities = 39/149 (26%), Positives = 63/149 (42%), Gaps = 23/149 (15%)
Query: 110 SAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSY 169
S I+ G + +S+ A N+ +++ LA S + S SP V PS
Sbjct: 673 SVITPGFSTSSASLAINSTVVSSSLAGYSFSTPES--------SPTTSTLVTSEAPSTV- 723
Query: 170 YINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQT 229
+ T AP ++ T S+ P+ S TE +SS+ S +A +++SS+
Sbjct: 724 ------SSMTTSAPFINNST---SARPSPSTASFITESTSSISSVPLASG--DVTSSLAA 772
Query: 230 ESVATDASISVPPESSPAEVSQVQVESQI 258
++ T S P SS VS+ S I
Sbjct: 773 HNLTT---FSAPSTSSAQLVSKSTTSSSI 798
>YL61_SCHPO (Q8TFG9) Hypothetical serine/threonine-rich protein
PB15E9.01c in chromosome I precursor
Length = 943
Score = 42.0 bits (97), Expect = 0.002
Identities = 48/197 (24%), Positives = 86/197 (43%), Gaps = 12/197 (6%)
Query: 73 VGTYPPRRKQGWTGYVEKDTAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAA 132
+ + P K+ T Y ++ ++ + A A S+++S ++ SS T+A
Sbjct: 49 LSSVPTLLKRATTSYNYNTSSASSSSLTSSSA---ASSSLTSSSSLASSSTNSTTSASPT 105
Query: 133 GLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELS 192
+L S +A SS L +S S S S + + + + + S+ S
Sbjct: 106 SSSLTSSSATSSSL---ASSSTTSSSLASSSITSSSLASSSITSSSLASSSTTSSSLASS 162
Query: 193 SSVPTESDVPTQTELSSSVQSESIAPAKTE---LSSSIQ--TESVATDASISVPPESSPA 247
S+ T S PT + SSS+ S + + + T SSS+ T + AT +S+S S+ A
Sbjct: 163 STNSTTSATPTSSATSSSLSSTAASNSATSSSLASSSLNSTTSATATSSSLSSTAASNSA 222
Query: 248 EVSQVQVESQIQTETSS 264
S + S + + TS+
Sbjct: 223 TSSSL-ASSSLNSTTSA 238
Score = 37.0 bits (84), Expect = 0.051
Identities = 39/161 (24%), Positives = 68/161 (42%), Gaps = 11/161 (6%)
Query: 113 SSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSYYIN 172
+S T TS + T+A + L ++ +A+S +S P + S+S
Sbjct: 354 ASSTPLTSVNSTTATSASSTPLTSVNSTSATSA------SSTPLTSANSTTSTSVSSTAP 407
Query: 173 EFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESV 232
+ T + SVS+ T LSS+ T + + T LSS + + + + T LSS T +
Sbjct: 408 SYNTSSVLPTSSVSS-TPLSSANSTTATSASSTPLSSVNSTTATSASSTPLSSVNSTTAT 466
Query: 233 ATDA----SISVPPESSPAEVSQVQVESQIQTETSSVDIVS 269
+ + S++ +S + V S T SS + S
Sbjct: 467 SASSTPLTSVNSTTATSASSTPLTSVNSTSATSASSTPLTS 507
Score = 36.2 bits (82), Expect = 0.086
Identities = 40/170 (23%), Positives = 69/170 (40%), Gaps = 8/170 (4%)
Query: 108 AESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSL 167
A S+ S TA ++S + + A+ + + A +SSI V ++P + S
Sbjct: 208 ATSSSLSSTAASNSATSSSLASSSLNSTTSATATSSSISSTVSSSTPLTSSNSTTAATSA 267
Query: 168 SYYINEFKTQEITQAPSVS-TETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSS 226
S + + + PS + + T LSS+ T + + T L+S + + + + T LSS
Sbjct: 268 SATSSSAQYNTSSLLPSSTPSSTPLSSANSTTATSASSTPLTSVNSTTTTSASSTPLSSV 327
Query: 227 IQTESVATDASISVPPES-------SPAEVSQVQVESQIQTETSSVDIVS 269
S ++ S P S S + V S T SS + S
Sbjct: 328 SSANSTTATSTSSTPLSSVNSTTATSASSTPLTSVNSTTATSASSTPLTS 377
Score = 35.4 bits (80), Expect = 0.15
