

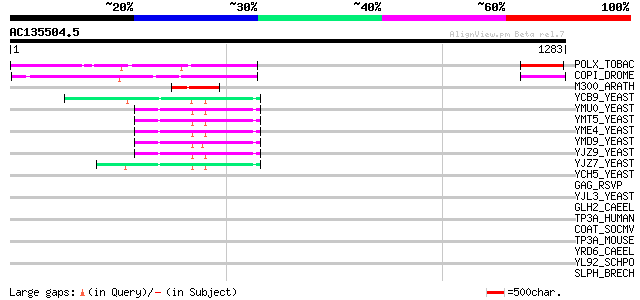
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135504.5 - phase: 0 /pseudo
(1283 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 350 2e-95
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 196 3e-49
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 97 2e-19
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 68 1e-10
YMU0_YEAST (Q04670) Transposon Ty1 protein B 62 8e-09
YMT5_YEAST (Q04214) Transposon Ty1 protein B 62 8e-09
YME4_YEAST (Q04711) Transposon Ty1 protein B 62 8e-09
YMD9_YEAST (Q03434) Transposon Ty1 protein B 62 8e-09
YJZ9_YEAST (P47100) Transposon Ty1 protein B 62 8e-09
YJZ7_YEAST (P47098) Transposon Ty1 protein B 62 1e-08
YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein 44 0.003
GAG_RSVP (P03322) Gag polyprotein [Contains: Core protein p19; C... 39 0.070
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 39 0.091
GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-... 38 0.16
TP3A_HUMAN (Q13472) DNA topoisomerase III alpha (EC 5.99.1.2) 37 0.26
COAT_SOCMV (P15627) Coat protein 37 0.26
TP3A_MOUSE (O70157) DNA topoisomerase III alpha (EC 5.99.1.2) 37 0.35
YRD6_CAEEL (Q09575) Hypothetical protein K02A2.6 in chromosome II 37 0.45
YL92_SCHPO (Q9HFF2) Hypothetical protein C683.02c in chromosome I 37 0.45
SLPH_BRECH (P38538) Surface layer protein precursor (Hexagonal w... 37 0.45
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 350 bits (897), Expect = 2e-95
Identities = 211/594 (35%), Positives = 318/594 (53%), Gaps = 41/594 (6%)
Query: 3 GSKWDIEKFTGSNLFGLWKVKMRAILIQEKCVEALKREAQMSAHLTPAEKTEMNDKAVSA 62
G K+++ KF G N F W+ +MR +LIQ+ + L +++ + + +++++A SA
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 63 IILCLGDKMLREVSRETTAVSMWNKLDLLYMTKSLAHRQCLKQQLYFYRMMESKPIMEQL 122
I L L D ++ + E TA +W +L+ LYM+K+L ++ LK+QLY M E + L
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 123 TEFNKIIDDLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQATLIT 182
FN +I LAN+ V +E+EDKA+ LL +LP S++N T+L+GK TI L++V + L+
Sbjct: 123 NVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKT-TIELKDVTSALLL 181
Query: 183 KELTKFKDLKVDDSGEGL-NVSRGRNQNRGKGKGKNSKSKSRSKGDGNKTKYKCFICHNP 241
E + K ++ G+ L RGR+ R S ++ +SK C+ C+ P
Sbjct: 182 NEKMRKKP---ENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSKSRVRNCYNCNQP 238
Query: 242 GHFKKDCPERKDNGG----------------GNPSVQL-ASKDEGCESAGALTVTSWEPE 284
GHFK+DCP + G N +V L +++E C PE
Sbjct: 239 GHFKRDCPNPRKGKGETSGQKNDDNTAAMVQNNDNVVLFINEEEECMHLSG-------PE 291
Query: 285 KGWVLDSGCSYHISPRKGYFETLELEEGGVVRLGNNKACKIQGIGTIRLKMFDDRDFLLK 344
WV+D+ S+H +P + F + G V++GN KI GIG I +K +LK
Sbjct: 292 SEWVVDTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVGCTLVLK 351
Query: 345 DVRYIPELRRNLISISMFDGLGYCTRIERGVMRISHGALVIAKGSKIHGLY-----ILEG 399
DVR++P+LR NLIS D GY + R++ G+LVIAKG LY I +G
Sbjct: 352 DVRHVPDLRMNLISGIALDRDGYESYFANQKWRLTKGSLVIAKGVARGTLYRTNAEICQG 411
