

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135505.8 + phase: 0
(454 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
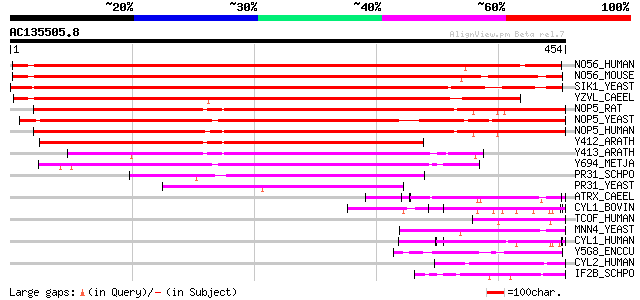
NO56_HUMAN (O00567) Nucleolar protein Nop56 (Nucleolar protein 5A) 481 e-135
NO56_MOUSE (Q9D6Z1) Nucleolar protein Nop56 (Nucleolar protein 5A) 474 e-133
SIK1_YEAST (Q12460) SIK1 protein (Nucleolar protein NOP56) 427 e-119
YZVL_CAEEL (Q21276) Hypothetical protein K07C5.4 in chromosome V 406 e-113
NOP5_RAT (Q9QZ86) Nucleolar protein NOP5 (Nucleolar protein 5) (... 318 2e-86
NOP5_YEAST (Q12499) Nucleolar protein NOP58 (Nucleolar protein N... 316 7e-86
NOP5_HUMAN (Q9Y2X3) Nucleolar protein NOP5 (Nucleolar protein 5)... 315 2e-85
Y412_ARATH (O04658) Hypothetical protein At5g27120 297 3e-80
Y413_ARATH (O04656) Hypothetical protein At5g27140 227 4e-59
Y694_METJA (Q58105) Hypothetical protein MJ0694 168 2e-41
PR31_SCHPO (O42904) Pre-mRNA splicing factor prp31 101 5e-21
PR31_YEAST (P49704) Pre-mRNA splicing factor PRP31 73 2e-12
ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog (X-li... 55 3e-07
CYL1_BOVIN (P35662) Cylicin I (Multiple-band polypeptide I) 54 8e-07
TCOF_HUMAN (Q13428) Treacle protein (Treacher Collins syndrome p... 52 3e-06
MNN4_YEAST (P36044) MNN4 protein 51 7e-06
CYL1_HUMAN (P35663) Cylicin I (Multiple-band polypeptide I) (Fra... 50 9e-06
Y5G8_ENCCU (Q8STA9) Hypothetical protein ECU05_1680/ECU11_0050 48 5e-05
CYL2_HUMAN (Q14093) Cylicin II (Multiple-band polypeptide II) 48 5e-05
IF2B_SCHPO (P56329) Probable eukaryotic translation initiation f... 48 6e-05
>NO56_HUMAN (O00567) Nucleolar protein Nop56 (Nucleolar protein 5A)
Length = 596
Score = 481 bits (1238), Expect = e-135
Identities = 254/452 (56%), Positives = 328/452 (72%), Gaps = 10/452 (2%)
Query: 3 DELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHF 62
++LR +LET+LP KK K LGV + KIG+ I E Q+ + E++RGVR HF
Sbjct: 74 EDLRLLLETHLPS----KKKKVLLGVGDPKIGAAIQEELGYNCQTGGVIAEILRGVRLHF 129
Query: 63 DKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRV 122
V L KAQLGL HSYSRAKVKFNVNRVDNM+IQ+I LLD LDKD+N+F+MRV
Sbjct: 130 HNLVKGLTDLSACKAQLGLGHSYSRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRV 189
Query: 123 REWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAK 182
REWY +HFPELVKI+NDN YC++A+FI ++ +L EDK+E L +L D KAK I++A++
Sbjct: 190 REWYGYHFPELVKIINDNATYCRLAQFIGNRRELNEDKLEKLEELTMDGAKAKAILDASR 249
Query: 183 ASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLI 242
+SMG D+S +DLIN+ F+ RV+ LS+YR+ L YL +KM+ +AP+L +L+G++VGARLI
Sbjct: 250 SSMGMDISAIDLINIESFSSRVVSLSEYRQSLHTYLRSKMSQVAPSLSALIGEAVGARLI 309
Query: 243 SHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMA 302
+HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++
Sbjct: 310 AHAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRIS 369
Query: 303 RYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVD 362
RYLANKCSIASRIDCFSE TS FGEKLREQVEERL FY+ G PRKN+DVMK A+ +
Sbjct: 370 RYLANKCSIASRIDCFSEVPTSVFGEKLREQVEERLSFYETGEIPRKNLDVMKEAMVQAE 429
Query: 363 NKDTEMETE---EVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKR 419
++ E E KK KK++K +A + + T + E+ + KKKKK+K
Sbjct: 430 AEEAAAEITRKLEKQEKKRLKKEKKRLAALALASSENSSSTPEECEEMSEKPKKKKKQK- 488
Query: 420 KLDQEVEVEDKVVEDGANADSSKKKKKKSKKK 451
QEV E+ + + + KKKK SK++
Sbjct: 489 --PQEVPQENGMEDPSISFSKPKKKKSFSKEE 518
>NO56_MOUSE (Q9D6Z1) Nucleolar protein Nop56 (Nucleolar protein 5A)
Length = 580
Score = 474 bits (1221), Expect = e-133
Identities = 252/452 (55%), Positives = 328/452 (71%), Gaps = 17/452 (3%)
Query: 3 DELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHF 62
++LR +LET LP KK K LGV + KIG+ I E Q+ + E++RGVR HF
Sbjct: 74 EDLRLLLETYLPS----KKKKVLLGVGDPKIGAAIQEELGYNCQTGGVIAEILRGVRLHF 129
Query: 63 DKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRV 122
V L KAQLGL HSYSRAKVKFNVNRVDNM+IQ+I LLD LDKD+N+F+MRV
Sbjct: 130 HNLVKGLTDLSACKAQLGLGHSYSRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRV 189
Query: 123 REWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAK 182
REWY +HFPELVKIVNDN YC++A+FI ++ +L E+K+E L ++ D KAK I++A++
Sbjct: 190 REWYGYHFPELVKIVNDNATYCRLAQFIGNRRELNEEKLEKLEEITMDGAKAKAILDASR 249
Query: 183 ASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLI 242
+SMG D+S +DLIN+ F+ RV+ LS+YR+ L YL +KM+ +AP+L +L+G++VGARLI
Sbjct: 250 SSMGMDISAIDLINIESFSSRVVSLSEYRQSLHTYLRSKMSQVAPSLSALIGEAVGARLI 309
Query: 243 SHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMA 302
+HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++
Sbjct: 310 AHAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRIS 369
Query: 303 RYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVD 362
RYLANKCSIASRIDCFSE TS FGEKLREQVEERL FY+ G PRKN+DVMK A+ +
Sbjct: 370 RYLANKCSIASRIDCFSEVPTSVFGEKLREQVEERLSFYETGEIPRKNLDVMKEAVVQAE 429
Query: 363 NKDTEM--ETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK 420
