

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
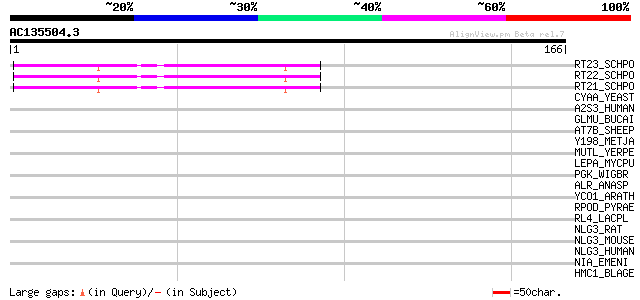
Query= AC135504.3 + phase: 0
(166 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protei... 54 1e-07
RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protei... 54 1e-07
RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protei... 54 1e-07
CYAA_YEAST (P08678) Adenylate cyclase (EC 4.6.1.1) (ATP pyrophos... 31 1.5
A2S3_HUMAN (O60296) Amyotrophic lateral sclerosis 2 chromosomal ... 30 3.3
GLMU_BUCAI (P57139) Bifunctional glmU protein [Includes: UDP-N-a... 29 4.3
AT7B_SHEEP (Q9XT50) Copper-transporting ATPase 2 (EC 3.6.3.4) (C... 29 4.3
Y198_METJA (Q57651) Hypothetical protein MJ0198 29 5.6
MUTL_YERPE (Q8ZIW4) DNA mismatch repair protein mutL 29 5.6
LEPA_MYCPU (Q98QW3) GTP-binding protein lepA 29 5.6
PGK_WIGBR (Q8D2P9) Phosphoglycerate kinase (EC 2.7.2.3) 28 7.3
ALR_ANASP (Q8YU96) Alanine racemase (EC 5.1.1.1) 28 7.3
YCO1_ARATH (P61430) Hypothetical protein At1g22275 28 9.6
RPOD_PYRAE (Q8ZYQ3) DNA-directed RNA polymerase subunit D (EC 2.... 28 9.6
RL4_LACPL (Q88XY5) 50S ribosomal protein L4 28 9.6
NLG3_RAT (Q62889) Neuroligin 3 precursor (Gliotactin homolog) 28 9.6
NLG3_MOUSE (Q8BYM5) Neuroligin 3 precursor (Gliotactin homolog) 28 9.6
NLG3_HUMAN (Q9NZ94) Neuroligin 3 precursor (Gliotactin homolog) 28 9.6
NIA_EMENI (P22945) Nitrate reductase [NADPH] (EC 1.7.1.3) (NR) 28 9.6
HMC1_BLAGE (P54961) Hydroxymethylglutaryl-CoA synthase 1 (EC 2.3... 28 9.6
>RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protein
type 3
Length = 1333
Score = 54.3 bits (129), Expect = 1e-07
Identities = 32/94 (34%), Positives = 52/94 (55%), Gaps = 5/94 (5%)
Query: 2 IREKMKASHSRQKSYHDKRRKALE-FQEGDHVFLRVTPMTGVRRALKSRKLTPKFIGPYQ 60
++E + ++ + K Y D + + +E FQ GD V ++ T TG KS KL P F GP+
Sbjct: 1167 VKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTK-TGFLH--KSNKLAPSFAGPFY 1223
Query: 61 ISERVGTVAYRVGLPTHLSNL-HDVFHVSQLQKY 93
+ ++ G Y + LP + ++ FHVS L+KY
Sbjct: 1224 VLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKY 1257
>RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protein
type 2
Length = 1333
Score = 54.3 bits (129), Expect = 1e-07
Identities = 32/94 (34%), Positives = 52/94 (55%), Gaps = 5/94 (5%)
Query: 2 IREKMKASHSRQKSYHDKRRKALE-FQEGDHVFLRVTPMTGVRRALKSRKLTPKFIGPYQ 60
++E + ++ + K Y D + + +E FQ GD V ++ T TG KS KL P F GP+
Sbjct: 1167 VKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTK-TGFLH--KSNKLAPSFAGPFY 1223
Query: 61 ISERVGTVAYRVGLPTHLSNL-HDVFHVSQLQKY 93
+ ++ G Y + LP + ++ FHVS L+KY
Sbjct: 1224 VLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKY 1257
>RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protein
type 1
Length = 1333
Score = 54.3 bits (129), Expect = 1e-07