Query: 400 STIIAHASVPSVDTLDITKLWHLRLGHVSERGLVELAKQGLLGNEKLNKLDFCDNCTLGK 459
A + SVD LWH R+GH+SE+GL LAK+ L+ K + CD C GK
Sbjct: 412 ELNAAQDEI-SVD------LWHKRMGHMSEKGLQILAKKSLISYAKGTTVKPCDYCLFGK 464
Query: 460 QHKVKFGVGVHKSSRPFE*VHSDLLGPA*VKTYGGGSYFTSIIDDYSRRVWVYILKNKSD 519
QH+V F + + V+SD+ GP +++ GG YF + IDD SR++WVYILK K
Sbjct: 465 QHRVSFQTSSERKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTKDQ 524
Query: 520 AFEKFKEWDILVENQIGTKLKVLRTDNGQEFVLEQFNEFCRKKGIKRHRIVAYT 573
F+ F+++ LVE + G KLK LR+DNG E+ +F E+C GI+ + V T
Sbjct: 525 VFQVFQKFHALVERETGRKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGT 578
Score = 103 bits (257), Expect = 3e-21
Identities = 44/100 (44%), Positives = 72/100 (72%)
Query: 1181 YIVWYNSNLEAIWLKGMIGEMGISQGCVKIHCDSQSAIHLANHQIYHERTKHINIRLHFV 1240
YI + E IWLK + E+G+ Q ++CDSQSAI L+ + +YH RTKHI++R H++
Sbjct: 1224 YIAATETGKEMIWLKRFLQELGLHQKEYVVYCDSQSAIDLSKNSMYHARTKHIDVRYHWI 1283
Query: 1241 RDMIETKEIKVEKVASEENPADIFTKSLPRSRFKHCLDLI 1280
R+M++ + +KV K+++ ENPAD+ TK +PR++F+ C +L+
Sbjct: 1284 REMVDDESLKVLKISTNENPADMLTKVVPRNKFELCKELV 1323
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 196 bits (499), Expect = 3e-49
Identities = 165/593 (27%), Positives = 270/593 (44%), Gaps = 41/593 (6%)
Query: 4 SKWDIEKFTGSNLFGLWKVKMRAILIQEKCVEALKREAQMSAHLTPAEKTEMNDKAV--- 60
+K +I+ F G + +WK ++RA+L ++ ++ + L P E + KA
Sbjct: 4 AKRNIKPFDGEK-YAIWKFRIRALLAEQDVLKVVDG-------LMPNEVDDSWKKAERCA 55
Query: 61 -SAIILCLGDKMLREVSRETTAVSMWNKLDLLYMTKSLAHRQCLKQQLYFYRMMESKPIM 119
S II L D L + + TA + LD +Y KSLA + L+++L ++ ++
Sbjct: 56 KSTIIEYLSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLL 115
Query: 120 EQLTEFNKIIDDLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAT 179
F+++I +L +E+ DK HLL LP ++ + E +TL V+
Sbjct: 116 SHFHIFDELISELLAAGAKIEEMDKISHLLITLPSCYDGIITAIETLSEENLTLAFVKNR 175
Query: 180 LITKELTKFKDLKVDDSGEGLNVSRGRNQNRGKG---KGKNSKSKSRSKGDGNKTKYKCF 236
L+ +E+ K K+ D S + +N N N K K + +K K KG+ +K K KC
Sbjct: 176 LLDQEI-KIKNDHNDTSKKVMNAIVHNNNNTYKNNLFKNRVTKPKKIFKGN-SKYKVKCH 233
Query: 237 ICHNPGHFKKDCPERK-----DNGGGNPSVQLASKDEGCESAGALTVTSWEPEKGWVLDS 291
C GH KKDC K N VQ A+ + TS G+VLDS
Sbjct: 234 HCGREGHIKKDCFHYKRILNNKNKENEKQVQTATSHGIAFMVKEVNNTSVMDNCGFVLDS 293
Query: 292 GCSYH-ISPRKGYFETLELEEG---GVVRLGNNKACKIQGIGTIRLKMFDDRDFLLKDVR 347
G S H I+ Y +++E+ V + G +GI +R +D + L+DV
Sbjct: 294 GASDHLINDESLYTDSVEVVPPLKIAVAKQGEFIYATKRGIVRLR----NDHEITLEDVL 349
Query: 348 YIPELRRNLISISMFDGLGYCTRIERGVMRISHGALVIAKGSKIHGLYILEGSTIIAHAS 407
+ E NL+S+ G ++ + IS L++ K S + + I A
Sbjct: 350 FCKEAAGNLMSVKRLQEAGMSIEFDKSGVTISKNGLMVVKNSGM----LNNVPVINFQAY 405
Query: 408 VPSVDTLDITKLWHLRLGHVSERGLVELAKQGLLGNEKL-NKLDF----CDNCTLGKQHK 462
+ + +LWH R GH+S+ L+E+ ++ + ++ L N L+ C+ C GKQ +
Sbjct: 406 SINAKHKNNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEICEPCLNGKQAR 465
Query: 463 VKFGVGVHKS--SRPFE*VHSDLLGPA*VKTYGGGSYFTSIIDDYSRRVWVYILKNKSDA 520
+ F K+ RP VHSD+ GP T +YF +D ++ Y++K KSD