E+ + E+ K+ KK+K++ A A+ A+ + E+ + +K KK+K+
Sbjct: 430 EAAAEITRKLEKQEKKRLKKEKKRLA-----ALALASSENSSTPEECEEVNEKSKKKKKL 484
Query: 421 LDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
QE +ED V K KKKK+ K++
Sbjct: 485 KPQENGMEDPPV------SLPKSKKKKAPKEE 510
>SIK1_YEAST (Q12460) SIK1 protein (Nucleolar protein NOP56)
Length = 504
Score = 427 bits (1098), Expect = e-119
Identities = 228/454 (50%), Positives = 311/454 (68%), Gaps = 25/454 (5%)
Query: 1 MTDELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATK-IPVQSNEFVGELIRGVR 59
+++ L+ +L+ NLPK KK +L +++ +G I E + SNE +LIRGVR
Sbjct: 74 VSESLKAILDLNLPKASS-KKKNITLAISDKNLGPSIKEEFPYVDCISNELAQDLIRGVR 132
Query: 60 QHFDKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFA 119
H +K L+ GDLE+AQLGL H+YSRAKVKF+V + DN +IQAI LLD LDKD+N+FA
Sbjct: 133 LHGEKLFKGLQSGDLERAQLGLGHAYSRAKVKFSVQKNDNHIIQAIALLDQLDKDINTFA 192
Query: 120 MRVREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDK-AKEIV 178
MRV+EWY WHFPEL K+V DNY + K+ FI+DK+ L +D + L L+ ++ A+ ++
Sbjct: 193 MRVKEWYGWHFPELAKLVPDNYTFAKLVLFIKDKASLNDDSLHDLAALLNEDSGIAQRVI 252
Query: 179 EAAKASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVG 238
+ A+ SMGQD+S D+ NV +FAQRV L+DYRR+L DYL KM+ +APNL L+G+ +G
Sbjct: 253 DNARISMGQDISETDMENVCVFAQRVASLADYRRQLYDYLCEKMHTVAPNLSELIGEVIG 312
Query: 239 ARLISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNK 298
ARLISHAGSLTNL+K +ST+QILGAEKALFRALKT+GNTPKYGLI+HS FI +ASA+NK
Sbjct: 313 ARLISHAGSLTNLSKQAASTVQILGAEKALFRALKTKGNTPKYGLIYHSGFISKASAKNK 372
Query: 299 GRMARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAI 358
GR++RYLANKCS+ASRID +SE+ ++ FG L++QVE+RL+FY+ G KN ++ A+
Sbjct: 373 GRISRYLANKCSMASRIDNYSEEPSNVFGSVLKKQVEQRLEFYNTGKPTLKNELAIQEAM 432
Query: 359 ESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEK 418
E + NKD E + +K KK+K D D + + K EKK KK++K
Sbjct: 433 E-LYNKDKPAAEVEETKEKESSKKRKLEDDD-------------EEKKEKKEKKSKKEKK 478
Query: 419 RKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
K +++ + E K D +KK KK K KD
Sbjct: 479 EKKEKKDKKEKK--------DKKEKKDKKKKSKD 504
>YZVL_CAEEL (Q21276) Hypothetical protein K07C5.4 in chromosome V
Length = 486
Score = 406 bits (1043), Expect = e-113
Identities = 213/419 (50%), Positives = 287/419 (67%), Gaps = 17/419 (4%)
Query: 4 ELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEA-TKIPVQSNEFVGELIRGVRQHF 62
+L L+ +LPK KK LG+ +SK+ + EA + + + E++RG R HF
Sbjct: 77 DLTNFLQKSLPK----KKKHVVLGINDSKLAGSLTEAFPDLKLVFGGVITEILRGTRVHF 132
Query: 63 DKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRV 122
++ +L L KAQL L HSYSR+KVKF+V+RVDNMVIQ+I LLD LDKD+N F MR+
Sbjct: 133 ERLAKNLPHHSLSKAQLSLGHSYSRSKVKFDVHRVDNMVIQSIALLDQLDKDINLFGMRI 192
Query: 123 REWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKI---ESLTDLVGDEDKAKEIVE 179
REWYS+H+PEL ++ D Y Y ++A I D++K+AE++ E L L D +K +I+E
Sbjct: 193 REWYSYHYPELFRLAPDQYKYSRLAVAILDRNKMAENENLENEILEILDNDSEKTAQIIE 252
Query: 180 AAKASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGA 239
AA+ SMG D+S +DL N+ FA RV L +YR++L +Y+ +M+ AP+L +L+G+ VGA
Sbjct: 253 AARTSMGMDISDLDLENIKRFAARVSSLMEYRQQLHEYIKDRMDHCAPSLSALIGEQVGA 312
Query: 240 RLISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKG 299
RLISHAGSLTNLAK P+ST+QILGAEKALFRALKTR NTPKYGL+FHSSFIG+A +NKG
Sbjct: 313 RLISHAGSLTNLAKYPASTVQILGAEKALFRALKTRSNTPKYGLLFHSSFIGKAGTKNKG 372
Query: 300 RMARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIE 359
R++RYLANKCSIA+R+DCFSE S +GE LR+QVE+RL+++ G P+KNIDVMK A E
Sbjct: 373 RVSRYLANKCSIAARVDCFSETPVSTYGEFLRQQVEDRLEYFTSGTVPKKNIDVMKEAEE 432
Query: 360 SVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEK 418
+ EV K KKKK+ A +A + + ++ + KKKKK K
Sbjct: 433 A---------AVEVKEKVIKKKKKAAKKAKRLAEESVTATAEAEVDEDAPKPKKKKKSK 482
>NOP5_RAT (Q9QZ86) Nucleolar protein NOP5 (Nucleolar protein 5)
(Nopp140 associated protein)
Length = 534
Score = 318 bits (814), Expect = 2e-86
Identities = 184/458 (40%), Positives = 279/458 (60%), Gaps = 28/458 (6%)
Query: 20 KKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQL 79
K+A L VA++K+G I E + + V EL+RG+R D + ++ ++ L
Sbjct: 81 KEAHEPLAVADAKLGGVIKEKLNLSCIHSPVVNELMRGIRSQMDGLIPGVEPREMAAMCL 140
Query: 80 GLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVND 139
GL HS SR ++KF+ ++VD M++QAI LLD LDK++N++ MR REWY WHFPEL KI++D
Sbjct: 141 GLAHSLSRYRLKFSADKVDTMIVQAISLLDDLDKELNNYIMRCREWYGWHFPELGKIISD 200
Query: 140 NYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHM 199
N YCK + + D+ A +L++ + +E +A E+ AA+ SMG ++S D+ N+
Sbjct: 201 NLTYCKCLQKVGDRKNYAS---ATLSEFLSEEVEA-EVKAAAEISMGTEVSEEDICNILH 256
Query: 200 FAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTL 259
+V+++S+YR +L +YL +M IAPN+ +VG+ VGARLI+HAGSL NLAK +ST+
Sbjct: 257 LCTQVIEISEYRTQLYEYLQNRMMAIAPNVTVMVGELVGARLIAHAGSLLNLAKHAASTV 316
Query: 260 QILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFS 319
QILGAEKALFRALK+R +TPKYGLI+H+S +G+ S ++KG+++R LA K +A R D F