Identities = 32/94 (34%), Positives = 52/94 (55%), Gaps = 5/94 (5%)
Query: 2 IREKMKASHSRQKSYHDKRRKALE-FQEGDHVFLRVTPMTGVRRALKSRKLTPKFIGPYQ 60
++E + ++ + K Y D + + +E FQ GD V ++ T TG KS KL P F GP+
Sbjct: 1167 VKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTK-TGFLH--KSNKLAPSFAGPFY 1223
Query: 61 ISERVGTVAYRVGLPTHLSNL-HDVFHVSQLQKY 93
+ ++ G Y + LP + ++ FHVS L+KY
Sbjct: 1224 VLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKY 1257
>CYAA_YEAST (P08678) Adenylate cyclase (EC 4.6.1.1) (ATP
pyrophosphate-lyase) (Adenylyl cyclase)
Length = 2026
Score = 30.8 bits (68), Expect = 1.5
Identities = 28/115 (24%), Positives = 44/115 (37%), Gaps = 25/115 (21%)
Query: 19 KRRKALEFQEGDHVF--LRVTPMTGVRRALKS--RKLTPKFIGPYQISERVGTVA----- 69
KR A+ D F L TP T V + + RK G +QIS +VG ++
Sbjct: 675 KRHYAIRIFNTDDTFTTLSCTPATTVEEIIPALKRKFNITAQGNFQISLKVGKLSKILRP 734
Query: 70 ----------------YRVGLPTHLSNLHDVFHVSQLQKYVADPSHVIPREDVQV 108
YR P H+ + D+ V + + PSH P ++ ++
Sbjct: 735 TSKPILIERKLLLLNGYRKSDPLHIMGIEDLSFVFKFLFHPVTPSHFTPEQEQRI 789
>A2S3_HUMAN (O60296) Amyotrophic lateral sclerosis 2 chromosomal
region candidate gene protein 3
Length = 914
Score = 29.6 bits (65), Expect = 3.3
Identities = 30/110 (27%), Positives = 52/110 (47%), Gaps = 23/110 (20%)
Query: 68 VAYRVGLPTHLSNLHDVFHVSQLQKYVADPS----HVIPREDVQVRDNLTVETSPLRIED 123
+ Y+ L + LS + D+ H +L+++V + H+ +D Q + LT+E L+ +
Sbjct: 278 IRYQEELSSLLSQIVDLQH--KLKEHVIEKEELKLHLQASKDAQRQ--LTMELHELQDRN 333
Query: 124 -----------REVKKLRGKDIPLVKVV----WGGVTGESLTWELESKMR 158
E+K+LR + P + +G TGESL E+E MR
Sbjct: 334 MECLGMLHESQEEIKELRSRSGPTAHLYFSQSYGAFTGESLAAEIEGTMR 383
>GLMU_BUCAI (P57139) Bifunctional glmU protein [Includes:
UDP-N-acetylglucosamine pyrophosphorylase (EC 2.7.7.23)
(N-acetylglucosamine-1-phosphate uridyltransferase);
Glucosamine-1-phosphate N-acetyltransferase (EC
2.3.1.157)]
Length = 459
Score = 29.3 bits (64), Expect = 4.3
Identities = 21/69 (30%), Positives = 30/69 (43%), Gaps = 11/69 (15%)
Query: 54 KFIGPYQISERVGTVAYRVGLPTHLSNLHDVFHVSQLQKY------VADPSHVIPREDVQ 107
K I P E +G + LSNL +F Q+ K + DPSH I R +Q
Sbjct: 213 KTIEPLNYEEILG-----INNKLQLSNLEKIFQKKQINKLLINGVTIKDPSHFIFRGTLQ 267
Query: 108 VRDNLTVET 116
N+ ++T
Sbjct: 268 HGQNVEIDT 276
>AT7B_SHEEP (Q9XT50) Copper-transporting ATPase 2 (EC 3.6.3.4)
(Copper pump 2) (Wilson disease-associated protein
homolog)
Length = 1505
Score = 29.3 bits (64), Expect = 4.3
Identities = 11/44 (25%), Positives = 23/44 (52%)
Query: 50 KLTPKFIGPYQISERVGTVAYRVGLPTHLSNLHDVFHVSQLQKY 93
K P+ IGP I + + + +R L + N H + H +++++
Sbjct: 648 KFDPEIIGPRDIVKLIEEIGFRASLAQRIPNAHHLDHKVEIKQW 691
>Y198_METJA (Q57651) Hypothetical protein MJ0198
Length = 314
Score = 28.9 bits (63), Expect = 5.6
Identities = 25/86 (29%), Positives = 39/86 (45%), Gaps = 14/86 (16%)