Sbjct: 466 LPFKQLKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKYKSDV 525
Query: 521 FEKFKEWDILVENQIGTKLKVLRTDNGQEFVLEQFNEFCRKKGIKRHRIVAYT 573
F F+++ E K+ L DNG+E++ + +FC KKGI H V +T
Sbjct: 526 FSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGISYHLTVPHT 578
Score = 75.9 bits (185), Expect = 7e-13
Identities = 37/104 (35%), Positives = 62/104 (59%), Gaps = 1/104 (0%)
Query: 1181 YIVWYNSNLEAIWLKGMIGEMGIS-QGCVKIHCDSQSAIHLANHQIYHERTKHINIRLHF 1239
Y+ + + EA+WLK ++ + I + +KI+ D+Q I +AN+ H+R KHI+I+ HF
Sbjct: 1298 YMALFEAVREALWLKFLLTSINIKLENPIKIYEDNQGCISIANNPSCHKRAKHIDIKYHF 1357
Query: 1240 VRDMIETKEIKVEKVASEENPADIFTKSLPRSRFKHCLDLINFI 1283
R+ ++ I +E + +E ADIFTK LP +RF D + +
Sbjct: 1358 AREQVQNNVICLEYIPTENQLADIFTKPLPAARFVELRDKLGLL 1401
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 97.4 bits (241), Expect = 2e-19
Identities = 46/112 (41%), Positives = 68/112 (60%), Gaps = 1/112 (0%)
Query: 374 GVMRISHGALVIAKGSKIHGLYILEGSTIIAHASVPSVDTLDITKLWHLRLGHVSERGLV 433
GV+++ G I KG++ LYIL+GS +++ D T+LWH RL H+S+RG+
Sbjct: 27 GVLKVLKGCRTILKGNRHDSLYILQGSVETGESNLAET-AKDETRLWHSRLAHMSQRGME 85
Query: 434 ELAKQGLLGNEKLNKLDFCDNCTLGKQHKVKFGVGVHKSSRPFE*VHSDLLG 485
L K+G L + K++ L FC++C GK H+V F G H + P + VHSDL G
Sbjct: 86 LLVKKGFLDSSKVSSLKFCEDCIYGKTHRVNFSTGQHTTKNPLDYVHSDLWG 137
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 68.2 bits (165), Expect = 1e-10
Identities = 104/482 (21%), Positives = 186/482 (38%), Gaps = 40/482 (8%)
Query: 128 IIDDLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQATLITKELTK 187
II L ++N+ D +L L F+ ++ Y + + L ++ A I +
Sbjct: 289 IIQRLKENNINVSDRLACQLILKGLSGDFKYLRNQ--YRTKTNMKLSQLFAE-IQLIYDE 345
Query: 188 FKDLKVDDSGEGLNVSRGRNQNRGKGKGKNSKSKSRSKGDGNKTKYKCFICHNPGHFKKD 247
K + ++ + S +N +R N+K +R+ N +K + HN K
Sbjct: 346 NKIMNLNKPSQYKQHSEYKNVSRTSPNTTNTKVTTRNYQRTNSSKPRAAKAHNIATSSKF 405
Query: 248 CPERKDN-GGGNPSVQLASKDEGC-------ESAGALTVTSWEPEKGWVL-DSGCSYHIS 298
D+ S Q S D ES T+ S + +L DSG S +
Sbjct: 406 SRVNNDHINESTVSSQYLSDDNELSLGQQQKESKPTHTIDSNDELPDHLLIDSGASQTLV 465
Query: 299 PRKGYFETLELEEGGVVRLGNNKACKIQGIGTIRLKMFDDRDFLLKDVRYIPELRRNLIS 358
Y + + I IG + + +K + + P + +L+S
Sbjct: 466 RSAHYLHHATPNSEINIVDAQKQDIPINAIGNLHFNFQNGTKTSIKAL-HTPNIAYDLLS 524
Query: 359 ISMFDGLGYCTRIERGVMRISHGALVIAKGSKIHGLYILEGSTII-AHASVPSVDTLDIT 417
+S R + S G V+A K Y L +I +H S +++ ++ +
Sbjct: 525 LSELANQNITACFTRNTLERSDGT-VLAPIVKHGDFYWLSKKYLIPSHISKLTINNVNKS 583
Query: 418 K--------LWHLRLGHVSERGLVELAKQGLLGNEKLNKLDF-------CDNCTLGKQHK 462
K L H LGH + R + + K+ + K + +++ C +C +GK K
Sbjct: 584 KSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYLKESDIEWSNASTYQCPDCLIGKSTK 643
Query: 463 VKFGVG----VHKSSRPFE*VHSDLLGPA*VKTYGGGSYFTSIIDDYSRRVWVYIL--KN 516
+ G +S PF+ +H+D+ GP SYF S D+ +R WVY L +
Sbjct: 644 HRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKSAPSYFISFTDEKTRFQWVYPLHDRR 703
Query: 517 KSDAFEKFKEWDILVENQIGTKLKVLRTDNGQEFVLEQFNEFCRKKGIKRHRIVAYTSTE 576