Sbjct: 317 QILGAEKALFRALKSRRDTPKYGLIYHASLVGQTSPKHKGKISRMLAAKTVLAIRYDAFG 376
Query: 320 EKGTSAFGEKLREQVEERLDFY-DKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKT 378
E +SA G + R ++E RL D+G+ + E ++K +E++T + S T
Sbjct: 377 EDSSSAMGAENRAKLEARLRILEDRGIRKISGTGKALAKAEKYEHK-SEVKTYDPSGDST 435
Query: 379 -----KKKKQKAADGDDMAVDKAA-------------EITNGD----AEDHKSEKKKKKK 416
KK+K + D +D +K A E+ + E+ + KKKKKK
Sbjct: 436 LPTCSKKRKIEEVDKEDEITEKKAKKAKIKIKAEVEEEMEEAEEEQVVEEEPTVKKKKKK 495
Query: 417 EKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+K+K +E + ++ S +KKKKK KKKDAE
Sbjct: 496 DKKKHIKEEPLSEEEPCTSTAVPSPEKKKKKKKKKDAE 533
>NOP5_YEAST (Q12499) Nucleolar protein NOP58 (Nucleolar protein
NOP5)
Length = 511
Score = 316 bits (810), Expect = 7e-86
Identities = 178/448 (39%), Positives = 279/448 (61%), Gaps = 27/448 (6%)
Query: 9 LETNLPKVKEGKKAKFSLGVAESKIGSHIHE-ATKIPVQSNEFVGELIRGVRQHFDKFVG 67
LE L ++K+ KK+ +L V+E+K+ + I++ V S+ ++ R ++++ + +
Sbjct: 72 LEKLLEEIKKDKKS--TLIVSETKLANAINKLGLNFNVVSDAVTLDIYRAIKEYLPELLP 129
Query: 68 DLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYS 127
+ DL K LGL HS R K+KF+ ++VD M+IQAI LLD LDK++N++AMR +EWY
Sbjct: 130 GMSDNDLSKMSLGLAHSIGRHKLKFSADKVDVMIIQAIALLDDLDKELNTYAMRCKEWYG 189
Query: 128 WHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQ 187
WHFPEL KIV D+ Y ++ + +SK +E + + E+ + + AA+ SMG
Sbjct: 190 WHFPELAKIVTDSVAYARIILTMGIRSKASETDLSEILP----EEIEERVKTAAEVSMGT 245
Query: 188 DLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGS 247
+++ DL N++ A+++++ + YR +LS+YLS +M IAPNL LVG+ VGARLI+H+GS
Sbjct: 246 EITQTDLDNINALAEQIVEFAAYREQLSNYLSARMKAIAPNLTQLVGELVGARLIAHSGS 305
Query: 248 LTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLAN 307
L +LAK P+ST+QILGAEKALFRALKT+ +TPKYGL++H+S +G+A+ +NKG++AR LA
Sbjct: 306 LISLAKSPASTIQILGAEKALFRALKTKHDTPKYGLLYHASLVGQATGKNKGKIARVLAA 365
Query: 308 KCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTE 367
K +++ R D + E+R D D G+ R ++ S +E D + T
Sbjct: 366 KAAVSLRYDALA---------------EDRDDSGDIGLESRAKVENRLSQLEGRDLRTTP 410
Query: 368 METEEVSAKKTKKKKQKAADGD-DMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVE 426
E AKK + + +A + D D A KAA + D++D + EKK+KK++KRK D + +
Sbjct: 411 KVVRE--AKKVEMTEARAYNADADTA--KAASDSESDSDDEEEEKKEKKEKKRKRDDDED 466
Query: 427 VEDKVVEDGANADSSKKKKKKSKKKDAE 454
+D D +KK+KK KK+ E
Sbjct: 467 SKDSKKAKKEKKDKKEKKEKKEKKEKKE 494
>NOP5_HUMAN (Q9Y2X3) Nucleolar protein NOP5 (Nucleolar protein 5)
(NOP58) (HSPC120)
Length = 529
Score = 315 bits (806), Expect = 2e-85
Identities = 181/454 (39%), Positives = 278/454 (60%), Gaps = 24/454 (5%)
Query: 20 KKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQL 79
K+A L VA++K+G I E + + V EL+RG+R D + ++ ++ L
Sbjct: 81 KEAHEPLAVADAKLGGVIKEKLNLSCIHSPVVNELMRGIRSQMDGLIPGVEPREMAAMCL 140
Query: 80 GLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVND 139
GL HS SR ++KF+ ++VD M++QAI LLD LDK++N++ MR REWY WHFPEL KI++D
Sbjct: 141 GLAHSLSRYRLKFSADKVDTMIVQAISLLDDLDKELNNYIMRCREWYGWHFPELGKIISD 200
Query: 140 NYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHM 199
N YCK + + D+ A K L++L+ +E +A E+ AA+ SMG ++S D+ N+
Sbjct: 201 NLTYCKCLQKVGDRKNYASAK---LSELLPEEVEA-EVKAAAEISMGTEVSEEDICNILH 256
Query: 200 FAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTL 259
+V+++S+YR +L +YL +M IAPN+ +VG+ VGARLI+HAGSL NLAK +ST+
Sbjct: 257 LCTQVIEISEYRTQLYEYLQNRMMAIAPNVTVMVGELVGARLIAHAGSLLNLAKHAASTV 316
Query: 260 QILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFS 319
QILGAEKALFRALK+R +TPKYGLI+H+S +G+ S ++KG+++R LA K +A R D F
Sbjct: 317 QILGAEKALFRALKSRRDTPKYGLIYHASLVGQTSPKHKGKISRMLAAKTVLAIRYDAFG 376
Query: 320 EKGTSAFGEKLREQVEERL-DFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKT 378
E +SA G + R ++E RL D+G+ + E ++K +E++T + S T
Sbjct: 377 EDSSSAMGVENRAKLEARLRTLEDRGIRKISGTGKALAKTEKYEHK-SEVKTYDPSGDST 435
Query: 379 -----KKKKQKAADGDDMAVDKAA-------------EITNGDAEDHKSEKKKKKKEKRK 420
KK+K + D +D +K A E + E+ +KKKK+ +K+
Sbjct: 436 LPTCSKKRKIEQVDKEDEITEKKAKKAKIKVKVEEEEEEKVAEEEETSVKKKKKRGKKKH 495
Query: 421 LDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+ +E E++ A A KKKKKK K+++ +
Sbjct: 496 IKEEPLSEEEPCTSTAIASPEKKKKKKKKRENED 529
>Y412_ARATH (O04658) Hypothetical protein At5g27120
Length = 439
Score = 297 bits (761), Expect = 3e-80
Identities = 157/314 (50%), Positives = 217/314 (69%), Gaps = 4/314 (1%)
Query: 25 SLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQLGLCHS 84
+L VA+SK+G+ I E KI N V EL+RG+R + + L DL LGL HS
Sbjct: 84 TLAVADSKLGNIIKEKLKIVCVHNNAVMELLRGIRSQLTELISGLGDQDLGPMSLGLSHS 143
Query: 85 YSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVNDNYLYC 144
+R K+KF+ ++VD M+IQAI LLD LDK++N++AMRVREW+ WHFPEL KIV DN LY
Sbjct: 144 LARYKLKFSSDKVDTMIIQAIGLLDDLDKELNTYAMRVREWFGWHFPELAKIVQDNILYA 203
Query: 145 KVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHMFAQRV 204
K K + ++ A+ ++++ DE +A E+ EAA SMG ++S +DL+++ +V
Sbjct: 204 KAVKLMGNRINAAK---LDFSEILADEIEA-ELKEAAVISMGTEVSDLDLLHIRELCDQV 259