Query: 76 THLSNLHDVFHVSQLQKYVADPSHVI-------PREDVQVRDNLTVETS---PLRIEDRE 125
T++S +D+ V+ ++ +AD +VI P D+ D+L ETS P
Sbjct: 28 TNISIGYDIIEVNNPEEAIADVENVIAPELIGYPATDIDFIDSLICETSVNNPTVAMGIS 87
Query: 126 VKKLRGK----DIPLVKVVWGGVTGE 147
+ R DIPL K + G +T E
Sbjct: 88 ISVARAASNSLDIPLFKFLGGALTTE 113
>MUTL_YERPE (Q8ZIW4) DNA mismatch repair protein mutL
Length = 635
Score = 28.9 bits (63), Expect = 5.6
Identities = 14/55 (25%), Positives = 27/55 (48%)
Query: 49 RKLTPKFIGPYQISERVGTVAYRVGLPTHLSNLHDVFHVSQLQKYVADPSHVIPR 103
+KL P+ +G E + L HL + H+V++VSQ + + + + P+
Sbjct: 556 QKLIPELLGYLSQHEEISPDTLATWLARHLGSEHEVWNVSQAIQLLTEVERLCPQ 610
>LEPA_MYCPU (Q98QW3) GTP-binding protein lepA
Length = 597
Score = 28.9 bits (63), Expect = 5.6
Identities = 21/65 (32%), Positives = 32/65 (48%), Gaps = 13/65 (20%)
Query: 104 EDVQVRDNLTVETSPLRIEDREVKKLR-----------GKDIPLVKVVWGGVT--GESLT 150
+DV V D LT+ +P + KK++ GKD P++K ++ S+T
Sbjct: 264 KDVSVGDTLTLVKNPTKEALPGYKKVKAVVFTGFYPIDGKDYPVLKESLEKISLSDSSIT 323
Query: 151 WELES 155
WELES
Sbjct: 324 WELES 328
>PGK_WIGBR (Q8D2P9) Phosphoglycerate kinase (EC 2.7.2.3)
Length = 394
Score = 28.5 bits (62), Expect = 7.3
Identities = 14/54 (25%), Positives = 35/54 (63%), Gaps = 4/54 (7%)
Query: 80 NLHDVFHVSQLQKYVADPSHVIPREDVQVRDNLTVETSPLRIEDREVKKLRGKD 133
+LHD ++S+++K++ + +IP+ D V DN+ +S +++ +K+++ +D
Sbjct: 233 SLHDKNNISKIKKFLYKKNIIIPK-DFIVTDNINKFSS---YKEKSIKEIKNED 282
>ALR_ANASP (Q8YU96) Alanine racemase (EC 5.1.1.1)
Length = 386
Score = 28.5 bits (62), Expect = 7.3
Identities = 21/80 (26%), Positives = 37/80 (46%), Gaps = 11/80 (13%)
Query: 54 KFIGPYQISERVGTVAYRVGLPTHLSNLHDVFHVSQLQKYVADPSHVIPREDVQVRDNLT 113
+FI P+++ V ++ Y G+P +LSN V Q +P+ D L
Sbjct: 281 QFIAPHEMRLAVVSIGYADGVPRNLSNKMQVLIRGQR----------VPQIGAITMDQLM 330
Query: 114 VETSPL-RIEDREVKKLRGK 132
++ S L +++ E+ L GK
Sbjct: 331 IDASALPDLQEGEIVTLLGK 350
>YCO1_ARATH (P61430) Hypothetical protein At1g22275
Length = 856
Score = 28.1 bits (61), Expect = 9.6
Identities = 15/51 (29%), Positives = 24/51 (46%), Gaps = 5/51 (9%)
Query: 47 KSRKLTPKFIGPYQISERVGTVAYRVGLPTHLSNLHDVFHVSQLQKYVADP 97
+S +LTPK + P I++ + H +N+ D+F L Y DP
Sbjct: 807 RSTRLTPKLMTPTIIAKETAMADH-----PHSANIGDLFSEGSLNPYADDP 852
>RPOD_PYRAE (Q8ZYQ3) DNA-directed RNA polymerase subunit D (EC
2.7.7.6)
Length = 260
Score = 28.1 bits (61), Expect = 9.6
Identities = 29/112 (25%), Positives = 48/112 (41%), Gaps = 14/112 (12%)
Query: 68 VAYRVGLPTHLSNLHDVFHVSQLQKYVADPSHVIPREDVQV--RDNLTVETSPLRIEDRE 125
+A+R+GL + L + +ADPS R +QV + T+ + L + +
Sbjct: 59 LAHRLGLIPLTTPLQSLPPYEDCVSGLADPSECSTRLTLQVTAEGDTTIYSGDLASDRPD 118
Query: 126 VKKLRGKDIPLVKVVWG-----------GVTGESLTWELESKMRDSYPESFV 166
V + KDIP+VK+V G GV + W+ + YP+ V
Sbjct: 119 VVPVY-KDIPIVKLVKGQSIVLEAYAKLGVAKDHAKWQAATASYYYYPKVVV 169