+ F ++NQ ++ V++ D G E+ + ++F +GI YT+T
Sbjct: 704 EESILNVFTSILAFIKNQFNARVLVIQMDRGSEYTNKTLHKFFTNRGI----TACYTTTA 759
Query: 577 RS 578
S
Sbjct: 760 DS 761
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 62.4 bits (150), Expect = 8e-09
Identities = 69/313 (22%), Positives = 128/313 (40%), Gaps = 28/313 (8%)
Query: 288 VLDSGCSYHISPRKGYFETLELEEGGVVRLGNNKACKIQGIGTIRLKMFDDRDFLLKDVR 347
+LDSG S + + + V + I IG ++ D+ +K V
Sbjct: 32 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIK-VL 90
Query: 348 YIPELRRNLISISMFDGLGYCTRIERGVMRISHGALVIAKGSKIHGLYILEGSTII-AHA 406
+ P + +L+S++ + + V+ S G V+A K Y + ++ ++
Sbjct: 91 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPSNI 149
Query: 407 SVPSVD---TLDITKLW-----HLRLGHVSERGLVELAKQGLLGNEKLNKLDF------- 451
SVP+++ T + T+ + H L H + + + K + + +D+
Sbjct: 150 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQ 209
Query: 452 CDNCTLGKQHKVKFGVG----VHKSSRPFE*VHSDLLGPA*VKTYGGGSYFTSIIDDYSR 507
C +C +GK K + G S PF+ +H+D+ GP SYF S D+ ++
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 508 RVWVYIL--KNKSDAFEKFKEWDILVENQIGTKLKVLRTDNGQEFVLEQFNEFCRKKGIK 565
WVY L + + + F ++NQ + V++ D G E+ ++F K GI
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGI- 328
Query: 566 RHRIVAYTSTERS 578
YT+T S
Sbjct: 329 ---TPCYTTTADS 338
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 62.4 bits (150), Expect = 8e-09
Identities = 69/313 (22%), Positives = 128/313 (40%), Gaps = 28/313 (8%)
Query: 288 VLDSGCSYHISPRKGYFETLELEEGGVVRLGNNKACKIQGIGTIRLKMFDDRDFLLKDVR 347
+LDSG S + + + V + I IG ++ D+ +K V
Sbjct: 32 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIK-VL 90
Query: 348 YIPELRRNLISISMFDGLGYCTRIERGVMRISHGALVIAKGSKIHGLYILEGSTII-AHA 406
+ P + +L+S++ + + V+ S G V+A K Y + ++ ++
Sbjct: 91 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPSNI 149
Query: 407 SVPSVD---TLDITKLW-----HLRLGHVSERGLVELAKQGLLGNEKLNKLDF------- 451
SVP+++ T + T+ + H L H + + + K + + +D+
Sbjct: 150 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQ 209
Query: 452 CDNCTLGKQHKVKFGVG----VHKSSRPFE*VHSDLLGPA*VKTYGGGSYFTSIIDDYSR 507
C +C +GK K + G S PF+ +H+D+ GP SYF S D+ ++
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 508 RVWVYIL--KNKSDAFEKFKEWDILVENQIGTKLKVLRTDNGQEFVLEQFNEFCRKKGIK 565
WVY L + + + F ++NQ + V++ D G E+ ++F K GI
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGI- 328
Query: 566 RHRIVAYTSTERS 578
YT+T S
Sbjct: 329 ---TPCYTTTADS 338
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 62.4 bits (150), Expect = 8e-09
Identities = 69/313 (22%), Positives = 128/313 (40%), Gaps = 28/313 (8%)
Query: 288 VLDSGCSYHISPRKGYFETLELEEGGVVRLGNNKACKIQGIGTIRLKMFDDRDFLLKDVR 347
+LDSG S + + + V + I IG ++ D+ +K V
Sbjct: 32 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIK-VL 90
Query: 348 YIPELRRNLISISMFDGLGYCTRIERGVMRISHGALVIAKGSKIHGLYILEGSTII-AHA 406
+ P + +L+S++ + + V+ S G V+A K Y + ++ ++
Sbjct: 91 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPSNI 149