Query: 205 MDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTLQILGA 264
+ L++YR +L DYL ++MN IAPNL +LVG+ VGARLISH GSL NLAK P ST+QILGA
Sbjct: 260 LSLAEYRAQLYDYLKSRMNTIAPNLTALVGELVGARLISHGGSLLNLAKQPGSTVQILGA 319
Query: 265 EKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEKGTS 324
EKALFRALKT+ TPKYGLIFH+S +G+A+ +NKG+++R LA K +A R D + +
Sbjct: 320 EKALFRALKTKHATPKYGLIFHASVVGQAAPKNKGKISRSLAAKSVLAIRCDALGDSQDN 379
Query: 325 AFGEKLREQVEERL 338
G + R ++E RL
Sbjct: 380 TMGVENRLKLEARL 393
>Y413_ARATH (O04656) Hypothetical protein At5g27140
Length = 435
Score = 227 bits (579), Expect = 4e-59
Identities = 139/355 (39%), Positives = 208/355 (58%), Gaps = 25/355 (7%)
Query: 48 NEFVGELIRGVRQHFDKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVD--------- 98
N+ V EL+RGVR + + L DL L L H +R K+K ++V
Sbjct: 52 NDAVMELLRGVRSQLTELLSGLDDNDLAPVSLELSHILARYKLKITSDKVVIEFSVFCFT 111
Query: 99 ---NMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSK 155
M+I +I LLD LDK++N++ V E Y HFPEL IV DN LY KV K + ++
Sbjct: 112 LAYTMIILSISLLDDLDKELNTYTTSVCELYGLHFPELANIVQDNILYAKVVKLMGNRIN 171
Query: 156 LAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLS 215
A ++++ DE +A E+ EA+ S ++S +DL+++ +V+ +++ + L
Sbjct: 172 AAT---LDFSEILADEVEA-ELKEASMVSTRTEVSDLDLMHIQELCDQVLSIAEDKTLLC 227
Query: 216 DYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTLQILGAEKALFRALKTR 275
D L KMN IAPNL +LVG+ VGARLISH GSL NL+K P ST+QILGAEK L++ALKT+
Sbjct: 228 DDLKNKMNKIAPNLTALVGELVGARLISHCGSLWNLSKLPWSTIQILGAEKTLYKALKTK 287
Query: 276 GNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEKGTSAFGEKLREQVE 335
TPKYGLI+H+ + +A+ NKG++AR LA K ++A R D F + G + R ++E
Sbjct: 288 QATPKYGLIYHAPLVRQAAPENKGKIARSLAAKSALAIRCDAFGNGQDNTMGVESRLKLE 347
Query: 336 ERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTK---KKKQKAAD 387
RL + G ++ + E V++KDT+ E ++ KT+ KK++K A+
Sbjct: 348 ARLRNLEGG-----DLGACEEE-EEVNDKDTKKEADDEEEPKTEECSKKRKKEAE 396
>Y694_METJA (Q58105) Hypothetical protein MJ0694
Length = 414
Score = 168 bits (426), Expect = 2e-41
Identities = 122/368 (33%), Positives = 185/368 (50%), Gaps = 17/368 (4%)
Query: 24 FSLGVAESKIGSHIHEA----TKIPVQSNE--FVGELIRGVRQHFDKFVGDLKQGDLEKA 77
F L +KI + E K+ S E +GE +R K +G D +
Sbjct: 42 FKLKTQPNKIADELKEEWGDEIKLETLSTEPFNIGEFLRNNLFKVGKELGYFNNYDEFRK 101
Query: 78 QLGLCHSYSRAKV-KFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKI 136
++ + KV K + D ++IQ + LDK +N + R+REWYS +FPEL +
Sbjct: 102 KMHYWSTELTKKVIKSYAQQKDKIIIQVAEAISDLDKTLNLLSERLREWYSLYFPELDHL 161
Query: 137 VNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLIN 196
VN + +Y + + + + +++ + A +I EAAK SMG +L DL
Sbjct: 162 VNKHEVYANLITKLGKRKNFTKSQLKKILP----SKLAGKIAEAAKNSMGGELEDYDLDV 217
Query: 197 VHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPS 256
+ FA+ + L + R+ L +YL MN+ APN+ L G S+GARLI AG L LAK P+
Sbjct: 218 IVKFAEEINHLYEKRKELYNYLEKLMNEEAPNITKLAGVSLGARLIGLAGGLEKLAKMPA 277
Query: 257 STLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRID 316
ST+Q+LGAEKALF L+ PK+G+I++ I + +G++AR LA K +IA+R D
Sbjct: 278 STIQVLGAEKALFAHLRMGVEPPKHGIIYNHPLIQGSPHWQRGKIARALACKLAIAARAD 337
Query: 317 CFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAK 376
+ EKL ++VEE Y K P+K K K+ + + E+ K
Sbjct: 338 YVGDYIADELLEKLNKRVEEIRRKYPK--PPKKK----KKEKPKAKKKEKKGKKEKSKKK 391
Query: 377 KTKKKKQK 384
K KKK +K
Sbjct: 392 KDKKKDKK 399
>PR31_SCHPO (O42904) Pre-mRNA splicing factor prp31
Length = 518
Score = 101 bits (251), Expect = 5e-21
Identities = 62/244 (25%), Positives = 118/244 (47%), Gaps = 11/244 (4%)
Query: 99 NMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVNDNYLYCKVAKFIE---DKSK 155
++++ + + +D ++ V+EWY FPEL +V + + YCK + D SK
Sbjct: 118 HLIVDSNSIAMEIDDEILRLHRLVKEWYHDRFPELSSLVLNAFDYCKTVSSLLNDLDNSK 177
Query: 156 LAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLS 215
+ S T +V I A ++G+ L + NV + + L + ++++
Sbjct: 178 TKLSFLPSATVMV--------IATTATTTVGKPLPDEMIKNVKNCCEAIQQLGEEKQKII 229
Query: 216 DYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTLQILGAEKALFRALKTR 275
+Y+ ++++ +APNL ++VG + A LI AG LT L K P+ L LG + +
Sbjct: 230 EYVQSRISVVAPNLSAVVGSTTAANLIGIAGGLTRLGKFPACNLPALGKRRLTTIGINNP 289
Query: 276 GNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEKGTSAFGEKLREQVE 335
+ YG ++ S + + + + R A K ++A+RID E +FG R++VE
Sbjct: 290 AVSGDYGFLYMSEIVQKTPPDVRKQAIRMTAAKVALAARIDSIHEYPDGSFGISARKEVE 349
Query: 336 ERLD 339
+++
Sbjct: 350 RKIE 353
>PR31_YEAST (P49704) Pre-mRNA splicing factor PRP31
Length = 494
Score = 72.8 bits (177), Expect = 2e-12
Identities = 52/204 (25%), Positives = 91/204 (44%), Gaps = 7/204 (3%)
Query: 126 YSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASM 185
YS FPEL ++ Y KV +E+++ + E L +E + SM
Sbjct: 120 YSRRFPELSSLIPSPLQYSKVISILENENYSKNESDELFFHLENKAKLTREQILVLTMSM 179
Query: 186 GQDLSPVDLINVHMFAQRVM------DLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGA 239
+ +++ Q + +L + + Y+++K++ IAPN+ LVG + A