Identities = 45/188 (23%), Positives = 73/188 (37%), Gaps = 30/188 (15%)
Query: 85 TGYVEKDTAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASS 144
T ++ T++ S P+ + +S + T A +T A +A +S+
Sbjct: 503 TPLTSANSTTSTSVSSTAPSYNTSSVLPTSSVSSTPLSSANSTTATSA---------SST 553
Query: 145 ILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQ 204
L V + + +++ + T E T S + + +S SV T PT
Sbjct: 554 PLTSVNSTTATSASSTPFGNSTITSSASG-STGEFTNTNSGNGD--VSGSVTT----PTS 606
Query: 205 TELSSSVQSESIAPAKTELSSSIQTESVATDASISVPPESSPA----------EVSQVQV 254
T LS+S ++AP T SS T S +S+S P +S A +S
Sbjct: 607 TPLSNS----TVAPTSTFTSSGFNTTSGLPTSSVSTPLSNSSAYPSSGSSTFSRLSSTLT 662
Query: 255 ESQIQTET 262
S I TET
Sbjct: 663 SSIIPTET 670
Score = 32.0 bits (71), Expect = 1.6
Identities = 26/90 (28%), Positives = 40/90 (43%), Gaps = 2/90 (2%)
Query: 180 TQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDASIS 239
T A +S+ L T + T + SSS+ S S A + SSS+ + S T+++ S
Sbjct: 44 TPATYLSSVPTLLKRATTSYNYNTSSASSSSLTSSSAASSSLTSSSSLASSS--TNSTTS 101
Query: 240 VPPESSPAEVSQVQVESQIQTETSSVDIVS 269
P SS S S + T+S + S
Sbjct: 102 ASPTSSSLTSSSATSSSLASSSTTSSSLAS 131
>ZAN_MOUSE (O88799) Zonadhesin precursor
Length = 5376
Score = 40.8 bits (94), Expect = 0.004
Identities = 48/197 (24%), Positives = 81/197 (40%), Gaps = 41/197 (20%)
Query: 94 GQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNS 153
G+T +Y+ P V + + + S + S+ +I +V S
Sbjct: 701 GETTLYTEVPTVPTEVTGVHTEVTNVSPEET-------------SVPTEETISTEVTTVS 747
Query: 154 PPQVQKVEYSGPSLSYYINEFKTQEIT---QAPSVSTE-----TELSSSVPTESDVPTQT 205
P + V P + T EIT + P+V TE TE+++ P E+ VPT+
Sbjct: 748 PEETT-VPTEVPIVLIEATASPTGEITLYTEVPTVPTEVTGVHTEVTNVSPEETSVPTEE 806
Query: 206 ELSSSVQSESIAPAKTELSSSIQTESV----ATDASISVPPESS------------PAEV 249
+S+ V +++P +T L + + T S + SVPPE + P EV
Sbjct: 807 TISTEV--TTVSPEETTLPTEVPTVSTEVTNVSPEETSVPPEETILTTLYTEVPTVPTEV 864
Query: 250 SQVQVE-SQIQTETSSV 265
+ V E + + E +SV
Sbjct: 865 TGVHTEVTNVSPEETSV 881
Score = 38.5 bits (88), Expect = 0.017
Identities = 27/89 (30%), Positives = 47/89 (52%), Gaps = 12/89 (13%)
Query: 180 TQAPSVSTE-----TELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESV-- 232
T+ P+V TE TE+++ P E+ VPT+ +S+ V +++P +T L + + T S
Sbjct: 855 TEVPTVPTEVTGVHTEVTNVSPEETSVPTEETISTEV--TTVSPEETTLPTEVPTVSTEV 912
Query: 233 --ATDASISVPPESS-PAEVSQVQVESQI 258
+ SVPPE + E++ V E +
Sbjct: 913 TNVSPEETSVPPEETILTEITTVSPEETV 941
Score = 33.1 bits (74), Expect = 0.73
Identities = 27/113 (23%), Positives = 48/113 (41%), Gaps = 2/113 (1%)
Query: 153 SPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQ 212
SP +V V + +E + P V+ TE+++ P E+ VPT+ + ++
Sbjct: 636 SPTEVTPVPTDVTAAYVEATNASPEETSVPPEVTILTEVTTVSPEETTVPTEVPI-VLIE 694
Query: 213 SESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSV 265
+ + +T L + + T + SP E S V E I TE ++V