Query: 407 SVPSVD---TLDITKLW-----HLRLGHVSERGLVELAKQGLLGNEKLNKLDF------- 451
SVP+++ T + T+ + H L H + + + K + + +D+
Sbjct: 150 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQ 209
Query: 452 CDNCTLGKQHKVKFGVG----VHKSSRPFE*VHSDLLGPA*VKTYGGGSYFTSIIDDYSR 507
C +C +GK K + G S PF+ +H+D+ GP SYF S D+ ++
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 508 RVWVYIL--KNKSDAFEKFKEWDILVENQIGTKLKVLRTDNGQEFVLEQFNEFCRKKGIK 565
WVY L + + + F ++NQ + V++ D G E+ ++F K GI
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGI- 328
Query: 566 RHRIVAYTSTERS 578
YT+T S
Sbjct: 329 ---TPCYTTTADS 338
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 62.4 bits (150), Expect = 8e-09
Identities = 69/313 (22%), Positives = 130/313 (41%), Gaps = 28/313 (8%)
Query: 288 VLDSGCSYHISPRKGYFETLELEEGGVVRLGNNKACKIQGIGTIRLKMFDDRDFLLKDVR 347
+LDSG S + + + V + I IG ++ D+ +K V
Sbjct: 32 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIK-VL 90
Query: 348 YIPELRRNLISISMFDGLGYCTRIERGVMRISHGALVIAKGSKIHGLYILEGSTII-AHA 406
+ P + +L+S++ + + V+ S G V+A K Y + ++ ++
Sbjct: 91 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPSNI 149
Query: 407 SVPSVD---TLDITKLW-----HLRLGHVSERGLVELAKQGLLGN------EKLNKLDF- 451
SVP+++ T + T+ + H L H + + + K + ++ + +D+
Sbjct: 150 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDRSSAIDYQ 209
Query: 452 CDNCTLGKQHKVKFGVG----VHKSSRPFE*VHSDLLGPA*VKTYGGGSYFTSIIDDYSR 507
C +C +GK K + G S PF+ +H+D+ GP SYF S D+ ++
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 508 RVWVYIL--KNKSDAFEKFKEWDILVENQIGTKLKVLRTDNGQEFVLEQFNEFCRKKGIK 565
WVY L + + + F ++NQ + V++ D G E+ ++F K GI
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGI- 328
Query: 566 RHRIVAYTSTERS 578
YT+T S
Sbjct: 329 ---TPCYTTTADS 338
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 62.4 bits (150), Expect = 8e-09
Identities = 69/313 (22%), Positives = 128/313 (40%), Gaps = 28/313 (8%)
Query: 288 VLDSGCSYHISPRKGYFETLELEEGGVVRLGNNKACKIQGIGTIRLKMFDDRDFLLKDVR 347
+LDSG S + + + V + I IG ++ D+ +K V
Sbjct: 459 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIK-VL 517
Query: 348 YIPELRRNLISISMFDGLGYCTRIERGVMRISHGALVIAKGSKIHGLYILEGSTII-AHA 406
+ P + +L+S++ + + V+ S G V+A K Y + ++ ++
Sbjct: 518 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPSNI 576
Query: 407 SVPSVD---TLDITKLW-----HLRLGHVSERGLVELAKQGLLGNEKLNKLDF------- 451
SVP+++ T + T+ + H L H + + + K + + +D+
Sbjct: 577 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQ 636
Query: 452 CDNCTLGKQHKVKFGVG----VHKSSRPFE*VHSDLLGPA*VKTYGGGSYFTSIIDDYSR 507
C +C +GK K + G S PF+ +H+D+ GP SYF S D+ ++
Sbjct: 637 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 696
Query: 508 RVWVYIL--KNKSDAFEKFKEWDILVENQIGTKLKVLRTDNGQEFVLEQFNEFCRKKGIK 565
WVY L + + + F ++NQ + V++ D G E+ ++F K GI
Sbjct: 697 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGI- 755
Query: 566 RHRIVAYTSTERS 578
YT+T S
Sbjct: 756 ---TPCYTTTADS 765
