

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135464.6 + phase: 0
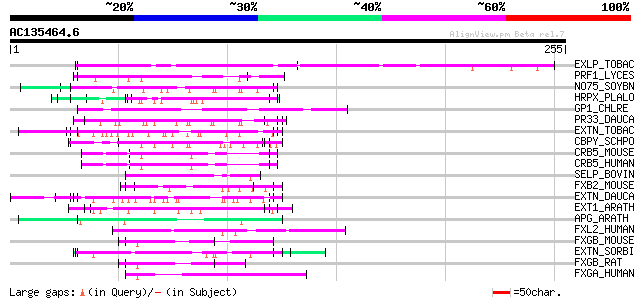
(255 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
EXLP_TOBAC (Q03211) Pistil-specific extensin-like protein precur... 87 5e-17
PRF1_LYCES (Q00451) 36.4 kDa proline-rich protein 60 5e-09
NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75) (NGM-75) 58 2e-08
HRPX_PLALO (P04929) Histidine-rich glycoprotein precursor 58 3e-08
GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor (H... 57 4e-08
PR33_DAUCA (P06600) Proline-rich 33 kDa extensin-related protein... 54 4e-07
EXTN_TOBAC (P13983) Extensin precursor (Cell wall hydroxyproline... 54 4e-07
CBPY_SCHPO (O13849) Carboxypeptidase Y precursor (EC 3.4.16.5) (... 54 4e-07
CRB5_MOUSE (Q8K1L0) cAMP response element binding protein 5 (CRE... 53 6e-07
CRB5_HUMAN (Q02930) cAMP response element binding protein 5 (CRE... 53 6e-07
SELP_BOVIN (P49907) Selenoprotein P-like protein precursor 52 1e-06
FXB2_MOUSE (Q64733) Forkhead box protein B2 (Transcription facto... 52 1e-06
EXTN_DAUCA (P06599) Extensin precursor 52 1e-06
EXT1_ARATH (Q38913) Extensin 1 precursor (AtExt1) (AtExt4) 52 1e-06
APG_ARATH (P40602) Anter-specific proline-rich protein APG precu... 52 1e-06
FXL2_HUMAN (P58012) Forkhead box protein L2 52 1e-06
FXGB_MOUSE (Q60987) Forkhead box protein G1B (Forkhead-related p... 52 1e-06
EXTN_SORBI (P24152) Extensin precursor (Proline-rich glycoprotein) 52 1e-06
FXGB_RAT (Q00939) Forkhead box protein G1B (Forkhead-related pro... 51 2e-06
FXGA_HUMAN (P55316) Forkhead box protein G1A (Forkhead-related p... 51 3e-06
>EXLP_TOBAC (Q03211) Pistil-specific extensin-like protein precursor
(PELP)
Length = 426
Score = 86.7 bits (213), Expect = 5e-17
Identities = 76/231 (32%), Positives = 105/231 (44%), Gaps = 20/231 (8%)
Query: 31 KPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSP 90
K P PP K+P P +PPT P S PL PP P P+ + SP
Sbjct: 195 KQPPPPPPPVKAPSPSPATQPPTKQPPPPPRAKKS--PLLPPPP--PVAYPPVMTPSPSP 250
Query: 91 APSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSSIAVEGVVYVKSCHHAGFDT 150
A P + PA PP + PP P +P + + I V G+VY KSC+ G T
Sbjct: 251 AAEPPIIAPFPSPPANPPLIPRRPAPPVVKP--LPPLGKPPI-VSGLVYCKSCNSYGVPT 307
Query: 151 LKGATLLFGAVVKFQCHNAKYKFVLKAKTNKEGYIYIGSSKNISSYASGHCNVVLESAPN 210
L A+LL GAVVK C+ K V A T+ +G I K++++ G C V L +PN
Sbjct: 308 LLNASLLQGAVVKLICYGKK-TMVQWATTDNKGEFRI-MPKSLTTADVGKCKVYLVKSPN 365
Query: 211 G--LKPSNLHGGLTGAHPKSV---KRIVSKGVSLIR------YTVDPLAFE 250
P+N +GG +G K + K+ ++ V ++ Y V P FE
Sbjct: 366 PNCNVPTNFNGGKSGGLLKPLLPPKQPITPAVVPVQPPMSDLYGVGPFIFE 416
Score = 45.1 bits (105), Expect = 2e-04
Identities = 30/101 (29%), Positives = 39/101 (37%), Gaps = 2/101 (1%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P P + SP+ KP PP+ S P PPP AK + +P+
Sbjct: 133 PVNQPKPSSPSPLVKPPPPPPSPCKPSPPDQSAKQPPQPPP--AKQPSPPPPPPPVKAPS 190
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSSI 132
PSP P P P K P+ PP P PR +S +
Sbjct: 191 PSPAKQPPPPPPPVKAPSPSPATQPPTKQPPPPPRAKKSPL 231
Score = 37.0 bits (84), Expect = 0.046
Identities = 21/71 (29%), Positives = 25/71 (34%)
Query: 51 PPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTL 110
PP + P P PPP P KP Q P P+ P P P K P+
Sbjct: 132 PPVNQPKPSSPSPLVKPPPPPPSPCKPSPPDQSAKQPPQPPPAKQPSPPPPPPPVKAPSP 191
Query: 111 HHHQHPPAHAP 121
+ PP P
Sbjct: 192 SPAKQPPPPPP 202
>PRF1_LYCES (Q00451) 36.4 kDa proline-rich protein
Length = 346
Score = 60.1 bits (144), Expect = 5e-09
Identities = 36/105 (34%), Positives = 44/105 (41%), Gaps = 12/105 (11%)
Query: 30 HKPTTPLHP--------PTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHH 81
+ P+TP HP P+ P H P H P H P P PP+ P T
Sbjct: 12 YPPSTPKHPKLPPKVKPPSTQPPHVKPPSTPKH-PKDPPHVKPPSTPKQPPYVKPPTTPK 70
Query: 82 HHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPR 126
H H P +P H P Q+P PP+ HH PP P H+PR
Sbjct: 71 HPPHV--KPPSTPKHPKHPPQKPCPPPS-HHGPKPPIVKPPHVPR 112
Score = 53.5 bits (127), Expect = 5e-07
Identities = 35/105 (33%), Positives = 44/105 (41%), Gaps = 24/105 (22%)
Query: 32 PTTPLHP-------PTKSPVHKPLAKPPT---HAPHHHHHHSPSHAPLPPPHPAKPLTHH 81
P+TP HP P +P P KPPT H PH +P H PP P P +HH
Sbjct: 39 PSTPKHPKDPPHVKPPSTPKQPPYVKPPTTPKHPPHVKPPSTPKHPKHPPQKPCPPPSHH 98
Query: 82 HHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPR 126
+ P P HVP +PP +H PP +P P+
Sbjct: 99 GPK----PPIVKPPHVP-------RPPIVH---PPPIVSPPSTPK 129
Score = 43.5 bits (101), Expect = 5e-04
Identities = 30/85 (35%), Positives = 34/85 (39%), Gaps = 15/85 (17%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHH----HHHSPSHAPLPPPHPAKPLTHHHHQHQH 87
PTTP HPP P P K P H P HH P + PPH +P H
Sbjct: 66 PTTPKHPPHVKPPSTP--KHPKHPPQKPCPPPSHHGPKPPIVKPPHVPRP------PIVH 117
Query: 88 HSPAPSPYHVPTPLQRP---AKPPT 109
P SP P P + P KPP+
Sbjct: 118 PPPIVSPPSTPKPPKTPPFTPKPPS 142
Score = 36.2 bits (82), Expect = 0.079
Identities = 26/95 (27%), Positives = 34/95 (35%), Gaps = 15/95 (15%)
Query: 30 HKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHH-----SPSHAPLPP---------PHPA 75
H P P PP+ P+ KPP H P H SP P PP P P
Sbjct: 86 HPPQKPCPPPSHHGPKPPIVKPP-HVPRPPIVHPPPIVSPPSTPKPPKTPPFTPKPPSPI 144
Query: 76 KPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTL 110
P+ + +P P H P + P+ P +
Sbjct: 145 PPIVSPPIVYPPITPTPPIVHPPVTPKPPSPTPPI 179
Score = 33.5 bits (75), Expect = 0.51
Identities = 32/112 (28%), Positives = 40/112 (35%), Gaps = 21/112 (18%)
Query: 29 HHKPTTPL----HPPTKSPVHKP-------LAKPPTHAPHHHHHHSPSHAPLPPPHPAKP 77
HH P P+ H P VH P KPP P SP + PP P
Sbjct: 97 HHGPKPPIVKPPHVPRPPIVHPPPIVSPPSTPKPPKTPPFTPKPPSPIPPIVSPPIVYPP 156
Query: 78 LTHH----HHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
+T H P+P+P V P+ P PT PP +P +P
Sbjct: 157 ITPTPPIVHPPVTPKPPSPTPPIVSPPIVYPPITPT------PPVVSPPIIP 202
>NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75) (NGM-75)
Length = 309
Score = 58.2 bits (139), Expect = 2e-08
Identities = 38/108 (35%), Positives = 44/108 (40%), Gaps = 18/108 (16%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHH--HHSPSHAPLPP----PHPAKPLTHHH 82