Sbjct: 180 KTSFKNKEPLDIKTRTQILEANSILENLWKLQEDIGQYIASKISIIAPNVCFLVGPEIAA 239
Query: 240 RLISHAGSLTNLAKCPSSTLQILGAEKALFRALKT-RGNTPKYGLIFHSSFIGRASARNK 298
+LI+HAG + ++ PS + +G K L L T + G +F S I +
Sbjct: 240 QLIAHAGGVLEFSRIPSCNIASIGKNKHLSHELHTLESGVRQEGYLFASDMIQKFPVSVH 299
Query: 299 GRMARYLANKCSIASRIDCFSEKG 322
+M R L K S+A+R+D + G
Sbjct: 300 KQMLRMLCAKVSLAARVDAGQKNG 323
>ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog
(X-linked nuclear protein-1)
Length = 1359
Score = 55.5 bits (132), Expect = 3e-07
Identities = 45/151 (29%), Positives = 64/151 (41%), Gaps = 20/151 (13%)
Query: 321 KGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKK 380
K S E E+ ++ K +K K S +++D++ E E+ S KK+KK
Sbjct: 87 KSESESDESDEEEDRKKSKSKKKVDQKKKEKSKKKRTTSSSEDEDSDEEREQKSKKKSKK 146
Query: 381 KKQ-----------------KAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K+ K+ + +V K AE + ED K KK KK K+K
Sbjct: 147 TKKQTSSESSEESEEERKVKKSKKNKEKSVKKRAETSEESDEDEKPSKKSKKGLKKKAKS 206
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
E E E ED SKKK KK KK++E
Sbjct: 207 ESESES---EDEKEVKKSKKKSKKVVKKESE 234
Score = 51.2 bits (121), Expect = 5e-06
Identities = 38/128 (29%), Positives = 56/128 (43%), Gaps = 5/128 (3%)
Query: 329 KLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKK--KQKAA 386
K R+ D D+ +PRK+ + +S D E E+ K+KKK ++K
Sbjct: 57 KKRKASSSEEDDDDEEESPRKSSKKSRKRAKSESESDESDEEEDRKKSKSKKKVDQKKKE 116
Query: 387 DGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKK 446
++E + D E + KKK KK K++ E E E+ SKK K+
Sbjct: 117 KSKKKRTTSSSEDEDSDEEREQKSKKKSKKTKKQTSSESSEES---EEERKVKKSKKNKE 173
Query: 447 KSKKKDAE 454
KS KK AE
Sbjct: 174 KSVKKRAE 181
Score = 47.0 bits (110), Expect = 1e-04
Identities = 32/124 (25%), Positives = 64/124 (50%), Gaps = 6/124 (4%)
Query: 328 EKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAAD 387
EK ++++E+ + K P+K + A S ++ D E E+ S+KK++K+ + ++
Sbjct: 33 EKRAQKLKEKREREGKP-PPKKRPAKKRKASSSEEDDDDEEESPRKSSKKSRKRAKSESE 91
Query: 388 GDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKK 447
D+ ++ + + + + +K+K KK++ E E D+ E SKKK KK
Sbjct: 92 SDESDEEEDRKKSKSKKKVDQKKKEKSKKKRTTSSSEDEDSDEERE-----QKSKKKSKK 146
Query: 448 SKKK 451
+KK+
Sbjct: 147 TKKQ 150
Score = 45.4 bits (106), Expect = 3e-04
Identities = 37/170 (21%), Positives = 70/170 (40%), Gaps = 14/170 (8%)
Query: 292 RASARNKGRMARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNI 351
+ S +NK + + A + + S+K +K + + E + K +
Sbjct: 166 KKSKKNKEKSVKKRAETSEESDEDEKPSKKSKKGLKKKAKSESESESE-------DEKEV 218
Query: 352 DVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEK 411
K + V K++E E E KKT+K+K+ ++ + + ++ + E K
Sbjct: 219 KKSKKKSKKVVKKESESEDEAPEKKKTEKRKRSKTSSEESSESEKSDEEEEEKESSPKPK 278
Query: 412 KKKKKEKRKLDQEVEVEDKVVE-------DGANADSSKKKKKKSKKKDAE 454
KKK +KL + E E+ VE GA S + +K +K ++E
Sbjct: 279 KKKPLAVKKLSSDEESEESDVEVLPQKKKRGAVTLISDSEDEKDQKSESE 328
Score = 39.7 bits (91), Expect = 0.016
Identities = 35/128 (27%), Positives = 57/128 (44%), Gaps = 10/128 (7%)
Query: 324 SAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQ 383
S+ EK E+ EE+ + P+K + + S D + E + E + KK +
Sbjct: 258 SSESEKSDEEEEEK----ESSPKPKKKKPLAVKKLSS-DEESEESDVEVLPQKKKRGAVT 312
Query: 384 KAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKK 443
+D +D D+ +E D E+ S+KK KK+E + + V N S KK
Sbjct: 313 LISDSEDEK-DQKSESEASDVEEKVSKKKAKKQESSESGSDSSEGSITV----NRKSKKK 367
Query: 444 KKKKSKKK 451
+K + KKK
Sbjct: 368 EKPEKKKK 375
Score = 38.9 bits (89), Expect = 0.028
Identities = 33/122 (27%), Positives = 60/122 (49%), Gaps = 8/122 (6%)
Query: 335 EERLDFYDKGVAPRKNIDVMKSAI-ESVDNKDTEMETE--EVSAKKTKKKKQKAADGDDM 391
+E + D V P+K + I +S D KD + E+E +V K +KKK +K +
Sbjct: 291 DEESEESDVEVLPQKKKRGAVTLISDSEDEKDQKSESEASDVEEKVSKKKAKKQESSESG 350
Query: 392 AVDKAAEITNGDAEDHKSEKKKK-KKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKK 450
+ IT + KS+KK+K +K+K+ + + K D A+ ++K+ + K+
Sbjct: 351 SDSSEGSIT----VNRKSKKKEKPEKKKKGIIMDSSKLQKETIDAERAEKERRKRLEKKQ 406
Query: 451 KD 452
K+
Sbjct: 407 KE 408
Score = 37.0 bits (84), Expect = 0.10
Identities = 26/101 (25%), Positives = 46/101 (44%), Gaps = 9/101 (8%)
Query: 361 VDNKDTEM------ETEEVSAKKTKKKKQKAADGDDM---AVDKAAEITNGDAEDHKSEK 411
++++D EM E +E A+K K+K+++ A + A + D +D +
Sbjct: 16 IEDEDLEMARQIENERKEKRAQKLKEKREREGKPPPKKRPAKKRKASSSEEDDDDEEESP 75
Query: 412 KKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
+K K+ RK + D+ E+ S KKK KKK+
Sbjct: 76 RKSSKKSRKRAKSESESDESDEEEDRKKSKSKKKVDQKKKE 116
>CYL1_BOVIN (P35662) Cylicin I (Multiple-band polypeptide I)
Length = 667
Score = 53.9 bits (128), Expect = 8e-07
Identities = 50/185 (27%), Positives = 79/185 (42%), Gaps = 20/185 (10%)
Query: 277 NTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSE---KGTSAFGEKLREQ 333
N+ GL+ H S + Y N ++ D + KG+ A E
Sbjct: 247 NSSNVGLMVHLGESDAESMEFDMWLKNYSQNNSKKPTKKDAKKDAKGKGSDA------ES 300
Query: 334 VEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAV 393