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 61.6 bits (148), Expect = 1e-08
Identities = 86/408 (21%), Positives = 159/408 (38%), Gaps = 35/408 (8%)
Query: 201 NVSRGRNQNRGKGKGKNSKSKSRSKGDGNKTKYKCFICHNPGHFKKDCPERKDNGGGNPS 260
N+S +N +R K +R+ N +K K HN D+ + +
Sbjct: 363 NLSDEKNDSRSYTNTTKPKVIARNPQKTNNSKSKTARAHNVSTSNNSPSTDNDSISKSTT 422
Query: 261 --VQLASK------DEGCESAGALTVTSWEPEKGWVL-DSGCSYHISPRKGYFETLELEE 311
+QL +K E ES T S + G +L DSG S + + +
Sbjct: 423 EPIQLNNKHDLTLGQELTESTVNHTNHSDDELPGHLLLDSGASRTLIRSAHHIHSASSNP 482
Query: 312 GGVVRLGNNKACKIQGIGTIRLKMFDDRDFLLKDVRYIPELRRNLISISMFDGLGYCTRI 371
V + I IG ++ D+ +K V + P + +L+S++ +
Sbjct: 483 DINVVDAQKRNIPINAIGDLQFHFQDNTKTSIK-VLHTPNIAYDLLSLNELAAVDITACF 541
Query: 372 ERGVMRISHGALVIAKGSKIHGLYILEGSTIIAHASVPSVD---TLDITKLW-----HLR 423
+ V+ S G ++ ++ + + ++ SVP+++ T + T+ + H
Sbjct: 542 TKNVLERSDGTVLAPIVQYGDFYWVSKRYLLPSNISVPTINNVHTSESTRKYPYPFIHRM 601
Query: 424 LGHVSERGLVELAKQGLLGNEKLNKLDF-------CDNCTLGKQHKVKFGVG----VHKS 472
L H + + + K + + +D+ C +C +GK K + G S
Sbjct: 602 LAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNS 661
Query: 473 SRPFE*VHSDLLGPA*VKTYGGGSYFTSIIDDYSRRVWVYIL--KNKSDAFEKFKEWDIL 530
PF+ +H+D+ GP SYF S D+ ++ WVY L + + + F
Sbjct: 662 YEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAF 721
Query: 531 VENQIGTKLKVLRTDNGQEFVLEQFNEFCRKKGIKRHRIVAYTSTERS 578
++NQ + V++ D G E+ ++F K GI YT+T S
Sbjct: 722 IKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGI----TPCYTTTADS 765
>YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein
Length = 146
Score = 43.9 bits (102), Expect = 0.003
Identities = 30/121 (24%), Positives = 49/121 (39%), Gaps = 15/121 (12%)
Query: 249 PERKDNGGGNPSVQLASKDEGCESAGALTVTSW----------EPEKGWVLDSGCSYHIS 298
P K + + SV + + ++AG +T SW W+ D+GC+ H+
Sbjct: 31 PNDKTSRSSSASVAIPDYETQGQTAGQITPKSWLCMLSSTVPATKSSEWIFDTGCTSHMC 90
Query: 299 PRKGYFETLELEEGGVVRLGNNKACKIQGIGTIRLKMFDDRDFLLKDVRYIPELRRNLIS 358
+ F + G + I G GT+ + L DV Y+P+L NLIS
Sbjct: 91 HDRSIFSSFTRSSRKDFVRGVGGSIPIMGSGTVNIGTVQ-----LHDVSYVPDLPVNLIS 145
Query: 359 I 359
+
Sbjct: 146 V 146
>GAG_RSVP (P03322) Gag polyprotein [Contains: Core protein p19; Core
protein p2A; Core protein p2B; Core protein p10; Capsid
protein p27; Inner coat protein p12; Protease p15 (EC
3.4.23.-)]
Length = 701
Score = 39.3 bits (90), Expect = 0.070
Identities = 12/48 (25%), Positives = 26/48 (54%)
Query: 218 SKSKSRSKGDGNKTKYKCFICHNPGHFKKDCPERKDNGGGNPSVQLAS 265
++ + G G + + C+ C +PGH++ CP+++ +G QL +
Sbjct: 492 NRERDGQTGSGGRARGLCYTCGSPGHYQAQCPKKRKSGNSRERCQLCN 539
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 38.9 bits (89), Expect = 0.091
Identities = 20/68 (29%), Positives = 35/68 (51%)
Query: 1213 DSQSAIHLANHQIYHERTKHINIRLHFVRDMIETKEIKVEKVASEENPADIFTKSLPRSR 1272
DS+ AI N + K I+ +++ I+ K IK+ K+ + N AD+ TK + S
Sbjct: 1721 DSKPAIQGLNRSYQQPKEKFTWIKTEIIKEKIKEKSIKLLKITGKGNIADLLTKPVSASD 1780
Query: 1273 FKHCLDLI 1280
FK + ++
Sbjct: 1781 FKRFIQVL 1788