H KP PP + P H+ PP H P H H P H PP PH P
Sbjct: 93 HEKPPPEYQPPHEKPPHEN--PPPEHQPPHEKPPEHQPPHEKPPPEYEPPHEKPP---PE 147
Query: 83 HQHQHHSPAPS---PYHVPTPLQRPA--KPPTLHH--HQHPPAHAPTH 123
+Q H P P P+ P P +P KPP H H+ PP H P H
Sbjct: 148 YQPPHEKPPPEYQPPHEKPPPEYQPPHEKPPPEHQPPHEKPPEHQPPH 195
Score = 53.1 bits (126), Expect = 6e-07
Identities = 40/126 (31%), Positives = 51/126 (39%), Gaps = 12/126 (9%)
Query: 6 LALIFLLLQITSFVVFAEELETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHH---HH 62
L L+ L + I + V A ++P PPT P P KPP + P HH +
Sbjct: 9 LLLLLLGVVILTTPVLANLKPRFFYEPPPIEKPPTYEP--PPFYKPPYYPPPVHHPPPEY 66
Query: 63 SPSHAP-----LPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPP 117
P H LPPPH KP + H+ P P H P + P P H+ PP
Sbjct: 67 QPPHEKTPPEYLPPPHE-KPPPEYLPPHEKPPPEYQPPHEKPPHENP-PPEHQPPHEKPP 124
Query: 118 AHAPTH 123
H P H
Sbjct: 125 EHQPPH 130
Score = 51.2 bits (121), Expect = 2e-06
Identities = 34/108 (31%), Positives = 41/108 (37%), Gaps = 18/108 (16%)
Query: 24 ELETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHH---HHSPSHAPLPPPHPAKPLTH 80
E E H KP PP + P PP + P H + P H PP H
Sbjct: 136 EYEPPHEKPPPEYQPPHEKP-------PPEYQPPHEKPPPEYQPPHEKPPPEHQPPHEKP 188
Query: 81 HHHQHQHHSPAPS---PYHVPTPLQRP--AKPPTLHHHQHPPAHAPTH 123
HQ H P P P+ P P +P KPP H + PP + P H
Sbjct: 189 PEHQPPHEKPPPEYQPPHEKPPPEYQPPQEKPP---HEKPPPEYQPPH 233
Score = 50.1 bits (118), Expect = 5e-06
Identities = 38/129 (29%), Positives = 48/129 (36%), Gaps = 31/129 (24%)
Query: 24 ELETLHHKPTTPLHPPTKSPVHK---PLAKPPTHAPHHHH---HHSPSHAPLPP------ 71
E + H KP PP + P + P KPP H P H + P H PP
Sbjct: 158 EYQPPHEKPPPEYQPPHEKPPPEHQPPHEKPPEHQPPHEKPPPEYQPPHEKPPPEYQPPQ 217
Query: 72 ---PHPAKPLTHH--------HHQHQHHSPAP---SPYHVPTPLQRP--AKPPTLHH--- 112
PH P + HQ H P P PY P P+ P KPP + +
Sbjct: 218 EKPPHEKPPPEYQPPHEKPPPEHQPPHEKPPPVYPPPYEKPPPVYEPPYEKPPPVVYPPP 277
Query: 113 HQHPPAHAP 121
H+ PP + P
Sbjct: 278 HEKPPIYEP 286
Score = 41.6 bits (96), Expect = 0.002
Identities = 31/107 (28%), Positives = 44/107 (40%), Gaps = 19/107 (17%)
Query: 24 ELETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHH----HHHSPSHAP---LPPPHPAK 76
E + H KP PP + P H+ PP + P H H P P PPP+
Sbjct: 201 EYQPPHEKPPPEYQPPQEKPPHEK--PPPEYQPPHEKPPPEHQPPHEKPPPVYPPPYEKP 258
Query: 77 PLTHHHHQHQHHSPAPSPYHVPTPLQRPA--KPPTLHHHQHPPAHAP 121
P ++ + P P Y P P ++P +PP L + PP + P
Sbjct: 259 PPV---YEPPYEKPPPVVY--PPPHEKPPIYEPPPL---EKPPVYNP 297
>HRPX_PLALO (P04929) Histidine-rich glycoprotein precursor
Length = 351
Score = 57.8 bits (138), Expect = 3e-08
Identities = 34/95 (35%), Positives = 36/95 (37%), Gaps = 24/95 (25%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHH 88
HH HP H L HAPHHHHHH HAP HHH H HH
Sbjct: 150 HHHHAAHHHPWFH---HHHLGYHHHHAPHHHHHHH--HAP------------HHHHHHHH 192
Query: 89 SPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
+P +H P HHH H P H H
Sbjct: 193 APHHHHHHHHAPHHH-------HHHHHAPHHHHHH 220
Score = 55.8 bits (133), Expect = 1e-07
Identities = 31/95 (32%), Positives = 33/95 (34%), Gaps = 3/95 (3%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHH 88
HH P H H A H HHHHHH+ H H HHH HH
Sbjct: 105 HHHPPHHHHHLGHHHHHHHAAHHHHHEEHHHHHHAAHHHHHEEHHHHHHAAHHHPWFHHH 164
Query: 89 SPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
+H P P HHH H HAP H
Sbjct: 165 HLGYHHHHAPHHHHHHHHAPHHHHHHH---HAPHH 196
Score = 53.5 bits (127), Expect = 5e-07
Identities = 25/71 (35%), Positives = 28/71 (39%), Gaps = 13/71 (18%)
Query: 54 HAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHH 113
H HHHHH H P H HHHH +HH P +H P HHH
Sbjct: 58 HEEHHHHHPEEHHEPHHEEH------HHHHPEEHHEPHHEEHHHHHPHPH-------HHH 104
Query: 114 QHPPAHAPTHM 124
H P H H+
Sbjct: 105 HHHPPHHHHHL 115
Score = 52.8 bits (125), Expect = 8e-07
Identities = 34/101 (33%), Positives = 38/101 (36%), Gaps = 15/101 (14%)
Query: 23 EELETLHHKPTTPLHPPTK-SPVHKPLAKPPTHAPHHHHHHSPSHAP-LPPPHPAKPLTH 80
EE HH+ HP P H+ H HHHHHH P H L H H
Sbjct: 67 EEHHEPHHEEHHHHHPEEHHEPHHEEHHHHHPHPHHHHHHHPPHHHHHLGHHHHHHHAAH 126
Query: 81 HHH--QHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
HHH +H HH A +H HHH H AH
Sbjct: 127 HHHHEEHHHHHHAAHHHHHEE-----------HHHHHHAAH 156
Score = 51.2 bits (121), Expect = 2e-06
Identities = 26/70 (37%), Positives = 27/70 (38%), Gaps = 12/70 (17%)
Query: 54 HAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHH 113
HAPHHHHHH H H HHHH H HH +H HHH
Sbjct: 212 HAPHHHHHHHHGHHHHHHHHHG----HHHHHHHHHGHHHHHHH--------HHDAHHHHH 259
Query: 114 QHPPAHAPTH 123
H AH H
Sbjct: 260 HHHDAHHHHH 269
Score = 50.1 bits (118), Expect = 5e-06
Identities = 32/92 (34%), Positives = 33/92 (35%), Gaps = 17/92 (18%)
Query: 36 LHPPTKSPVHKPLAKPPTHAPHH--HHHHSPS--HAPLPPPHPAKPLTHHHHQHQHHSPA 91
+HP H H PHH HHHH P H P H HHHH H HH
Sbjct: 52 VHPEHLHEEHHHHHPEEHHEPHHEEHHHHHPEEHHEPHHEEH------HHHHPHPHHHHH 105
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
P H L HHH H AH H
Sbjct: 106 HHPPHHHHHLGH-------HHHHHHAAHHHHH 130
Score = 48.5 bits (114), Expect = 2e-05
Identities = 28/71 (39%), Positives = 29/71 (40%), Gaps = 4/71 (5%)
Query: 54 HAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQH-HSPAPSPYHVPTPLQRPAKPPTLHH 112
HAPHHHHHH HAP H HHHH H H H +H HH
Sbjct: 192 HAPHHHHHHH--HAP-HHHHHHHHAPHHHHHHHHGHHHHHHHHHGHHHHHHHHHGHHHHH 248
Query: 113 HQHPPAHAPTH 123
H H AH H
Sbjct: 249 HHHHDAHHHHH 259
Score = 48.1 bits (113), Expect = 2e-05
Identities = 24/69 (34%), Positives = 26/69 (36%), Gaps = 14/69 (20%)
Query: 55 APHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQ 114
A HHHHHH +H H A HHHH HH +H HHH
Sbjct: 274 AHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHDAHH--------------HHHH 319
Query: 115 HPPAHAPTH 123
H AH H
Sbjct: 320 HHDAHHHHH 328
Score = 48.1 bits (113), Expect = 2e-05
Identities = 24/69 (34%), Positives = 26/69 (36%), Gaps = 14/69 (20%)
Query: 55 APHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQ 114
A HHHHHH +H H A HHH H HH +H HHH