Sbjct: 695 ATAFPTGETTLYTEVPTVPTEVTGVHTEVTNVSPEETS-VPTEETISTEVTTV 746
Score = 31.2 bits (69), Expect = 2.8
Identities = 33/111 (29%), Positives = 55/111 (48%), Gaps = 10/111 (9%)
Query: 165 PSLSYYINEFKTQEITQAP---SVSTE--TELSSSVPTE-SDVPTQTELSSSVQSESIAP 218
P++S + T+ T P S+ TE T L +S+P E + PT+ + ++ A
Sbjct: 975 PTVSTEMTGVHTEVTTVFPEETSIPTEVATVLPASIPPEETTTPTEVTTTPPEETTIPAE 1034
Query: 219 AKTELSSSIQTESVATDASI-SVPPE--SSPAEVSQVQVE-SQIQTETSSV 265
T +SI E A+ + + PPE ++P EV+ V E + I TE ++V
Sbjct: 1035 VTTVPPASIPPEETASLTEVTTTPPEETTTPTEVTTVPPEKTTIPTEVTTV 1085
>Y741_CHLMU (Q9PJT6) Hypothetical protein TC0741 precursor
Length = 1007
Score = 40.8 bits (94), Expect = 0.004
Identities = 43/176 (24%), Positives = 72/176 (40%), Gaps = 34/176 (19%)
Query: 96 TNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPP 155
+N S P V V+ + SSG+ GTS+ + T+ A A + +A++SI+ G+N
Sbjct: 6 SNSPSSIPTVTVSTTTASSGSLGTSTVSSTTTSTSVAQTATTTSSASTSIIQSSGEN--- 62
Query: 156 QVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSES 215
Q T PS T + +S+S P+ T + SS+V +
Sbjct: 63 --------------------IQSTTGTPSPIT-SSVSTSAPSPKASATANKTSSAVSGKI 101
Query: 216 IAPAKTELSSSIQTESVATDASIS-------VPPESSPAEVSQVQVESQIQTETSS 264
E S +T++ +D +S P SS + V + + Q +T S
Sbjct: 102 ---TSQETSEESETQATTSDGEVSSNYDDVDTPTNSSDSTVDSDYQDVETQYKTIS 154
>MSB2_YEAST (P32334) MSB2 protein (Multicopy suppressor of bud
emergence 2)
Length = 1306
Score = 40.8 bits (94), Expect = 0.004
Identities = 45/160 (28%), Positives = 71/160 (44%), Gaps = 17/160 (10%)
Query: 99 YSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAA-----GLALISIAAASSILLQVGKNS 153
+S + S+ISS + + + N+A+ ++ ++S + +S+ +S
Sbjct: 654 FSTSSVLVQMPSSISSEFSPSQTTTQMNSASSSSQYTISSTGILSQVSDTSVSYTTSSSS 713
Query: 154 PPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVP-TQTELSSSVQ 212
QV S +SY + +++ P T + SSSV SD P + T SSSV
Sbjct: 714 VSQV-----SDTPVSYTTSSSSVSQVSDTPVSYTTS--SSSVSQVSDTPVSYTTSSSSVS 766
Query: 213 SESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQV 252
S P SSS SV+ + SVP SS + VSQV
Sbjct: 767 QVSDTPVSYTTSSS----SVSQVSDTSVPSTSSRSSVSQV 802
Score = 37.0 bits (84), Expect = 0.051
Identities = 37/175 (21%), Positives = 75/175 (42%), Gaps = 6/175 (3%)
Query: 97 NIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQ 156
N+ S P+ S++ S + TS+ +++++ + AS+ + ++ +
Sbjct: 565 NLQSQPPST----SSLLSESQATSTSAVLASSSVSTTSPYTTAGGASTEASSLISSTSAE 620
Query: 157 VQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESI 216
+V YS + + + F + T+ S++ +SSV + +E S S + +
Sbjct: 621 TSQVSYSQSTTALQTSSFASSSTTEGSETSSQGFSTSSVLVQMPSSISSEFSPSQTTTQM 680
Query: 217 APAKTELSSSIQTESVATDAS-ISVPPESSPAEVSQV-QVESQIQTETSSVDIVS 269
A + +I + + + S SV +S + VSQV T +SSV VS
Sbjct: 681 NSASSSSQYTISSTGILSQVSDTSVSYTTSSSSVSQVSDTPVSYTTSSSSVSQVS 735
Score = 33.1 bits (74), Expect = 0.73
Identities = 47/174 (27%), Positives = 75/174 (43%), Gaps = 31/174 (17%)