Score = 38.5 bits (88), Expect = 0.12
Identities = 68/323 (21%), Positives = 118/323 (36%), Gaps = 50/323 (15%)
Query: 288 VLDSGCSYHISPRKGYFETLELEEGGV--VRLGNNKACKIQGIGTIRLKM-FDDRDFLLK 344
++D+G +I+ K E +G N + ++G G I++K ++ D
Sbjct: 413 IIDTGSGVNITNDKTLLHNYEDSNRSTRFFGIGKNSSVSVKGYGYIKIKNGHNNTDNKCL 472
Query: 345 DVRYIPELRRNLISI------------SMFDGLG-----YCTRIERGVMRISHGALVI-- 385
Y+PE +IS + LG T+I GV+ + L+
Sbjct: 473 LTYYVPEEESTIISCYDLAKKTKMVLSRKYTRLGNKIIKIKTKIVNGVIHVKMNELIERP 532
Query: 386 AKGSKIHGLYILEGSTI-IAHASVPSVDTLDITKLWHLRLGHVSERGLVELAKQGLLGNE 444
+ SKI+ + + S+ D H R+GH ++ + + N
Sbjct: 533 SDDSKINAIKPTSSPGFKLNKRSITLEDA-------HKRMGHTG----IQQIENSIKHNH 581
Query: 445 KLNKLD--------FCDNCTLGKQHKVKFGVG-VHKSSRPFE*VHS---DLLGPA*VKTY 492
LD +C C + K K G ++ S E S D+ GP
Sbjct: 582 YEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNHSTDHEPGSSWCMDIFGPVSSSNA 641
Query: 493 GGGSYFTSIIDDYSRRVWV--YILKNKSDAFEKFKEWDILVENQIGTKLKVLRTDNGQEF 550
Y ++D+ +R + KN + ++ VE Q K++ + +D G EF
Sbjct: 642 DTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQYVETQFDRKVREINSDRGTEF 701
Query: 551 VLEQFNEFCRKKGIKRHRIVAYT 573
+Q E+ KGI H I+ T
Sbjct: 702 TNDQIEEYFISKGI--HHILTST 722
>GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-)
(Germline helicase-2)
Length = 974
Score = 38.1 bits (87), Expect = 0.16
Identities = 20/60 (33%), Positives = 25/60 (41%), Gaps = 4/60 (6%)
Query: 197 GEGLNVSRGRNQNRGKGKGKNSKSKSRSKGDGNKTKYK----CFICHNPGHFKKDCPERK 252
G G + G + G G G S G G + + + CF C PGH DCPE K
Sbjct: 217 GGGKSGGFGGGNSGGSGFGSGGNSNGFGSGGGGQDRGERNNNCFNCQQPGHRSNDCPEPK 276
Score = 36.2 bits (82), Expect = 0.59
Identities = 23/63 (36%), Positives = 24/63 (37%), Gaps = 5/63 (7%)
Query: 195 DSGEGLNVSRGRNQNRGKGKGK---NSKSKSRSKG--DGNKTKYKCFICHNPGHFKKDCP 249
DSG G N G G G NS G D + CF C PGH DCP
Sbjct: 328 DSGLTNGFGSGNNGESGFGSGGFGGNSNGFGSGGGGQDRGERNNNCFNCQQPGHRSNDCP 387
Query: 250 ERK 252
E K
Sbjct: 388 EPK 390
Score = 32.3 bits (72), Expect = 8.5
Identities = 21/64 (32%), Positives = 28/64 (42%), Gaps = 8/64 (12%)
Query: 203 SRGRNQNRGKGKGK--NSKSKSRSKGDGNKTKYK-----CFICHNPGHFKKDCP-ERKDN 254
S G Q+RG+ N + D + K + C+ C PGH +DCP ERK
Sbjct: 359 SGGGGQDRGERNNNCFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHNSRDCPEERKPR 418
Query: 255 GGGN 258
G N
Sbjct: 419 EGRN 422
Score = 32.3 bits (72), Expect = 8.5
Identities = 21/64 (32%), Positives = 28/64 (42%), Gaps = 8/64 (12%)
Query: 203 SRGRNQNRGKGKGK--NSKSKSRSKGDGNKTKYK-----CFICHNPGHFKKDCP-ERKDN 254
S G Q+RG+ N + D + K + C+ C PGH +DCP ERK
Sbjct: 245 SGGGGQDRGERNNNCFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHNSRDCPEERKPR 304
Query: 255 GGGN 258
G N
Sbjct: 305 EGRN 308
>TP3A_HUMAN (Q13472) DNA topoisomerase III alpha (EC 5.99.1.2)
Length = 1001
Score = 37.4 bits (85), Expect = 0.26
Identities = 16/49 (32%), Positives = 24/49 (48%), Gaps = 1/49 (2%)
Query: 204 RGRNQNRGKGKGKNSKSKSRSKGDGNKTKYKCFICHNPGHFKKDCPERK 252
RGR + + K ++ S G K KC +CH PGH + CP+ +