Sbjct: 284 AHHHHHHHHDAHHHHHHHHDAHHHHHHHDAHHHHHHHHDAHH--------------HHHH 329
Query: 115 HPPAHAPTH 123
H AH H
Sbjct: 330 HHDAHHHHH 338
Score = 47.8 bits (112), Expect = 3e-05
Identities = 24/69 (34%), Positives = 27/69 (38%), Gaps = 4/69 (5%)
Query: 55 APHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQ 114
A HHHHHH +H H A HHHH H H + +H HHH
Sbjct: 254 AHHHHHHHHDAHHHHHHHHDA----HHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHH 309
Query: 115 HPPAHAPTH 123
H AH H
Sbjct: 310 HHDAHHHHH 318
Score = 47.4 bits (111), Expect = 3e-05
Identities = 34/107 (31%), Positives = 35/107 (31%), Gaps = 16/107 (14%)
Query: 20 VFAEELETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHH-HHHSPSHAPLPPPHPAKPL 78
V E L HH H H H PHH HHH H P H P
Sbjct: 52 VHPEHLHEEHHHHHPEEHHEPHHEEHHHHHPEEHHEPHHEEHHHHHPH-PHHHHHHHPPH 110
Query: 79 THHH--HQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
HHH H H HH A +H HHH H AH H
Sbjct: 111 HHHHLGHHHHHHHAAHHHHHEE------------HHHHHHAAHHHHH 145
Score = 46.6 bits (109), Expect = 6e-05
Identities = 24/68 (35%), Positives = 26/68 (37%), Gaps = 15/68 (22%)
Query: 57 HHHHHHSPSHAPLPPPHPAKPLTHHHHQ-HQHHSPAPSPYHVPTPLQRPAKPPTLHHHQH 115
HHHHHH +H H A HHHH H HH +H HHH H
Sbjct: 246 HHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHH--------------HHHHH 291
Query: 116 PPAHAPTH 123
AH H
Sbjct: 292 HDAHHHHH 299
Score = 44.7 bits (104), Expect = 2e-04
Identities = 30/95 (31%), Positives = 31/95 (32%), Gaps = 13/95 (13%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHH 88
HH P H H P H HHHHHH H H HHHH H HH
Sbjct: 198 HHHHHAPHHHHHHH--HAPHHHHHHHHGHHHHHH---HHHGHHHHHHHHHGHHHHHHHHH 252
Query: 89 SPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
+H HHH H AH H
Sbjct: 253 DAHHHHHH--------HHDAHHHHHHHHDAHHHHH 279
Score = 44.3 bits (103), Expect = 3e-04
Identities = 24/70 (34%), Positives = 27/70 (38%), Gaps = 23/70 (32%)
Query: 54 HAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHH 113
H HHHHHH +H H A HHHH H H + +H HHH
Sbjct: 302 HDAHHHHHHHDAHHHHHHHHDA----HHHHHHHHDA-----HH--------------HHH 338
Query: 114 QHPPAHAPTH 123
H AH H
Sbjct: 339 HHHDAHHHHH 348
Score = 42.0 bits (97), Expect = 0.001
Identities = 23/79 (29%), Positives = 27/79 (34%), Gaps = 18/79 (22%)
Query: 48 LAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKP 107
L + H H H H HHHH +HH P +H P +
Sbjct: 47 LVEDTVHPEHLHEEH-----------------HHHHPEEHHEPHHEEHHHHHPEEHHEPH 89
Query: 108 PTLHHHQHP-PAHAPTHMP 125
HHH HP P H H P
Sbjct: 90 HEEHHHHHPHPHHHHHHHP 108
Score = 40.4 bits (93), Expect = 0.004
Identities = 24/71 (33%), Positives = 25/71 (34%), Gaps = 18/71 (25%)
Query: 54 HAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQ-HQHHSPAPSPYHVPTPLQRPAKPPTLHH 112
H HHHH H H H A HHHH H HH +H HH
Sbjct: 236 HHHHHHHGHHHHH---HHHHDAHHHHHHHHDAHHHHHHHHDAHH--------------HH 278
Query: 113 HQHPPAHAPTH 123
H H AH H
Sbjct: 279 HHHHDAHHHHH 289
Score = 38.5 bits (88), Expect = 0.016
Identities = 16/33 (48%), Positives = 17/33 (51%), Gaps = 4/33 (12%)
Query: 55 APHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQH 87
A HHHHHH +H H A HHHH H H
Sbjct: 323 AHHHHHHHHDAHHHHHHHHDA----HHHHHHHH 351
>GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor
(Hydroxyproline-rich glycoprotein 1)
Length = 555
Score = 57.0 bits (136), Expect = 4e-08
Identities = 43/124 (34%), Positives = 56/124 (44%), Gaps = 20/124 (16%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P TP PP+ SP P PP+ AP PS AP PPP PA P SP+
Sbjct: 299 PPTPPTPPSPSP---PSPVPPSPAPVPPSPAPPSPAPSPPPSPAPPTP---------SPS 346
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSSIAVEGVVYVKSCHHAGFDTL 151
PSP P+P P+ P+ P+ P+ P+ S S +AV+ V + FD L
Sbjct: 347 PSPSPSPSPSPSPSPSPS-----PSPSPIPSPSPKPSPSPVAVK---LVWADDAIAFDDL 398
Query: 152 KGAT 155
G +
Sbjct: 399 NGTS 402
Score = 42.4 bits (98), Expect = 0.001
Identities = 31/100 (31%), Positives = 39/100 (39%), Gaps = 5/100 (5%)
Query: 31 KPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSP 90
KP P PP+ P P P + P SP +P PP P P + SP
Sbjct: 260 KPPAPPPPPSPPPPPPPRPPFPANTPMPPSPPSPPPSPAPPTPPTPP-SPSPPSPVPPSP 318
Query: 91 APSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRS 130
AP P P P+ PP+ PP +P+ P S S
Sbjct: 319 APVPPSPAPPSPAPSPPPS----PAPPTPSPSPSPSPSPS 354
Score = 40.8 bits (94), Expect = 0.003
Identities = 31/96 (32%), Positives = 41/96 (42%), Gaps = 6/96 (6%)
Query: 38 PPTKSPVHKPLAKPPTHAPHHHHHHSPS-HAPLP----PPHPAKPLTHHHHQHQHHSPAP 92
PP+ +P P PP+ AP SP+ +P P PP PA P P+P
Sbjct: 70 PPSPAPPSPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPPSPAPPSP 129
Query: 93 SPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVS 128
SP P+P P+ P L PP+ +P P S
Sbjct: 130 SPPAPPSP-SPPSPAPPLPPSPAPPSPSPPVPPSPS 164
Score = 40.0 bits (92), Expect = 0.005
Identities = 31/91 (34%), Positives = 38/91 (41%), Gaps = 9/91 (9%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSH-APLPPPHPAKPLTHHHHQHQHHSP 90
P +P PP+ SP P PP+ AP SP AP PPP P P ++P
Sbjct: 228 PPSPA-PPSPSPPAPPSPVPPSPAPPSPAPPSPKPPAPPPPPSPPPP-PPPRPPFPANTP 285
Query: 91 APSPYHVPTPLQRPAKPPTLHHHQHPPAHAP 121
P P P P PPT PP+ +P
Sbjct: 286 MPPSPPSPPPSPAPPTPPT------PPSPSP 310
Score = 39.3 bits (90), Expect = 0.009
Identities = 30/90 (33%), Positives = 40/90 (44%), Gaps = 11/90 (12%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P+ P+ PP+ SP P PP+ P PS +P PP PA P P+
Sbjct: 155 PSPPV-PPSPSPPVPPSPAPPSPTP-------PSPSPPVPPSPAPPSPAPPVPPSPAPPS 206
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQHPPAHAP 121
P+P P+P P PP+ PP+ AP
Sbjct: 207 PAPPVPPSP--APPSPPS-PAPPSPPSPAP 233
Score = 38.5 bits (88), Expect = 0.016
Identities = 32/100 (32%), Positives = 41/100 (41%), Gaps = 7/100 (7%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSP-SHAPLPPPHPAKPLTHHHHQHQHHSP 90
P +P PP+ +P P PP+ +P SP S AP PP PA P P
Sbjct: 108 PPSPA-PPSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSPAPP-----SPSPPVPP 161
Query: 91 APSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRS 130
+PSP P+P PP+ P P+ P V S