V+ + DK A + K ES D + + + + K++KK K+K D
Sbjct: 301 VDSKDAKKDKKGATKDTKKGAKKDTESTDAESGDSKDAKKGKKESKKDKKK-----DAKK 355
Query: 394 DKAAEITNGDAEDHKSEKKK----KKKEKRKLDQEVEVEDKVVEDGANADSSK--KKKKK 447
D A++ +GD++D K + KK KK+ +K D + + E E G + D+ K KK KK
Sbjct: 356 DAASDAESGDSKDAKKDSKKGKKDSKKDNKKKDAKKDAESTDAESGDSKDAKKDSKKGKK 415
Query: 448 SKKKD 452
KKD
Sbjct: 416 DSKKD 420
Score = 52.0 bits (123), Expect = 3e-06
Identities = 33/102 (32%), Positives = 52/102 (50%), Gaps = 3/102 (2%)
Query: 356 SAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKK 415
S ES D+KD + ++++ K K+K A D + D + + +D K KK K
Sbjct: 359 SDAESGDSKDAKKDSKKGKKDSKKDNKKKDAKKDAESTDAESGDSKDAKKDSKKGKKDSK 418
Query: 416 KEKRKLDQEVEVEDKVVEDGANADS---SKKKKKKSKKKDAE 454
K+ +K D + + E E G + ++ SKK KK KKKDA+
Sbjct: 419 KDDKKKDAKKDAESTDAESGDSKNAKKDSKKGKKDDKKKDAK 460
Score = 50.1 bits (118), Expect = 1e-05
Identities = 37/117 (31%), Positives = 55/117 (46%), Gaps = 7/117 (5%)
Query: 343 KGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKK-----KQKAADGDDMAVDKAA 397
KG K D K A + ++ D E + +AKK KK K+K A D ++ D +
Sbjct: 412 KGKKDSKKDDKKKDAKKDAESTDAE-SGDSKNAKKDSKKGKKDDKKKDAKKDAVSTDADS 470
Query: 398 EITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
E + GDA+ K + KK KK+ +K DQ+ +D KK K+ KKD +
Sbjct: 471 E-SEGDAKKSKKDSKKDKKDLKKDDQKKPAMKSKESTETESDWESKKVKRDSKKDTK 526
Score = 48.5 bits (114), Expect = 3e-05
Identities = 39/115 (33%), Positives = 56/115 (47%), Gaps = 8/115 (6%)
Query: 343 KGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVD--KAAEIT 400
KG K + K A + ++ D E + AKK KK +K + DD D K AE T
Sbjct: 375 KGKKDSKKDNKKKDAKKDAESTDAE-SGDSKDAKKDSKKGKKDSKKDDKKKDAKKDAEST 433
Query: 401 N---GDAEDHKSEKKKKKKEKRKLDQEVEV--EDKVVEDGANADSSKKKKKKSKK 450
+ GD+++ K + KK KK+ +K D + + D E +A SKK KK KK
Sbjct: 434 DAESGDSKNAKKDSKKGKKDDKKKDAKKDAVSTDADSESEGDAKKSKKDSKKDKK 488
Score = 40.4 bits (93), Expect = 0.009
Identities = 36/116 (31%), Positives = 50/116 (43%), Gaps = 8/116 (6%)
Query: 343 KGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNG 402
KG K D K A+ + + D+E E + +KK KK +K DD K A +
Sbjct: 449 KGKKDDKKKDAKKDAVST--DADSESEGDAKKSKKDSKKDKKDLKKDDQ--KKPAMKSKE 504
Query: 403 DAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANAD-SSKKKKKKS---KKKDAE 454
E + KK K K D + + GA +D SSK+ KK+ K DAE
Sbjct: 505 STETESDWESKKVKRDSKKDTKKTAKKATESSGAESDVSSKRYLKKTEMFKSSDAE 560
>TCOF_HUMAN (Q13428) Treacle protein (Treacher Collins syndrome
protein)
Length = 1411
Score = 52.0 bits (123), Expect = 3e-06
Identities = 33/86 (38%), Positives = 47/86 (54%), Gaps = 10/86 (11%)
Query: 379 KKKKQKAADGDDMAVDKAAE--------ITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDK 430
K+KK K + G A D+ E + GD + KS+K+KKK +KRK D+E + + K
Sbjct: 1323 KEKKGKGSLGSQGAKDEPEEELQKGMGTVEGGDQSNPKSKKEKKKSDKRKKDKEKKEKKK 1382
Query: 431 VVEDGANADSS--KKKKKKSKKKDAE 454
+ + DS +KKKK KKK AE
Sbjct: 1383 KAKKASTKDSESPSQKKKKKKKKTAE 1408
Score = 33.1 bits (74), Expect = 1.5
Identities = 31/134 (23%), Positives = 58/134 (43%), Gaps = 29/134 (21%)
Query: 292 RASARNKGRMARYLANKCSIASRIDCFSEKGTSAFGEK-LREQVEERLDFYDKGVAPRKN 350
+ S +KG+ R A+ D +KG + G + +++ EE L KG+
Sbjct: 1304 KTSTTSKGKAKRDKASG-------DVKEKKGKGSLGSQGAKDEPEEELQ---KGMG---- 1349
Query: 351 IDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSE 410
+E D + + + E+ + K KK K+K +K + +D +S
Sbjct: 1350 ------TVEGGDQSNPKSKKEKKKSDKRKKDKEKK--------EKKKKAKKASTKDSESP 1395
Query: 411 KKKKKKEKRKLDQE 424
+KKKK+K+K ++
Sbjct: 1396 SQKKKKKKKKTAEQ 1409
>MNN4_YEAST (P36044) MNN4 protein
Length = 1178
Score = 50.8 bits (120), Expect = 7e-06
Identities = 36/138 (26%), Positives = 67/138 (48%), Gaps = 7/138 (5%)
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTE---METEEVSAK 376
EK EK +++ EE+ ++ ++ + K E K+ E + EE K
Sbjct: 1041 EKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKQEEEEKKK 1100
Query: 377 KTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGA 436
K +++K+K +G+ M + N D E K+E+++KKK++ K + + E K E+
Sbjct: 1101 KEEEEKKKQEEGEKMKNEDEENKKNEDEEKKKNEEEEKKKQEEKNKKNEDEEKKKQEE-- 1158
Query: 437 NADSSKKKKKKSKKKDAE 454
+ KK +++ KKK E
Sbjct: 1159 --EEKKKNEEEEKKKQEE 1174
Score = 42.4 bits (98), Expect = 0.002
Identities = 30/137 (21%), Positives = 60/137 (42%), Gaps = 4/137 (2%)
Query: 322 GTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKK 381
G F L + + + +DK +E ++ + + EE KK +++
Sbjct: 998 GLDLFAPTLSDVNRKGIQMFDKDPIIVYEDYAYAKLLEERKRREKKKKEEEEKKKKEEEE 1057
Query: 382 KQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKK----EKRKLDQEVEVEDKVVEDGAN 437
K+K + + ++ + + E K E+++KKK EK+K ++E + + + E N
Sbjct: 1058 KKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKQEEEEKKKKEEEEKKKQEEGEKMKN 1117
Query: 438 ADSSKKKKKKSKKKDAE 454
D KK + +KK E
Sbjct: 1118 EDEENKKNEDEEKKKNE 1134
>CYL1_HUMAN (P35663) Cylicin I (Multiple-band polypeptide I)
(Fragment)
Length = 598
Score = 50.4 bits (119), Expect = 9e-06