>RL4_LACPL (Q88XY5) 50S ribosomal protein L4
Length = 207
Score = 28.1 bits (61), Expect = 9.6
Identities = 15/40 (37%), Positives = 25/40 (62%), Gaps = 2/40 (5%)
Query: 103 REDVQVRDNLTVETSPLRI--EDREVKKLRGKDIPLVKVV 140
++ V V +NL V+T L + ED E L G+++P VK++
Sbjct: 137 KDFVNVLNNLNVDTKTLVLVEEDNEKAALAGRNLPNVKIL 176
>NLG3_RAT (Q62889) Neuroligin 3 precursor (Gliotactin homolog)
Length = 848
Score = 28.1 bits (61), Expect = 9.6
Identities = 17/45 (37%), Positives = 21/45 (45%), Gaps = 5/45 (11%)
Query: 64 RVGTVAYRVGLPTHLSNLHDVFHVSQLQKYVADP-----SHVIPR 103
R VA+ L HL NLHD+FH + V P SH+ R
Sbjct: 617 RATKVAFWKHLVPHLYNLHDMFHYTSTTTKVPPPDTTHSSHITRR 661
>NLG3_MOUSE (Q8BYM5) Neuroligin 3 precursor (Gliotactin homolog)
Length = 825
Score = 28.1 bits (61), Expect = 9.6
Identities = 17/45 (37%), Positives = 21/45 (45%), Gaps = 5/45 (11%)
Query: 64 RVGTVAYRVGLPTHLSNLHDVFHVSQLQKYVADP-----SHVIPR 103
R VA+ L HL NLHD+FH + V P SH+ R
Sbjct: 594 RATKVAFWKHLVPHLYNLHDMFHYTSTTTKVPPPDTTHSSHITRR 638
>NLG3_HUMAN (Q9NZ94) Neuroligin 3 precursor (Gliotactin homolog)
Length = 848
Score = 28.1 bits (61), Expect = 9.6
Identities = 17/45 (37%), Positives = 21/45 (45%), Gaps = 5/45 (11%)
Query: 64 RVGTVAYRVGLPTHLSNLHDVFHVSQLQKYVADP-----SHVIPR 103
R VA+ L HL NLHD+FH + V P SH+ R
Sbjct: 617 RATKVAFWKHLVPHLYNLHDMFHYTSTTTKVPPPDTTHSSHITRR 661
>NIA_EMENI (P22945) Nitrate reductase [NADPH] (EC 1.7.1.3) (NR)
Length = 873
Score = 28.1 bits (61), Expect = 9.6
Identities = 11/24 (45%), Positives = 19/24 (78%)
Query: 113 TVETSPLRIEDREVKKLRGKDIPL 136
T++TS +R+E++E+ +L DIPL
Sbjct: 16 TLKTSQIRVEEQEITELDTADIPL 39
>HMC1_BLAGE (P54961) Hydroxymethylglutaryl-CoA synthase 1 (EC
2.3.3.10) (HMG-CoA synthase 1)
(3-hydroxy-3-methylglutaryl coenzyme A synthase 1)
Length = 453
Score = 28.1 bits (61), Expect = 9.6
Identities = 28/98 (28%), Positives = 46/98 (46%), Gaps = 13/98 (13%)
Query: 63 ERVGTVAYRVGLPTHLSNLH---DVFHVSQLQKYVADPSHVIPREDVQVRDNLTVETSPL 119
+RV +Y GL + + +L D S LQ+ V++ SH+ P+ D +R ++ E
Sbjct: 350 KRVALFSYGSGLASSMFSLRISSDASAKSSLQRLVSNLSHIKPQLD--LRHKVSPEEFAQ 407
Query: 120 RIEDREVKKLRGKDIP--LVKVVWGGVTGESLTWELES 155
+E RE + P + V++ G TW LES
Sbjct: 408 TMETREHNHHKAPYTPEGSIDVLFPG------TWYLES 439
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,146,736
Number of Sequences: 164201
Number of extensions: 745382
Number of successful extensions: 1882
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 1876
Number of HSP's gapped (non-prelim): 22
length of query: 166
length of database: 59,974,054
effective HSP length: 102
effective length of query: 64
effective length of database: 43,225,552
effective search space: 2766435328
effective search space used: 2766435328
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135504.3