Sbjct: 162 SPSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPPS 201
Score = 38.5 bits (88), Expect = 0.016
Identities = 29/95 (30%), Positives = 38/95 (39%), Gaps = 3/95 (3%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLP-PPHPAKPLTHHHHQHQHHSP 90
P P+ PP+ +P P PP+ SP P P PP PA P + + +P
Sbjct: 207 PAPPV-PPSPAPPSPPSPAPPSPPSPAPPSPSPPAPPSPVPPSPAPP-SPAPPSPKPPAP 264
Query: 91 APSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
P P P P RP P PP+ P+ P
Sbjct: 265 PPPPSPPPPPPPRPPFPANTPMPPSPPSPPPSPAP 299
Score = 38.1 bits (87), Expect = 0.021
Identities = 30/94 (31%), Positives = 37/94 (38%), Gaps = 19/94 (20%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P +P PP+ +P PP+ AP PS AP PP PA P SPA
Sbjct: 168 PPSPA-PPSPTPPSPSPPVPPSPAP-------PSPAPPVPPSPAPP-----------SPA 208
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
P P P P+ P PP+ +P P
Sbjct: 209 PPVPPSPAPPSPPSPAPPSPPSPAPPSPSPPAPP 242
Score = 37.4 bits (85), Expect = 0.035
Identities = 30/92 (32%), Positives = 36/92 (38%), Gaps = 10/92 (10%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P +P PP+ SP P PP+ AP P PP PA P+ SPA
Sbjct: 173 PPSPT-PPSPSPPVPPSPAPPSPAP-------PVPPSPAPPSPAPPVPPSPAPPSPPSPA 224
Query: 92 P--SPYHVPTPLQRPAKPPTLHHHQHPPAHAP 121
P P P PA P + PP+ AP
Sbjct: 225 PPSPPSPAPPSPSPPAPPSPVPPSPAPPSPAP 256
Score = 37.4 bits (85), Expect = 0.035
Identities = 33/95 (34%), Positives = 39/95 (40%), Gaps = 18/95 (18%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P +P PP+ +P P PP+ AP PS AP PP PA P SPA
Sbjct: 45 PPSPA-PPSPAP---PSPAPPSPAPPSPG--PPSPAPPSPPSPAPP-----------SPA 87
Query: 92 PSPYHVPTPL-QRPAKPPTLHHHQHPPAHAPTHMP 125
P P+P PA P PP+ AP P
Sbjct: 88 PPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPP 122
Score = 36.6 bits (83), Expect = 0.060
Identities = 28/94 (29%), Positives = 38/94 (39%), Gaps = 9/94 (9%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P +P PP+ +P PP+ AP PS +P PP P+ P P+
Sbjct: 103 PPSPA-PPSPAPPSPAPPSPPSPAP-------PSPSPPAPPSPSPPSPAPPLPPSPAPPS 154
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
PSP P+P P P PP+ +P P
Sbjct: 155 PSPPVPPSP-SPPVPPSPAPPSPTPPSPSPPVPP 187
Score = 31.6 bits (70), Expect = 1.9
Identities = 23/72 (31%), Positives = 29/72 (39%), Gaps = 9/72 (12%)
Query: 51 PPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPL-QRPAKPPT 109
PP+ AP SP+ PP PA P P+P+P P+P PA P
Sbjct: 40 PPSPAPPSPAPPSPAPPSPAPPSPAPP--------SPGPPSPAPPSPPSPAPPSPAPPSP 91
Query: 110 LHHHQHPPAHAP 121
PP+ AP
Sbjct: 92 APPSPAPPSPAP 103
>PR33_DAUCA (P06600) Proline-rich 33 kDa extensin-related protein
precursor (Fragment)
Length = 211
Score = 53.9 bits (128), Expect = 4e-07
Identities = 38/102 (37%), Positives = 45/102 (43%), Gaps = 24/102 (23%)
Query: 30 HKPTTPLHPPT---KSPVHKPLA--KPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQ 84
H P P+H P + PVHKP KPP H P H SP + P P KP+ H
Sbjct: 122 HYPAHPIHKPQPIHRPPVHKPPTEHKPPVHEPATEHKPSPVYQP---PKTEKPVPEH--- 175
Query: 85 HQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPP-AHAPTHMP 125
P H+P + RP PPT H +PP H P H P
Sbjct: 176 --------KPPHLPPIVVRP--PPT--HKPNPPYGHHPRHPP 205
Score = 51.2 bits (121), Expect = 2e-06
Identities = 36/109 (33%), Positives = 49/109 (44%), Gaps = 21/109 (19%)
Query: 35 PLHPPT--KSPVHKPLA--KPPTHAPHHHH---HHSPS----HAPLPPPHPAKPLTH--- 80
P+H P K PV KP KPP H P H H++PS H P P H +P+
Sbjct: 80 PIHKPPVYKPPVQKPAPEHKPPVHKPPIHKPPVHNTPSVTDDHYPAHPIHKPQPIHRPPV 139
Query: 81 HHHQHQHHSPA--PSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRV 127
H +H P P+ H P+P+ +P K + P H P H+P +
Sbjct: 140 HKPPTEHKPPVHEPATEHKPSPVYQPPKT-----EKPVPEHKPPHLPPI 183
Score = 48.5 bits (114), Expect = 2e-05
Identities = 36/113 (31%), Positives = 43/113 (37%), Gaps = 30/113 (26%)
Query: 30 HKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPA--KPLTHH----HH 83
HKP P+H P P+HKP H+ P H P P P KP T H H
Sbjct: 98 HKP--PVHKP---PIHKPPVHNTPSVTDDHYPAHPIHKPQPIHRPPVHKPPTEHKPPVHE 152
Query: 84 QHQHHSPAP--------------SPYHVPTPLQRP-----AKPPTLHHHQHPP 117
H P+P P H+P + RP PP HH +HPP
Sbjct: 153 PATEHKPSPVYQPPKTEKPVPEHKPPHLPPIVVRPPPTHKPNPPYGHHPRHPP 205
Score = 43.1 bits (100), Expect = 6e-04
Identities = 34/100 (34%), Positives = 43/100 (43%), Gaps = 15/100 (15%)
Query: 30 HKPTTPLHPPTKSPVHKP-LAKPPTHAPHHHHH--HSPSHAPLPPPHPAKPLTHHHHQHQ 86
HKP P K P+HKP + PP H P + H P PP +T H+
Sbjct: 23 HKPPVYTPPVHKPPIHKPPVYTPPVHKPPVYTPPVHKPPSEYKPPVEATNSVTEDHYPI- 81
Query: 87 HHSPAPSPYHVPTPLQRPA---KPPTLHHHQHPPAHAPTH 123
H P P + P P+Q+PA KPP H+ P P H
Sbjct: 82 -HKP---PVYKP-PVQKPAPEHKPPV---HKPPIHKPPVH 113
Score = 36.2 bits (82), Expect = 0.079
Identities = 20/50 (40%), Positives = 26/50 (52%), Gaps = 10/50 (20%)
Query: 30 HKPTTPLHPP-TKSPV--HKP-------LAKPPTHAPHHHHHHSPSHAPL 69
HKP+ PP T+ PV HKP + PPTH P+ + H P H P+
Sbjct: 157 HKPSPVYQPPKTEKPVPEHKPPHLPPIVVRPPPTHKPNPPYGHHPRHPPV 206
Score = 36.2 bits (82), Expect = 0.079
Identities = 27/96 (28%), Positives = 35/96 (36%), Gaps = 23/96 (23%)
Query: 45 HKPLAKPPTHAPHHHH---HHSPSHAP-------------LPPPHPAKPLTHHHHQHQHH 88
H P+ KPP + P H H P + P PP P+ + + H
Sbjct: 19 HPPIHKPPVYTPPVHKPPIHKPPVYTPPVHKPPVYTPPVHKPPSEYKPPVEATNSVTEDH 78
Query: 89 SPAPSPYHVPTPLQRPA---KPPTLHHHQHPPAHAP 121
P P P+Q+PA KPP PP H P
Sbjct: 79 YPIHKPPVYKPPVQKPAPEHKPPV----HKPPIHKP 110
>EXTN_TOBAC (P13983) Extensin precursor (Cell wall
hydroxyproline-rich glycoprotein)
Length = 620
Score = 53.9 bits (128), Expect = 4e-07
Identities = 41/137 (29%), Positives = 57/137 (40%), Gaps = 23/137 (16%)
Query: 5 ILALIFLLLQITSFVVFAEELETLHH----KPTTPLHPPTK---SPVHKPLAKPPT---- 53
+ AL+ LL + A E T + P T PP+ SP P PP+
Sbjct: 6 LFALVLLLQSTAILSLVAAEATTQYGGYLPPPVTSQPPPSSIGLSPPSAPTTTPPSRGHV 65
Query: 54 ----HAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVP-TPLQRPAKPP 108
HAP H + PSH LPP P H+ H P ++ P +P+ P+ PP
Sbjct: 66 PSPRHAPPRHAYPPPSHGHLPPSVGGPP------PHRGHLPPSRGFNPPPSPVISPSHPP 119
Query: 109 TLHHHQHPPAHAPTHMP 125
+ PP+H P H+P
Sbjct: 120 P-SYGAPPPSHGPGHLP 135
Score = 51.2 bits (121), Expect = 2e-06
Identities = 40/130 (30%), Positives = 51/130 (38%), Gaps = 31/130 (23%)