Sbjct: 954 RGRTLE-SEARSKRPRASSSDMGSTAKKPRKCSLCHQPGHTRPFCPQNR 1001
>COAT_SOCMV (P15627) Coat protein
Length = 441
Score = 37.4 bits (85), Expect = 0.26
Identities = 17/54 (31%), Positives = 27/54 (49%), Gaps = 1/54 (1%)
Query: 201 NVSRGRNQNRGKGKGKNSKSKSRSKGDGNKTKYKCFICHNPGHFKKDCPERKDN 254
N + NQ + K K + + K K +C++CH GH+ +CP +KDN
Sbjct: 349 NFWKWNNQRKKKTFRKKRPFRKQQTCPTGKKKCQCWLCHEEGHYANECP-KKDN 401
>TP3A_MOUSE (O70157) DNA topoisomerase III alpha (EC 5.99.1.2)
Length = 1003
Score = 37.0 bits (84), Expect = 0.35
Identities = 17/49 (34%), Positives = 25/49 (50%), Gaps = 1/49 (2%)
Query: 204 RGRNQNRGKGKGKNSKSKSRSKGDGNKTKYKCFICHNPGHFKKDCPERK 252
RG+ Q R + K ++ S G K KC +CH PGH + CP+ +
Sbjct: 956 RGKAQ-RPEAASKRPRAGSSDAGSTVKKPRKCSLCHQPGHTRTFCPQNR 1003
>YRD6_CAEEL (Q09575) Hypothetical protein K02A2.6 in chromosome II
Length = 1268
Score = 36.6 bits (83), Expect = 0.45
Identities = 26/92 (28%), Positives = 42/92 (45%), Gaps = 11/92 (11%)
Query: 186 TKFKDLKVDDSGEGLNVSRGRNQNRGKGKGKNSKSKSRSKGDGNKTKYK---CFICHNPG 242
+K +K S + ++V++ + NR K K K K +K K + CF C+ G
Sbjct: 190 SKIVSVKPSKSSQQVDVNKV-DTNRSKKKKKPIPRKPEKSSQDSKKKGEIPTCFYCNKKG 248
Query: 243 HFKKDCPE--RKDNGGGNPSVQLASKDEGCES 272
H+ +C + N GGN K +GC+S
Sbjct: 249 HYATNCRSNPKTGNQGGN-----KGKSKGCDS 275
>YL92_SCHPO (Q9HFF2) Hypothetical protein C683.02c in chromosome I
Length = 218
Score = 36.6 bits (83), Expect = 0.45
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 1/41 (2%)
Query: 214 KGKNSKSKSRSKGDGNKTKYKCFICHNPGHFKKDCPERKDN 254
K ++ + R N+ K+ CF C GH +DCPE KDN
Sbjct: 59 KKRSEYRRLRRINQRNRDKF-CFACRQQGHIVQDCPEAKDN 98
>SLPH_BRECH (P38538) Surface layer protein precursor (Hexagonal wall
protein) (HWP)
Length = 1116
Score = 36.6 bits (83), Expect = 0.45
Identities = 35/140 (25%), Positives = 55/140 (39%), Gaps = 10/140 (7%)
Query: 110 YRMMESKPIMEQLTEFNKIIDDLANI---------DVNLEDEDKALHLLCALPRSFENFK 160
YR+ E I T FN +D L+ I V L D ++ +L ++ +
Sbjct: 381 YRLTEDTKITYNFTRFNDPVDALSKIYKDNDTFGVKVVLNDNNEVAYLHIIDDQTIDKSV 440
Query: 161 DTMLYGKEGTITLEEVQATLITKELTKFKDLKVDDSGEGLNVSRGRNQNRGKGKGKNSKS 220
+ YG + ++ + + + +KF DL+ D G+ V Q G K S
Sbjct: 441 KGVKYGSKVISKIDADKKKITNLDNSKFSDLEDQDEGKDFLVFLD-GQPAKLGDLKESDV 499
Query: 221 KSRSKGDGNKTKYKCFICHN 240
S DG+K KY F N
Sbjct: 500 YSVYYADGDKDKYLVFANRN 519
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.343 0.153 0.518
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 140,757,046
Number of Sequences: 164201
Number of extensions: 5784596
Number of successful extensions: 24781
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 24632
Number of HSP's gapped (non-prelim): 121
length of query: 1283
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1161
effective length of database: 39,941,532
effective search space: 46372118652
effective search space used: 46372118652
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.5 bits)
S2: 72 (32.3 bits)
Medicago: description of AC135504.5