Identities = 37/112 (33%), Positives = 61/112 (54%), Gaps = 10/112 (8%)
Query: 349 KNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDM-AVDKAAEITNGDAEDH 407
KN D K A + + D+E E E ++K +KK +K + D+ +V E T+ D+E
Sbjct: 339 KNDDKKKDAKKITFSTDSESELESKESQKDEKKDKKDSKTDNKKSVKNDEESTDADSEP- 397
Query: 408 KSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSS--------KKKKKKSKKK 451
K + KK KK+++K ++ + +DK + NA+S+ KK KK SK+K
Sbjct: 398 KGDSKKGKKDEKKGKKDSKKDDKKKDAKKNAESTEMESDLELKKDKKHSKEK 449
Score = 49.7 bits (117), Expect = 2e-05
Identities = 32/106 (30%), Positives = 53/106 (49%), Gaps = 7/106 (6%)
Query: 356 SAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKK-- 413
S ES D+KD + ++++V K K+K D + D + + + +D K +KKK
Sbjct: 243 SDAESEDSKDAKKDSKKVKKNVKKDDKKKDVKKDTESTDAESGDSKDERKDTKKDKKKLK 302
Query: 414 ---KKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKS--KKKDAE 454
KKK+ +K + + E +D N + KK K+ KKKDA+
Sbjct: 303 KDDKKKDTKKYPESTDTESGDAKDARNDSRNLKKASKNDDKKKDAK 348
Score = 44.3 bits (103), Expect = 7e-04
Identities = 31/134 (23%), Positives = 65/134 (48%), Gaps = 6/134 (4%)
Query: 319 SEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKT 378
+E S +K ++V++ + DK +K D + ES D+KD +T++ K
Sbjct: 245 AESEDSKDAKKDSKKVKKNVKKDDKKKDVKK--DTESTDAESGDSKDERKDTKKDKKKLK 302
Query: 379 KKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANA 438
K K+K D ++ + +GDA+D +++ + KK + D++ + + + +
Sbjct: 303 KDDKKK----DTKKYPESTDTESGDAKDARNDSRNLKKASKNDDKKKDAKKITFSTDSES 358
Query: 439 DSSKKKKKKSKKKD 452
+ K+ +K +KKD
Sbjct: 359 ELESKESQKDEKKD 372
Score = 43.9 bits (102), Expect = 9e-04
Identities = 30/105 (28%), Positives = 49/105 (46%), Gaps = 9/105 (8%)
Query: 350 NIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKS 409
N D ++K+ ++ + + K TKK +K++D + +E + +D K
Sbjct: 207 NFDAWLRNYSQNNSKNYSLKYTKYTKKDTKKNAKKSSDAE-------SEDSKDAKKDSKK 259
Query: 410 EKKKKKKEKRKLDQEVEVEDKVVEDGANADSSK--KKKKKSKKKD 452
KK KK+ +K D + + E E G + D K KK KK KKD
Sbjct: 260 VKKNVKKDDKKKDVKKDTESTDAESGDSKDERKDTKKDKKKLKKD 304
Score = 39.7 bits (91), Expect = 0.016
Identities = 30/121 (24%), Positives = 53/121 (43%), Gaps = 4/121 (3%)
Query: 328 EKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAAD 387
E Q +E+ D D +K++ K+ ES D D+E + + KK +KK +K +
Sbjct: 361 ESKESQKDEKKDKKDSKTDNKKSV---KNDEESTD-ADSEPKGDSKKGKKDEKKGKKDSK 416
Query: 388 GDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKK 447
DD D + + E KK KK K K + +++ +D + D+ + K
Sbjct: 417 KDDKKKDAKKNAESTEMESDLELKKDKKHSKEKKGSKKDIKKDARKDTESTDAEFDESSK 476
Query: 448 S 448
+
Sbjct: 477 T 477
Score = 39.7 bits (91), Expect = 0.016
Identities = 32/139 (23%), Positives = 61/139 (43%), Gaps = 7/139 (5%)
Query: 319 SEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKT 378
+E G S K ++ +++L DK +K + + ES D KD ++ +
Sbjct: 282 AESGDSKDERKDTKKDKKKLKKDDKKKDTKKYPE--STDTESGDAKDARNDSRNLKKASK 339
Query: 379 KKKKQKAADGDDMAVDKAAEITNGDAE-DHKSEKKKKKKEKRKL----DQEVEVEDKVVE 433
K+K A + D +E+ + +++ D K +KK K + +K ++ + + +
Sbjct: 340 NDDKKKDAKKITFSTDSESELESKESQKDEKKDKKDSKTDNKKSVKNDEESTDADSEPKG 399
Query: 434 DGANADSSKKKKKKSKKKD 452
D +KK KK KKD
Sbjct: 400 DSKKGKKDEKKGKKDSKKD 418
Score = 37.7 bits (86), Expect = 0.061
Identities = 36/137 (26%), Positives = 57/137 (41%), Gaps = 5/137 (3%)
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTK 379
E+ T A E + + + D KG K D K A ++ ++ TEME++ K K
Sbjct: 388 EESTDADSEPKGDSKKGKKD-EKKGKKDSKKDDKKKDAKKNAES--TEMESDLELKKDKK 444
Query: 380 KKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGA--N 437
K+K D+ D + + DAE +S K K + + E E+ + + GA
Sbjct: 445 HSKEKKGSKKDIKKDARKDTESTDAEFDESSKTGFKTSTKIKGSDTESEESLYKPGAKKK 504
Query: 438 ADSSKKKKKKSKKKDAE 454
D S SK + E
Sbjct: 505 IDESDGTSANSKMEGLE 521
>Y5G8_ENCCU (Q8STA9) Hypothetical protein ECU05_1680/ECU11_0050
Length = 612
Score = 48.1 bits (113), Expect = 5e-05
Identities = 38/138 (27%), Positives = 67/138 (48%), Gaps = 26/138 (18%)
Query: 315 IDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVS 374
+D E G G+ RE V++ L +K ++ + I V N E + E+
Sbjct: 262 LDAHREHG----GDVTRELVKQML-------LGKKGDEIDRRYINKVANVVKERQRREME 310
Query: 375 AKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVED 434
K+ +KKK++ +K E E+ + E+KKKKKE++K +++ + E+K E
Sbjct: 311 KKEEEKKKEE---------EKKKE------EEKRKEEKKKKKEEKKEEKKKKKEEKKEEK 355
Query: 435 GANADSSKKKKKKSKKKD 452
KK++KK +KK+
Sbjct: 356 KEEKKEEKKEEKKEEKKE 373
Score = 42.7 bits (99), Expect = 0.002
Identities = 29/99 (29%), Positives = 46/99 (46%), Gaps = 6/99 (6%)
Query: 359 ESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEK 418
E K+ E EE KK +KK++K ++ +K E E+ K EKK++KKE+
Sbjct: 319 EEEKKKEEEKRKEEKKKKKEEKKEEKKKKKEEKKEEKKEEKKEEKKEEKKEEKKEEKKEE 378
Query: 419 ------RKLDQEVEVEDKVVEDGANADSSKKKKKKSKKK 451
R+ + E E VE G + KK + +K+
Sbjct: 379 KSGKSLREGEASEEAEMPSVEVGGARRKTGKKSEGGRKR 417
>CYL2_HUMAN (Q14093) Cylicin II (Multiple-band polypeptide II)
Length = 348
Score = 48.1 bits (113), Expect = 5e-05