Query: 27 TLHHKPTTPLH--------PPTKSP---VH-KPLAKPPTHA----PHHHHHHSPSHAPLP 70
T P TP++ PPT SP H +P PP+ P H H P+H P
Sbjct: 182 TYAQPPPTPIYSPSPQVQPPPTYSPPPPTHVQPTPSPPSRGHQPQPPTHRHAPPTHRHAP 241
Query: 71 PPHPAKPLTH----HHHQHQ-----------HHSPAPSPYHVPTPLQRPAKPPTLHHHQH 115
P H PL H Q Q SP PSP + P P PP+ +
Sbjct: 242 PTHQPSPLRHLPPSPRRQPQPPTYSPPPPAYAQSPQPSPTYSPPPPTYSPPPPSPIYSPP 301
Query: 116 PPAHAPTHMP 125
PPA++P+ P
Sbjct: 302 PPAYSPSPPP 311
Score = 47.8 bits (112), Expect = 3e-05
Identities = 40/140 (28%), Positives = 50/140 (35%), Gaps = 45/140 (32%)
Query: 29 HHKPTTPLHPPTK---SPVHKPL---AKPPTHAPHHHHHH-----SPSHAPLP------- 70
H P+ +PP SP H P A PP+H P H H SPSH P
Sbjct: 98 HLPPSRGFNPPPSPVISPSHPPPSYGAPPPSHGPGHLPSHGQRPPSPSHGHAPPSGGHTP 157
Query: 71 -----PPHPAKPLTHHHHQH----QHHSPAPSPYHVPTPLQRP---------------AK 106
PP +P H H + P P+P + P+P +P
Sbjct: 158 PRGQHPPSHRRPSPPSRHGHPPPPTYAQPPPTPIYSPSPQVQPPPTYSPPPPTHVQPTPS 217
Query: 107 PPTLHHHQHPPAH---APTH 123
PP+ H PP H PTH
Sbjct: 218 PPSRGHQPQPPTHRHAPPTH 237
Score = 45.4 bits (106), Expect = 1e-04
Identities = 33/100 (33%), Positives = 43/100 (43%), Gaps = 13/100 (13%)
Query: 32 PTTPLHPPTKSP----VHKPLAKPPTHAPHHHHHHSPSHAPL--PPPHPAKPLTHHHHQH 85
PT PPT SP +P PPT++P + P +P+ PPP +PL
Sbjct: 446 PTYSPPPPTYSPPPPAYAQPPPPPPTYSPPPPAYSPPPPSPIYSPPPPQVQPL-----PP 500
Query: 86 QHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
P P H+P P R +PPT + Q P PT P
Sbjct: 501 TFSPPPPRRIHLPPPPHRQPRPPTPTYGQ--PPSPPTFSP 538
Score = 45.4 bits (106), Expect = 1e-04
Identities = 29/97 (29%), Positives = 39/97 (39%), Gaps = 12/97 (12%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPH---PAKPLTHHHHQHQHH 88
PT PP PL PPT++P P+++P PP + P P T+ +
Sbjct: 387 PTYEQSPPPPPAYSPPLPAPPTYSPP-----PPTYSPPPPTYAQPPPLPPTYSPPPPAYS 441
Query: 89 SPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
P P Y P P P P + PP PT+ P
Sbjct: 442 PPPPPTYSPPPPTYSPPPPA----YAQPPPPPPTYSP 474
Score = 43.1 bits (100), Expect = 6e-04
Identities = 28/90 (31%), Positives = 37/90 (41%), Gaps = 15/90 (16%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
PT PPT SP A+PP P + P+++P PPP +SP
Sbjct: 407 PTYSPPPPTYSPPPPTYAQPPP-LPPTYSPPPPAYSPPPPP--------------TYSPP 451
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQHPPAHAP 121
P Y P P PP + PPA++P
Sbjct: 452 PPTYSPPPPAYAQPPPPPPTYSPPPPAYSP 481
Score = 42.4 bits (98), Expect = 0.001
Identities = 27/96 (28%), Positives = 40/96 (41%), Gaps = 8/96 (8%)
Query: 34 TPLHPPTKSPVHKP----LAKPPTHAPHHHHH----HSPSHAPLPPPHPAKPLTHHHHQH 85
+P PPT +P P + PPT++P + SP ++P PP + P +
Sbjct: 306 SPSPPPTPTPTFSPPPPAYSPPPTYSPPPPTYLPLPSSPIYSPPPPVYSPPPPPSYSPPP 365
Query: 86 QHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAP 121
+ P P P P P P P PPA++P
Sbjct: 366 PTYLPPPPPSSPPPPSFSPPPPTYEQSPPPPPAYSP 401
Score = 41.6 bits (96), Expect = 0.002
Identities = 33/99 (33%), Positives = 38/99 (38%), Gaps = 16/99 (16%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P T L PP S P PP P + P PP PA P +SP
Sbjct: 365 PPTYLPPPPPSSPPPPSFSPP---PPTYEQSPPPPPAYSPPLPAPPT---------YSPP 412
Query: 92 PSPYHVPTPL--QRPAKPPTLHHHQHPPAHAPTHMPRVS 128
P Y P P Q P PPT + PPA++P P S
Sbjct: 413 PPTYSPPPPTYAQPPPLPPT--YSPPPPAYSPPPPPTYS 449
Score = 40.8 bits (94), Expect = 0.003
Identities = 32/110 (29%), Positives = 44/110 (39%), Gaps = 18/110 (16%)
Query: 32 PTTPLHPPT-----KSPVHKPLAKPPTHAPHHHHHHSPSHAP-LPPPHPAKP----LTHH 81
PT PPT SP++ P PP ++P +SP LPPP P+ P +
Sbjct: 328 PTYSPPPPTYLPLPSSPIYSP--PPPVYSPPPPPSYSPPPPTYLPPPPPSSPPPPSFSPP 385
Query: 82 HHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHA------PTHMP 125
++ P P Y P P PP + PP +A PT+ P
Sbjct: 386 PPTYEQSPPPPPAYSPPLPAPPTYSPPPPTYSPPPPTYAQPPPLPPTYSP 435
Score = 39.7 bits (91), Expect = 0.007
Identities = 33/105 (31%), Positives = 47/105 (44%), Gaps = 21/105 (20%)
Query: 28 LHHKPTTPLH---PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPH-PAKPLTHHHH 83
L H P +P PPT SP PP +A SP+++P PP + P P +
Sbjct: 249 LRHLPPSPRRQPQPPTYSP------PPPAYA--QSPQPSPTYSPPPPTYSPPPPSPIYSP 300
Query: 84 QHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHA---PTHMP 125
+SP+P P PTP P P + PP ++ PT++P
Sbjct: 301 PPPAYSPSPPP--TPTPTFSPPPPA----YSPPPTYSPPPPTYLP 339
Score = 38.5 bits (88), Expect = 0.016
Identities = 32/113 (28%), Positives = 52/113 (45%), Gaps = 18/113 (15%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHH----HHHHSPSHAPLPPPHPA---KPLTHHHHQ 84
P P SP + P PPT++P + P+++P PPP P P +
Sbjct: 269 PPAYAQSPQPSPTYSP--PPPTYSPPPPSPIYSPPPPAYSPSPPPTPTPTFSPPPPAYSP 326
Query: 85 HQHHSPAPSPYHVP---TPLQRPAKPPTLHHHQHPPAHA---PTHMPRVSRSS 131
+SP P P ++P +P+ P PP ++ PP+++ PT++P SS
Sbjct: 327 PPTYSP-PPPTYLPLPSSPIYSP--PPPVYSPPPPPSYSPPPPTYLPPPPPSS 376
Score = 37.7 bits (86), Expect = 0.027
Identities = 32/103 (31%), Positives = 40/103 (38%), Gaps = 24/103 (23%)
Query: 31 KPTTPLH-----PPTKSP-----VHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTH 80
+P TP + PPT SP +H P PP H + P PP A P
Sbjct: 521 RPPTPTYGQPPSPPTFSPPPPRQIHSP---PPPHWQPRTPTPTYGQPPSPPTFSAPP--- 574
Query: 81 HHHQHQHHSPAPSPYHVPTP----LQRPAKPPTLHHHQHPPAH 119
Q HSP P P+ P P +P PPT + PP +
Sbjct: 575 ---PRQIHSP-PPPHRQPRPPTPTYGQPPSPPTTYSPPSPPPY 613
Score = 37.7 bits (86), Expect = 0.027
Identities = 34/120 (28%), Positives = 42/120 (34%), Gaps = 24/120 (20%)
Query: 30 HKPTTPLH-PPTKSPVHKPLAK-----PPTHAP----HHHHHHSPSHAPLPPPH-----P 74
+ P P + PP SP++ P PPT +P H P P PP P
Sbjct: 472 YSPPPPAYSPPPPSPIYSPPPPQVQPLPPTFSPPPPRRIHLPPPPHRQPRPPTPTYGQPP 531
Query: 75 AKPLTHHHHQHQHHSPAPSPYHVPTPLQ---RPAKPPTLHH------HQHPPAHAPTHMP 125
+ P Q HSP P + TP +P PPT H PP H P
Sbjct: 532 SPPTFSPPPPRQIHSPPPPHWQPRTPTPTYGQPPSPPTFSAPPPRQIHSPPPPHRQPRPP 591
>CBPY_SCHPO (O13849) Carboxypeptidase Y precursor (EC 3.4.16.5)
(CPY)
Length = 1002
Score = 53.9 bits (128), Expect = 4e-07
Identities = 38/109 (34%), Positives = 45/109 (40%), Gaps = 21/109 (19%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTH-----HHH 83
HH+P + PP P+H KP H P HH P PPP +P H HH