Query: 101 VEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKV 160
V P+V S SS A TS+ + +++ + S+ A S + P +
Sbjct: 485 VSPSVSFVPSQSSSDVASTSAP-----SVVSSSFSYTSLQAGGSSMTN------PSSSTI 533
Query: 161 EYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAK 220
YS + S E + A + S SSS ++ +Q +SS+ SES A +
Sbjct: 534 VYSSSTGS--------SEESAASTASATLSGSSSTYMAGNLQSQPPSTSSLLSESQATST 585
Query: 221 TEL--SSSIQTES-------VATDASISVPPESSPAEVSQVQV-ESQIQTETSS 264
+ + SSS+ T S +T+AS + S+ AE SQV +S +TSS
Sbjct: 586 SAVLASSSVSTTSPYTTAGGASTEASSLI--SSTSAETSQVSYSQSTTALQTSS 637
Score = 33.1 bits (74), Expect = 0.73
Identities = 36/177 (20%), Positives = 74/177 (41%), Gaps = 12/177 (6%)
Query: 91 DTAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVG 150
D + + + S+ A +A S+ + + + +T + ++ A S+ LQ
Sbjct: 292 DGSSASPVVSMSAAGQIASSSSTDNPTMSETFSLTSTEVDGSDVSSTVSALLSAPFLQTS 351
Query: 151 KNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSS 210
++ + PS+S+ +Q + S ST +SSS SD+P QT S S
Sbjct: 352 TSNSFSIVS-----PSVSFV----PSQSSSDVASSSTANVVSSSF---SDIPPQTSTSGS 399
Query: 211 VQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSVDI 267
V S + + + SS + + +++S ++ + + + S + SS D+
Sbjct: 400 VVSVAQSASALAFQSSTEVYGASASSTMSSLLSTTSLQSTTLDSSSLASSSASSSDL 456
Score = 32.3 bits (72), Expect = 1.2
Identities = 45/230 (19%), Positives = 96/230 (41%), Gaps = 17/230 (7%)
Query: 38 VRASSESDCNDEECAPDKEVGKVSMEWLAEEKTKVVGTYPPRRKQGWTGYVEKDTAGQTN 97
++ SS + + E + G + L + + + + P Q T ++ Q
Sbjct: 633 LQTSSFASSSTTEGSETSSQGFSTSSVLVQMPSSISSEFSP--SQTTTQMNSASSSSQYT 690
Query: 98 IYSVEPAVYVAESAISSGTAGTSSDGAQNT-AAIAAGLALISIAAASSILLQVGKNSPPQ 156
I S V+++++S T+ +S +T + + +S + + + +S Q
Sbjct: 691 ISSTGILSQVSDTSVSYTTSSSSVSQVSDTPVSYTTSSSSVSQVSDTPVSYTTSSSSVSQ 750
Query: 157 VQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQ-SES 215
V S +SY + +++ P T + S S +++ VP+ + SS Q S++
Sbjct: 751 V-----SDTPVSYTTSSSSVSQVSDTPVSYTTSSSSVSQVSDTSVPSTSSRSSVSQVSDT 805
Query: 216 IAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSV 265
P+ + SS QT S S+ P ++ ++ + + +E+SSV
Sbjct: 806 PVPSTSSRSSVSQTSS-------SLQPTTTSSQRFTISTHGAL-SESSSV 847
>YB1E_SCHPO (P87179) Serine-rich protein C30B4.01c precursor
Length = 374
Score = 40.0 bits (92), Expect = 0.006
Identities = 42/161 (26%), Positives = 74/161 (45%), Gaps = 14/161 (8%)
Query: 107 VAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPS 166
V+ S++SS T+ +SS +++ S +++SS +S S S
Sbjct: 127 VSSSSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSS-------SSSSSSSSSSSSSSS 179
Query: 167 LSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSS 226
S + + + + S S+ + SSSVP S + SSS S S + ++ SSS
Sbjct: 180 SSSSSSSSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHSSSSSSSSSSSSSSRPSSSSS 239
Query: 227 -IQTESVATDAS-ISVPPESSPAEVSQVQVESQIQTETSSV 265
I T S +T S ++V P SS + S S++ + T+++