Identities = 31/107 (28%), Positives = 53/107 (48%), Gaps = 4/107 (3%)
Query: 348 RKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDH 407
+K+ D K A + ++KD + + +E+ KK KK K+K + D + D + + DA
Sbjct: 242 KKDEDGKKDANKGDESKDAKKDAKEI--KKGKKDKKKPSSTDSDSKDDVKKESKKDATKD 299
Query: 408 KSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+ KK EK D + + + +D +KK KK +KKDA+
Sbjct: 300 AKKVAKKDTEKESADSKKDAKKNAKKDAKK--DAKKNAKKDEKKDAK 344
Score = 43.1 bits (100), Expect = 0.001
Identities = 36/117 (30%), Positives = 53/117 (44%), Gaps = 14/117 (11%)
Query: 352 DVMKSAIESVDNKDTEMETEEVSA--KKTKKKKQKAADGDDM--AVDKAAEITNG----- 402
D K +S KD+ +E + V A KK + K+ A GD+ A A EI G
Sbjct: 216 DSKKGKKDSKKGKDSAIELQAVKADEKKDEDGKKDANKGDESKDAKKDAKEIKKGKKDKK 275
Query: 403 -----DAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
D++ KK+ KK+ K ++V +D E + +KK KK KKDA+
Sbjct: 276 KPSSTDSDSKDDVKKESKKDATKDAKKVAKKDTEKESADSKKDAKKNAKKDAKKDAK 332
Score = 43.1 bits (100), Expect = 0.001
Identities = 36/97 (37%), Positives = 47/97 (48%), Gaps = 8/97 (8%)
Query: 359 ESVDNKDTEMETEE-VSAKKTKKKKQKAAD-GDDMAVDKAAEITNGDAEDHKSEKKKKKK 416
+S KD E EE + AKK KK +K A+ G D A + E G +D+K +KK K
Sbjct: 140 DSKKGKDIEKGKEEKLDAKKDSKKGKKDAEKGKDSATESEDE-KGGAKKDNKKDKKDSNK 198
Query: 417 EKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDA 453
K D E E + + G DS K KK K KD+
Sbjct: 199 GK---DSATESEGE--KGGTEKDSKKGKKDSKKGKDS 230
Score = 42.7 bits (99), Expect = 0.002
Identities = 41/134 (30%), Positives = 60/134 (44%), Gaps = 21/134 (15%)
Query: 321 KGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKK 380
K S G+ + + EE+LD +K+ K E + TE E E+ AKK K
Sbjct: 138 KKDSKKGKDIEKGKEEKLD-------AKKDSKKGKKDAEKGKDSATESEDEKGGAKKDNK 190
Query: 381 KKQKAAD-GDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANAD 439
K +K ++ G D A + E G E K KK KK K+ D +E++ A
Sbjct: 191 KDKKDSNKGKDSATESEGE--KGGTE--KDSKKGKKDSKKGKDSAIELQ---------AV 237
Query: 440 SSKKKKKKSKKKDA 453
+ +KK + KKDA
Sbjct: 238 KADEKKDEDGKKDA 251
Score = 40.4 bits (93), Expect = 0.009
Identities = 43/140 (30%), Positives = 69/140 (48%), Gaps = 9/140 (6%)
Query: 319 SEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKT 378
S+KG EK ++ E D +KG A + N K + + D+ TE E E+ +K
Sbjct: 161 SKKGKKD-AEKGKDSATESED--EKGGAKKDNKKDKKDSNKGKDSA-TESEGEKGGTEKD 216
Query: 379 KKK-KQKAADGDDMAVD-KAAEITNGDAEDHKSEKKK--KKKEKRKLDQEVEVEDKVVED 434
KK K+ + G D A++ +A + ED K + K + K+ +K +E++ K +
Sbjct: 217 SKKGKKDSKKGKDSAIELQAVKADEKKDEDGKKDANKGDESKDAKKDAKEIKKGKKDKKK 276
Query: 435 GANADS-SKKKKKKSKKKDA 453
++ DS SK KK KKDA
Sbjct: 277 PSSTDSDSKDDVKKESKKDA 296
Score = 30.8 bits (68), Expect = 7.5
Identities = 25/77 (32%), Positives = 35/77 (44%), Gaps = 5/77 (6%)
Query: 377 KTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGA 436
+T + KAA+ DK + D+E K+ KK K+ D E E+K+ A
Sbjct: 103 RTVEVDSKAAEIGKKGEDKTTQKDTTDSESEL--KQGKKDSKKGKDIEKGKEEKL---DA 157
Query: 437 NADSSKKKKKKSKKKDA 453
DS K KK K KD+
Sbjct: 158 KKDSKKGKKDAEKGKDS 174
>IF2B_SCHPO (P56329) Probable eukaryotic translation initiation
factor 2 beta subunit (eIF-2-beta)
Length = 321
Score = 47.8 bits (112), Expect = 6e-05
Identities = 44/132 (33%), Positives = 57/132 (42%), Gaps = 19/132 (14%)
Query: 332 EQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDM 391
E V +++D G P K K +SV D E + VS + A D DD
Sbjct: 5 EAVVDQIDL--NGALPEKK----KKPKKSVAFDD---EVDAVSKDGPEASSAGATDEDDS 55
Query: 392 -----AVDKAAEITNGDAEDHK----SEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSK 442
A D +T G+ ++ K S KKKKK +K E + ED E G D S
Sbjct: 56 ERKSSAKDLTTPVTEGEVDELKDMFSSMKKKKKSKKSSASAEEQTEDITTESG-ELDFSS 114
Query: 443 KKKKKSKKKDAE 454
KKKK KKK A+
Sbjct: 115 MKKKKKKKKSAD 126
Score = 38.5 bits (88), Expect = 0.036
Identities = 29/99 (29%), Positives = 48/99 (48%), Gaps = 11/99 (11%)
Query: 355 KSAIESVDNKDTEMETEEV-----SAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKS 409
KS+ + + TE E +E+ S KK KK K+ +A ++ D IT E S
Sbjct: 58 KSSAKDLTTPVTEGEVDELKDMFSSMKKKKKSKKSSASAEEQTED----ITTESGELDFS 113
Query: 410 EKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKS 448
KKKKK+K+ D + +K +E + D++ K++
Sbjct: 114 SMKKKKKKKKSAD--LSAFEKELEASSTGDATSDLSKQT 150
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.130 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 49,451,192
Number of Sequences: 164201
Number of extensions: 2118759
Number of successful extensions: 18554
Number of sequences better than 10.0: 870
Number of HSP's better than 10.0 without gapping: 279
Number of HSP's successfully gapped in prelim test: 615
Number of HSP's that attempted gapping in prelim test: 11567
Number of HSP's gapped (non-prelim): 3722
length of query: 454
length of database: 59,974,054
effective HSP length: 114
effective length of query: 340
effective length of database: 41,255,140
effective search space: 14026747600
effective search space used: 14026747600
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC135505.8