Sbjct: 200 HHEPGEHMPPP---PMHH---KPGEHMPPPPMHHEPGEHMPPPPMHHEPGEHMPPPPMHH 253
Query: 84 QHQHHSPAPSPYHVPTPLQRPAKPPTLHH----HQHPPA--HAP-THMP 125
+ H P P +H P PP +HH H PP H P HMP
Sbjct: 254 EPGEHMPPPPMHHEP---GEHMPPPPMHHEPGEHMPPPPMHHEPGEHMP 299
Score = 50.8 bits (120), Expect = 3e-06
Identities = 33/101 (32%), Positives = 42/101 (40%), Gaps = 17/101 (16%)
Query: 28 LHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHH-----H 82
+HH+P + PP P+H +P H P HH P PPP +P H H
Sbjct: 264 MHHEPGEHMPPP---PMHH---EPGEHMPPPPMHHEPGEHMPPPPMHHEPGEHMPPPPMH 317
Query: 83 HQHQHHSPAPSPYHVPTPLQRPAKPPTLHH----HQHPPAH 119
H+ H P P +H P P PP HH H+ P H
Sbjct: 318 HEPGEHMPPPPMHHEPGEHMPP--PPFKHHELEEHEGPEHH 356
Score = 50.4 bits (119), Expect = 4e-06
Identities = 32/99 (32%), Positives = 41/99 (41%), Gaps = 18/99 (18%)
Query: 28 LHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTH-----HH 82
+HH+P + PP P+H +P H P HH P PPP +P H H
Sbjct: 251 MHHEPGEHMPPP---PMHH---EPGEHMPPPPMHHEPGEHMPPPPMHHEPGEHMPPPPMH 304
Query: 83 HQHQHHSPAPSPYHVPTPLQRPAKPPTLHH----HQHPP 117
H+ H P P +H P PP +HH H PP
Sbjct: 305 HEPGEHMPPPPMHHEP---GEHMPPPPMHHEPGEHMPPP 340
Score = 50.4 bits (119), Expect = 4e-06
Identities = 43/133 (32%), Positives = 53/133 (39%), Gaps = 41/133 (30%)
Query: 28 LHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPS-HAPLPPPH-------PAKPLT 79
+HHKP + PP P+H +P H P HH P H P PP H P P+
Sbjct: 212 MHHKPGEHMPPP---PMHH---EPGEHMPPPPMHHEPGEHMPPPPMHHEPGEHMPPPPMH 265
Query: 80 H-----------HHHQHQHHSPAPSPY----HVPTP--LQRPAK---PPTLHH----HQH 115
H HH +H P P + H+P P P + PP +HH H
Sbjct: 266 HEPGEHMPPPPMHHEPGEHMPPPPMHHEPGEHMPPPPMHHEPGEHMPPPPMHHEPGEHMP 325
Query: 116 PPA--HAP-THMP 125
PP H P HMP
Sbjct: 326 PPPMHHEPGEHMP 338
Score = 45.1 bits (105), Expect = 2e-04
Identities = 26/90 (28%), Positives = 34/90 (36%), Gaps = 7/90 (7%)
Query: 28 LHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQH 87
+HH+P + PP P+H +P H P HH P PPP L H H
Sbjct: 303 MHHEPGEHMPPP---PMHH---EPGEHMPPPPMHHEPGEHMPPPPFKHHELEEHEGPEHH 356
Query: 88 HSPAPSPYHV-PTPLQRPAKPPTLHHHQHP 116
P +H P + P HH+ P
Sbjct: 357 RGPEDKEHHKGPKDKEHHKGPKDKEHHKGP 386
Score = 43.5 bits (101), Expect = 5e-04
Identities = 32/95 (33%), Positives = 36/95 (37%), Gaps = 26/95 (27%)
Query: 51 PPTHAPHHH------HHHSPSHAPLPPPH-------PAKPLTHHHHQHQHHSPAPSPYHV 97
P H P H HH H P PP H P P+ HH+ H P P +H
Sbjct: 198 PAHHEPGEHMPPPPMHHKPGEHMPPPPMHHEPGEHMPPPPM---HHEPGEHMPPPPMHHE 254
Query: 98 PTPLQRPAKPPTLHH----HQHPPA--HAP-THMP 125
P PP +HH H PP H P HMP
Sbjct: 255 P---GEHMPPPPMHHEPGEHMPPPPMHHEPGEHMP 286
Score = 42.4 bits (98), Expect = 0.001
Identities = 31/92 (33%), Positives = 34/92 (36%), Gaps = 25/92 (27%)
Query: 56 PHHHH----------HHSPSHAPLPPPHPAKPLTHH-----HHQHQHHSPAPSPYHVPTP 100
P HH HH P PPP KP H HH+ H P P +H P
Sbjct: 185 PKHHEFDDEDREFPAHHEPGEHMPPPPMHHKPGEHMPPPPMHHEPGEHMPPPPMHHEP-- 242
Query: 101 LQRPAKPPTLHH----HQHPPA--HAP-THMP 125
PP +HH H PP H P HMP
Sbjct: 243 -GEHMPPPPMHHEPGEHMPPPPMHHEPGEHMP 273
Score = 42.0 bits (97), Expect = 0.001
Identities = 27/96 (28%), Positives = 37/96 (38%), Gaps = 15/96 (15%)
Query: 28 LHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTH------- 80
+HH+P + PP P+H +P H P HH P PPP +P H
Sbjct: 290 MHHEPGEHMPPP---PMHH---EPGEHMPPPPMHHEPGEHMPPPPMHHEPGEHMPPPPFK 343
Query: 81 HHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHP 116
HH +H P + P + P HH+ P
Sbjct: 344 HHELEEHE--GPEHHRGPEDKEHHKGPKDKEHHKGP 377
>CRB5_MOUSE (Q8K1L0) cAMP response element binding protein 5
(CRE-BPa) (Fragment)
Length = 353
Score = 53.1 bits (126), Expect = 6e-07
Identities = 28/69 (40%), Positives = 29/69 (41%), Gaps = 12/69 (17%)
Query: 56 PHHH-HHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQ 114
PHHH H H H LPP HP H HQH H P P P+H P H H
Sbjct: 115 PHHHLHSHPHQHQTLPPHHPYP----HQHQHPAHHPHPQPHH-------QQNHPHHHSHS 163
Query: 115 HPPAHAPTH 123
H AH H
Sbjct: 164 HLHAHPAHH 172
Score = 48.9 bits (115), Expect = 1e-05
Identities = 31/87 (35%), Positives = 37/87 (41%), Gaps = 20/87 (22%)
Query: 34 TPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHH-HQHQHHSPAP 92
TP H P H+ PP H P+ H H P+H P P PH + HHH H H H PA
Sbjct: 114 TPHHHLHSHP-HQHQTLPPHH-PYPHQHQHPAHHPHPQPHHQQNHPHHHSHSHLHAHPA- 170
Query: 93 SPYHVPTPLQRPAKPPTLHHHQHPPAH 119
+H +P HPP H
Sbjct: 171 --HHQTSP--------------HPPLH 181
Score = 37.0 bits (84), Expect = 0.046
Identities = 25/67 (37%), Positives = 27/67 (39%), Gaps = 16/67 (23%)
Query: 29 HHKPTTPLHPPT----KSPVHKPLAKPPTHAPH-HHHHHSPSHAPLPPPHPAKPLTHHHH 83
H T P H P + P H P +P H HHH HS HA HPA HH
Sbjct: 124 HQHQTLPPHHPYPHQHQHPAHHPHPQPHHQQNHPHHHSHSHLHA-----HPA------HH 172
Query: 84 QHQHHSP 90
Q H P
Sbjct: 173 QTSPHPP 179
>CRB5_HUMAN (Q02930) cAMP response element binding protein 5
(CRE-BPa)
Length = 508
Score = 53.1 bits (126), Expect = 6e-07
Identities = 28/69 (40%), Positives = 29/69 (41%), Gaps = 12/69 (17%)
Query: 56 PHHH-HHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQ 114
PHHH H H H LPP HP H HQH H P P P+H P H H
Sbjct: 270 PHHHMHSHPHQHQTLPPHHPYP----HQHQHPAHHPHPQPHH-------QQNHPHHHSHS 318
Query: 115 HPPAHAPTH 123
H AH H
Sbjct: 319 HLHAHPAHH 327
Score = 49.3 bits (116), Expect = 9e-06
Identities = 31/87 (35%), Positives = 37/87 (41%), Gaps = 20/87 (22%)
Query: 34 TPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHH-HQHQHHSPAP 92
TP H P H+ PP H P+ H H P+H P P PH + HHH H H H PA
Sbjct: 269 TPHHHMHSHP-HQHQTLPPHH-PYPHQHQHPAHHPHPQPHHQQNHPHHHSHSHLHAHPA- 325
Query: 93 SPYHVPTPLQRPAKPPTLHHHQHPPAH 119
+H +P HPP H
Sbjct: 326 --HHQTSP--------------HPPLH 336
Score = 37.0 bits (84), Expect = 0.046
Identities = 25/67 (37%), Positives = 27/67 (39%), Gaps = 16/67 (23%)
Query: 29 HHKPTTPLHPPT----KSPVHKPLAKPPTHAPH-HHHHHSPSHAPLPPPHPAKPLTHHHH 83
H T P H P + P H P +P H HHH HS HA HPA HH
Sbjct: 279 HQHQTLPPHHPYPHQHQHPAHHPHPQPHHQQNHPHHHSHSHLHA-----HPA------HH 327
Query: 84 QHQHHSP 90
Q H P
Sbjct: 328 QTSPHPP 334
>SELP_BOVIN (P49907) Selenoprotein P-like protein precursor
Length = 402