Sbjct: 240 FITTMSSSTFISTVTVTPSSSSSSTS-----SEVPSSTAAL 275
Score = 37.4 bits (85), Expect = 0.039
Identities = 29/84 (34%), Positives = 41/84 (48%), Gaps = 5/84 (5%)
Query: 186 STETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDASISVPPESS 245
ST + SSS P+ S T T SSS S S + + + SSS + S ++ +S S SS
Sbjct: 134 STTSSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 193
Query: 246 PAEVSQVQVESQIQTETSSVDIVS 269
+ S S + +SSV I S
Sbjct: 194 SSSSS-----SSSSSSSSSVPITS 212
Score = 32.3 bits (72), Expect = 1.2
Identities = 25/94 (26%), Positives = 43/94 (45%)
Query: 176 TQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATD 235
T + + S S+ + SSS + S + + SSS S S + + + SSS + SV
Sbjct: 152 TTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSVPIT 211
Query: 236 ASISVPPESSPAEVSQVQVESQIQTETSSVDIVS 269
+S S SS + S S+ + +S + +S
Sbjct: 212 SSTSSSHSSSSSSSSSSSSSSRPSSSSSFITTMS 245
Score = 30.8 bits (68), Expect = 3.6
Identities = 33/165 (20%), Positives = 71/165 (43%), Gaps = 8/165 (4%)
Query: 84 WTGYVEKDTAGQTNIYSVEPAVYVAESAISSG--------TAGTSSDGAQNTAAIAAGLA 135
W+ Y+ + QT + S + + S+ SS T+ +SS + ++++ ++ +
Sbjct: 113 WSVYLTGNGVLQTTVSSSSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSS 172
Query: 136 LISIAAASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSV 195
S +++SS +S S S S + + + + S S+ + SSS
Sbjct: 173 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHSSSSSSSSSSSSSS 232
Query: 196 PTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDASISV 240
S T +SSS ++ + SSS +E ++ A++++
Sbjct: 233 RPSSSSSFITTMSSSTFISTVTVTPSSSSSSTSSEVPSSTAALAL 277
Score = 29.6 bits (65), Expect = 8.1
Identities = 27/111 (24%), Positives = 49/111 (43%), Gaps = 3/111 (2%)
Query: 157 VQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSS---VQS 213
V + S S+S + + + + + +T + SSS + S + + SSS S
Sbjct: 122 VLQTTVSSSSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSS 181
Query: 214 ESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSS 264
S + + + SSS + S ++ +S SVP SS + S + +SS
Sbjct: 182 SSSSSSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHSSSSSSSSSSSSSS 232
>FLO9_YEAST (P39712) Flocculation protein FLO9 precursor
Length = 1322
Score = 39.7 bits (91), Expect = 0.008
Identities = 45/183 (24%), Positives = 80/183 (43%), Gaps = 15/183 (8%)
Query: 84 WTGY-----VEKDTAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALIS 138
WTG E T TN + V + ++ ++ ++ SS Q T+ I + +I+
Sbjct: 823 WTGTFTSTSTEMTTITGTNGQPTDETVIIVKTPTTAISSSLSSSSGQITSFITSARPIIT 882
Query: 139 ----IAAASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSS 194
S I V +S + S S + + +P +S+ T S+S
Sbjct: 883 PFYPSNGTSVISSSVISSSDTSSLVISSSVTSSLVTSSPVISSSFISSPVISSTTT-SAS 941
Query: 195 VPTESD----VPTQTELSSSVQSESIAPAKTELSSSIQTES-VATDASISVPPESSPAEV 249
+ +ES +PT + S S +SE+ + + SSSI +ES +T +S S+PP +S