Score = 52.4 bits (124), Expect = 1e-06
Identities = 28/64 (43%), Positives = 31/64 (47%), Gaps = 6/64 (9%)
Query: 54 HAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPT--LH 111
H PH H H P P P PHP H H++ H S +P P TP P PPT LH
Sbjct: 205 HHPHPHSHPHPHPHPHPHPHPHPHHGHQLHENAHLSESPKP---DTP-DTPENPPTSGLH 260
Query: 112 HHQH 115
HH H
Sbjct: 261 HHHH 264
Score = 41.2 bits (95), Expect = 0.002
Identities = 32/97 (32%), Positives = 37/97 (37%), Gaps = 14/97 (14%)
Query: 24 ELETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHH------HHSPSHAPLPPPHPAKP 77
E HH P HP P P P H PHH H H S S P P P P
Sbjct: 199 EASQRHHHPHPHSHP---HPHPHPHPHPHPH-PHHGHQLHENAHLSESPKPDTPDTPENP 254
Query: 78 LT----HHHHQHQHHSPAPSPYHVPTPLQRPAKPPTL 110
T HHHH+H+ + TPL + P+L
Sbjct: 255 PTSGLHHHHHRHKGPQRQGHSDNCDTPLGSESLQPSL 291
Score = 35.0 bits (79), Expect = 0.18
Identities = 15/41 (36%), Positives = 18/41 (43%)
Query: 75 AKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQH 115
A+ HHH H H P P P+ P P P LH + H
Sbjct: 198 AEASQRHHHPHPHSHPHPHPHPHPHPHPHPHHGHQLHENAH 238
>FXB2_MOUSE (Q64733) Forkhead box protein B2 (Transcription factor
FKH-4)
Length = 428
Score = 52.4 bits (124), Expect = 1e-06
Identities = 27/63 (42%), Positives = 35/63 (54%), Gaps = 9/63 (14%)
Query: 54 HAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPT-PLQRP---AKPPT 109
HA HHHHHH P P PP P + + HQ +PAP P H+P+ P Q+P ++PP
Sbjct: 153 HAAHHHHHHHPPQPP--PPPPPHMVPYF---HQQPAPAPQPPHLPSQPAQQPQPQSQPPQ 207
Query: 110 LHH 112
H
Sbjct: 208 TSH 210
Score = 52.0 bits (123), Expect = 1e-06
Identities = 30/71 (42%), Positives = 31/71 (43%), Gaps = 13/71 (18%)
Query: 52 PTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYH-VPTPLQRPAKPPTL 110
P HA HHHHHH H A HHHH H P P P H VP Q+PA P
Sbjct: 141 PHHAHHHHHHHH---------HAAH---HHHHHHPPQPPPPPPPHMVPYFHQQPAPAPQP 188
Query: 111 HHHQHPPAHAP 121
H PA P
Sbjct: 189 PHLPSQPAQQP 199
Score = 49.3 bits (116), Expect = 9e-06
Identities = 29/75 (38%), Positives = 33/75 (43%), Gaps = 7/75 (9%)
Query: 58 HHHHHSPSHAPLPPP----HPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTL--H 111
H H S AP P HP P HHH H HH A +H P P PP + +
Sbjct: 118 HLHSGSSKGAPGTGPGGHLHPHHPHHAHHHHHHHHHAAHHHHHHHPPQPPPPPPPHMVPY 177
Query: 112 HHQHP-PAHAPTHMP 125
HQ P PA P H+P
Sbjct: 178 FHQQPAPAPQPPHLP 192
Score = 40.8 bits (94), Expect = 0.003
Identities = 24/77 (31%), Positives = 27/77 (34%), Gaps = 30/77 (38%)
Query: 51 PPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTL 110
P T H H H P HA HHHH H HH+ +H
Sbjct: 128 PGTGPGGHLHPHHPHHA------------HHHHHHHHHAA----HH-------------- 157
Query: 111 HHHQHPPAHAPTHMPRV 127
HHH HPP P P +
Sbjct: 158 HHHHHPPQPPPPPPPHM 174
Score = 33.5 bits (75), Expect = 0.51
Identities = 25/79 (31%), Positives = 30/79 (37%), Gaps = 7/79 (8%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPH--HHHHHSPSHAPLPPPHPAKPLTHHHHQHQ 86
HH H H P PP PH + H P+ AP PP P++P Q Q
Sbjct: 145 HHHHHHHHHAAHHHHHHHPPQPPPPPPPHMVPYFHQQPAPAPQPPHLPSQPA----QQPQ 200
Query: 87 HHSPAPSPYHVPTPLQRPA 105
S P H P +Q A
Sbjct: 201 PQSQPPQTSH-PGKMQEAA 218
>EXTN_DAUCA (P06599) Extensin precursor
Length = 306
Score = 52.4 bits (124), Expect = 1e-06
Identities = 35/106 (33%), Positives = 53/106 (49%), Gaps = 21/106 (19%)
Query: 30 HKPTTPLH------PPTKSPVHKPLAKPPT-HAPHHHHHHSPSHAPLPPPHPAKPLTHHH 82
H P P+H PP +PV+K + PP H+P HH+ +P PP H P HH
Sbjct: 121 HSPA-PVHHYKYKSPPPPTPVYKYKSPPPPKHSPAPEHHYKYK-SPPPPKH--FPAPEHH 176
Query: 83 HQHQHHSPAPSPYHVPTPLQR---PAKPPTLHHHQHPPAHAPTHMP 125
+++++ SP P PTP+ + P P ++ ++ PP P H P
Sbjct: 177 YKYKYKSPPP-----PTPVYKYKSPPPPTPVYKYKSPP--PPKHSP 215
Score = 50.8 bits (120), Expect = 3e-06
Identities = 33/113 (29%), Positives = 48/113 (42%), Gaps = 16/113 (14%)
Query: 29 HHKPTTPLH-PPTKSPVHKPLAKPPT-HAPHHHHHHS----PSHAPLPP--------PHP 74
+ P P H PP +PV+K + PP H+P +H P H+P PP P P
Sbjct: 59 YESPPPPKHSPPPPTPVYKYKSPPPPMHSPPPPYHFESPPPPKHSPPPPTPVYKYKSPPP 118
Query: 75 AK--PLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
K P HH++++ P Y +P P HH+++ P H P
Sbjct: 119 PKHSPAPVHHYKYKSPPPPTPVYKYKSPPPPKHSPAPEHHYKYKSPPPPKHFP 171
Score = 50.4 bits (119), Expect = 4e-06
Identities = 36/107 (33%), Positives = 52/107 (47%), Gaps = 21/107 (19%)
Query: 32 PTTPLH----PPTKSPVHKPLAKPPT-HAP---HHHHHHSPS------HAPLPPPHPAKP 77
P TP++ PP +PV+K + PP H+P HH+ + SP +P PP H P
Sbjct: 186 PPTPVYKYKSPPPPTPVYKYKSPPPPKHSPAPVHHYKYKSPPPPTPVYKSPPPPEHSPPP 245
Query: 78 LTHHHHQHQHHSPAPSPYH---VPTPLQRPAKPPTLHHHQHPPAHAP 121
T +++ SP P P H PTP+ + PP H PP ++P
Sbjct: 246 PT---PVYKYKSP-PPPMHSPPPPTPVYKYKSPPPPMHSPPPPVYSP 288
Score = 47.4 bits (111), Expect = 3e-05
Identities = 39/132 (29%), Positives = 58/132 (43%), Gaps = 25/132 (18%)
Query: 1 MAKIILALIFLLLQITSFVVFAEELETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHH 60
M+ +I++L+ +L+ + A E + + P PP SP PP H+P +
Sbjct: 10 MSSLIVSLLVVLVSLN----LASETTAKYTYSSPP--PPEHSP------PPPEHSPPPPY 57
Query: 61 HHS----PSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHV---PTPLQRPAKPPTLHHH 113
H+ P H+P PPP P + HSP P PYH P P P P ++ +
Sbjct: 58 HYESPPPPKHSP-PPPTPV--YKYKSPPPPMHSP-PPPYHFESPPPPKHSPPPPTPVYKY 113
Query: 114 QHPPAHAPTHMP 125
+ PP P H P
Sbjct: 114 KSPP--PPKHSP 123
Score = 45.8 bits (107), Expect = 1e-04
Identities = 29/91 (31%), Positives = 40/91 (43%), Gaps = 17/91 (18%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P TP++ P H P PPT + P H+P PPP P +++ SP
Sbjct: 228 PPTPVYKSPPPPEHSP--PPPTPVYKYKSPPPPMHSP-PPPTPV---------YKYKSPP 275
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQH---PPAH 119
P + P P+ P PP HH+ + PP H
Sbjct: 276 PPMHSPPPPVYSP--PPPKHHYSYTSPPPPH 304
Score = 44.3 bits (103), Expect = 3e-04
Identities = 31/111 (27%), Positives = 45/111 (39%), Gaps = 9/111 (8%)
Query: 22 AEELETLHHKPTTPLHPPTKSPVHKPLAK-PPTHAPHHHHHHSPSHAPL-----PPPHPA 75
A E + P P H P +K K PP P + + P P+ PPP
Sbjct: 154 APEHHYKYKSPPPPKHFPAPEHHYKYKYKSPPPPTPVYKYKSPPPPTPVYKYKSPPPPKH 213
Query: 76 KPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPT-LHHHQHPPAHAPTHMP 125