Sbjct: 942 ILSESSKSSVIPTSSSTSGSSESETGSASSASSSSSISSESPKSTYSSSSLPPVTSATTS 1001
Query: 250 SQV 252
++
Sbjct: 1002 QEI 1004
Score = 32.0 bits (71), Expect = 1.6
Identities = 43/188 (22%), Positives = 73/188 (37%), Gaps = 20/188 (10%)
Query: 92 TAGQTNIYSVEPAVYVAESAISSGTAGTSSD--GAQNTAAIAAGLALISIAAASSILLQV 149
++GQ + + S+GT+ SS + +T+++ ++ S SS ++
Sbjct: 866 SSGQITSFITSARPIITPFYPSNGTSVISSSVISSSDTSSLVISSSVTSSLVTSSPVISS 925
Query: 150 GKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETE--------LSSSVPTESDV 201
S P + S LS K+ I + S S +E SSS S+
Sbjct: 926 SFISSPVISSTTTSASILS---ESSKSSVIPTSSSTSGSSESETGSASSASSSSSISSES 982
Query: 202 PTQTELSSSVQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTE 261
P T SSS+ + A E++SS+ + + V+ ES + TE
Sbjct: 983 PKSTYSSSSLPPVTSATTSQEITSSLPPVT-------TTKTSEQTTLVTVTSCESHVCTE 1035
Query: 262 TSSVDIVS 269
+ S IVS
Sbjct: 1036 SISSAIVS 1043
Score = 29.6 bits (65), Expect = 8.1
Identities = 43/162 (26%), Positives = 76/162 (46%), Gaps = 15/162 (9%)
Query: 106 YVAESAISSGTAGTS--SDGAQN----TAAIAAGLALISIAAASSILLQVGKNSPPQVQK 159
+++ ISS T S S+ +++ T++ +G + +ASS +S + K
Sbjct: 927 FISSPVISSTTTSASILSESSKSSVIPTSSSTSGSSESETGSASSASSSSSISS--ESPK 984
Query: 160 VEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPA 219
YS SL + +QEIT + T T+ S ++ + T T S V +ESI+ A
Sbjct: 985 STYSSSSLPPVTSATTSQEITSSLPPVTTTKTSE----QTTLVTVTSCESHVCTESISSA 1040
Query: 220 KTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTE 261
+S++ T S AT + P S+ E+++ E+ QT+
Sbjct: 1041 --IVSTATVTVSGATTEYTTWCPIST-TEITKQTTETTKQTK 1079
>TIR1_YEAST (P10863) Cold shock induced protein TIR1 precursor
(Serine-rich protein 1)
Length = 254
Score = 39.3 bits (90), Expect = 0.010
Identities = 31/85 (36%), Positives = 41/85 (47%), Gaps = 3/85 (3%)
Query: 182 APSVSTETELSSSVPTESDVPTQTELSSS--VQSESIAPAKTELSSSIQTESVATDASIS 239
APS S+ SS+ P+ S PT + SSS +S S AP+ +E SS S + S S
Sbjct: 118 APS-SSAAPTSSAAPSSSAAPTSSAASSSSEAKSSSAAPSSSEAKSSSAAPSSSEAKSSS 176
Query: 240 VPPESSPAEVSQVQVESQIQTETSS 264
P SS A+ S S TS+
Sbjct: 177 AAPSSSEAKSSSAAPSSTEAKITSA 201
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.303 0.118 0.308
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,299,071
Number of Sequences: 164201
Number of extensions: 969427
Number of successful extensions: 4514
Number of sequences better than 10.0: 231
Number of HSP's better than 10.0 without gapping: 53
Number of HSP's successfully gapped in prelim test: 184
Number of HSP's that attempted gapping in prelim test: 3287
Number of HSP's gapped (non-prelim): 785
length of query: 269
length of database: 59,974,054
effective HSP length: 108
effective length of query: 161
effective length of database: 42,240,346
effective search space: 6800695706
effective search space used: 6800695706
T: 11
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC135566.8