P HH++++ P Y P P + PPT ++ ++ PP P H P
Sbjct: 214 SPAPVHHYKYKSPPPPTPVYKSPPPPEHSPPPPTPVYKYKSPP--PPMHSP 262
Score = 43.5 bits (101), Expect = 5e-04
Identities = 29/102 (28%), Positives = 41/102 (39%), Gaps = 10/102 (9%)
Query: 29 HHKPTTPLHPPTKSPVHK-PLAKPPTHAPHHHHHHSPSHAPLPPPHPA---KPLTHHHHQ 84
+ P P H P +K PP H P HH+ + PPP P K
Sbjct: 143 YKSPPPPKHSPAPEHHYKYKSPPPPKHFPAPEHHYKYKYKSPPPPTPVYKYKSPPPPTPV 202
Query: 85 HQHHSPAPSPYHVPTPL-----QRPAKPPTLHHHQHPPAHAP 121
+++ SP P P H P P+ + P P ++ PP H+P
Sbjct: 203 YKYKSP-PPPKHSPAPVHHYKYKSPPPPTPVYKSPPPPEHSP 243
Score = 38.9 bits (89), Expect = 0.012
Identities = 23/78 (29%), Positives = 36/78 (45%), Gaps = 13/78 (16%)
Query: 28 LHHKPTTPLH-PPTKSPVHK------PLAKPPTHAPHHHHHH--SPSHAPLPPPHPAKPL 78
++ P P H PP +PV+K P+ PP P + + P H+P PP + P
Sbjct: 232 VYKSPPPPEHSPPPPTPVYKYKSPPPPMHSPPPPTPVYKYKSPPPPMHSPPPPVYSPPPP 291
Query: 79 THHHHQHQHHSPAPSPYH 96
HH+ ++ P P+H
Sbjct: 292 KHHY----SYTSPPPPHH 305
>EXT1_ARATH (Q38913) Extensin 1 precursor (AtExt1) (AtExt4)
Length = 373
Score = 52.4 bits (124), Expect = 1e-06
Identities = 38/124 (30%), Positives = 54/124 (42%), Gaps = 31/124 (25%)
Query: 28 LHHKPTTPLH---PPT-----KSPVHKPLAKPPT--HAPHHHHHHSPS----HAPLPPPH 73
++H P P+H PP PVH + PP H+P H+SP H+P PP H
Sbjct: 254 VYHSPPPPVHYSPPPVVYHSPPPPVH--YSPPPVVYHSPPPPVHYSPPPVVYHSPPPPVH 311
Query: 74 PAKPLTHHH-----HQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAH--APTHMPR 126
+ P +H +H + P P H PPT++H PP H +P H P
Sbjct: 312 YSPPPVVYHSPPPPKKHYEYKSPPPPVHY--------SPPTVYHSPPPPVHHYSPPHQPY 363
Query: 127 VSRS 130
+ +S
Sbjct: 364 LYKS 367
Score = 52.4 bits (124), Expect = 1e-06
Identities = 28/84 (33%), Positives = 37/84 (43%), Gaps = 2/84 (2%)
Query: 38 PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQ--HHSPAPSPY 95
PP K P+ K P H+ H+P PP H + P +H H+SP P Y
Sbjct: 228 PPVKYYSPPPVYKSPPPPVHYSPPPVVYHSPPPPVHYSPPPVVYHSPPPPVHYSPPPVVY 287
Query: 96 HVPTPLQRPAKPPTLHHHQHPPAH 119
H P P + PP ++H PP H
Sbjct: 288 HSPPPPVHYSPPPVVYHSPPPPVH 311
Score = 51.6 bits (122), Expect = 2e-06
Identities = 31/91 (34%), Positives = 43/91 (47%), Gaps = 10/91 (10%)
Query: 35 PLHPPTKSPVHKPLAKPPT--HAPHHHHHHSPS----HAPLPPPHPAKPLTHHHHQHQ-- 86
P++ PVH + PP H+P H+SP H+P PP H + P +H
Sbjct: 237 PVYKSPPPPVH--YSPPPVVYHSPPPPVHYSPPPVVYHSPPPPVHYSPPPVVYHSPPPPV 294
Query: 87 HHSPAPSPYHVPTPLQRPAKPPTLHHHQHPP 117
H+SP P YH P P + PP ++H PP
Sbjct: 295 HYSPPPVVYHSPPPPVHYSPPPVVYHSPPPP 325
Score = 47.4 bits (111), Expect = 3e-05
Identities = 29/102 (28%), Positives = 46/102 (44%), Gaps = 14/102 (13%)
Query: 28 LHHKPTTPLHPPTKSPVHKPLAKPPT--HAPHHHHHHSPS----HAPLPPPHPAKPLTHH 81
+H+ P ++ PVH + PP H+P H+SP H+P PP H + P
Sbjct: 246 VHYSPPPVVYHSPPPPVH--YSPPPVVYHSPPPPVHYSPPPVVYHSPPPPVHYSPPPV-- 301
Query: 82 HHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
+HSP P ++ P P+ + PP H+++ P H
Sbjct: 302 ----VYHSPPPPVHYSPPPVVYHSPPPPKKHYEYKSPPPPVH 339
Score = 47.0 bits (110), Expect = 4e-05
Identities = 29/87 (33%), Positives = 36/87 (41%), Gaps = 8/87 (9%)
Query: 38 PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAK-----PLTHHHHQHQHHSPAP 92
PP K P+ K P P H+SP PP P K P+ H+SP P
Sbjct: 196 PPVKYYSPPPVYKSP---PPPVKHYSPPPVYKSPPPPVKYYSPPPVYKSPPPPVHYSPPP 252
Query: 93 SPYHVPTPLQRPAKPPTLHHHQHPPAH 119
YH P P + PP ++H PP H
Sbjct: 253 VVYHSPPPPVHYSPPPVVYHSPPPPVH 279
Score = 40.0 bits (92), Expect = 0.005
Identities = 24/85 (28%), Positives = 35/85 (40%), Gaps = 4/85 (4%)
Query: 38 PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPP---HPAKPLTHHHHQHQHHSPAPSP 94
PP K P+ K P P ++ P + PPP P P+ H+ + SP P
Sbjct: 60 PPVKHYSPPPVYKSPP-PPVKYYSPPPVYKSPPPPVYKSPPPPVKHYSPPPVYKSPPPPV 118
Query: 95 YHVPTPLQRPAKPPTLHHHQHPPAH 119
H P + PP + H+ PP +
Sbjct: 119 KHYSPPPVYKSPPPPVKHYSPPPVY 143
Score = 39.3 bits (90), Expect = 0.009
Identities = 27/96 (28%), Positives = 40/96 (41%), Gaps = 10/96 (10%)
Query: 28 LHHKPTTPLHPPTKSPVHK----PLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHH 83
++ P P+ + PV+K P+ K P P H+SP PP P K H+
Sbjct: 70 VYKSPPPPVKYYSPPPVYKSPPPPVYKSP---PPPVKHYSPPPVYKSPPPPVK---HYSP 123
Query: 84 QHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
+ SP P H P + PP + H+ PP +
Sbjct: 124 PPVYKSPPPPVKHYSPPPVYKSPPPPVKHYSPPPVY 159
Score = 38.9 bits (89), Expect = 0.012
Identities = 28/108 (25%), Positives = 45/108 (40%), Gaps = 18/108 (16%)
Query: 28 LHHKPTTPLH----PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPP----------- 72
++ P P++ PP K P+ K P P H+ P + PPP
Sbjct: 86 VYKSPPPPVYKSPPPPVKHYSPPPVYKSPP-PPVKHYSPPPVYKSPPPPVKHYSPPPVYK 144
Query: 73 HPAKPLTHHHHQHQHHSPAPS-PYHVPTPLQRPAKPPTLHHHQHPPAH 119
P P+ H+ + SP P Y+ P P+ + + PP + H+ PP +
Sbjct: 145 SPPPPVKHYSPPPVYKSPPPPVKYYSPPPVYK-SPPPPVKHYSPPPVY 191
Score = 38.1 bits (87), Expect = 0.021
Identities = 26/94 (27%), Positives = 39/94 (40%), Gaps = 14/94 (14%)
Query: 38 PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPP-----------HPAKPLTHHHHQHQ 86
PP K P+ K P P H+ P + PPP P P+ H+
Sbjct: 132 PPVKHYSPPPVYKSPP-PPVKHYSPPPVYKSPPPPVKYYSPPPVYKSPPPPVKHYSPPPV 190
Query: 87 HHSPAPS-PYHVPTPLQRPAKPPTLHHHQHPPAH 119
+ SP P Y+ P P+ + + PP + H+ PP +
Sbjct: 191 YKSPPPPVKYYSPPPVYK-SPPPPVKHYSPPPVY 223
Score = 37.4 bits (85), Expect = 0.035
Identities = 27/101 (26%), Positives = 39/101 (37%), Gaps = 20/101 (19%)
Query: 38 PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPP-----------HPAKPLTHHHHQHQ 86
PP K P+ K P P H+ P + PPP P P+ H+
Sbjct: 164 PPVKYYSPPPVYKSPP-PPVKHYSPPPVYKSPPPPVKYYSPPPVYKSPPPPVKHYSPPPV 222
Query: 87 HHSPAPS-PYHVPTPLQRP-------AKPPTLHHHQHPPAH 119
+ SP P Y+ P P+ + + PP ++H PP H
Sbjct: 223 YKSPPPPVKYYSPPPVYKSPPPPVHYSPPPVVYHSPPPPVH 263
Score = 37.0 bits (84), Expect = 0.046
Identities = 26/85 (30%), Positives = 36/85 (41%), Gaps = 14/85 (16%)
Query: 28 LHHKPTTPLH---PPT-----KSPVHKPLAKPPT--HAPHHHHHHSPSHAPLPPPHPAKP 77
++H P P+H PP PVH + PP H+P H +P PP H + P
Sbjct: 286 VYHSPPPPVHYSPPPVVYHSPPPPVH--YSPPPVVYHSPPPPKKHYEYKSPPPPVHYSPP 343
Query: 78 LTHHHHQH--QHHSPAPSPYHVPTP 100
+H H+SP PY +P
Sbjct: 344 TVYHSPPPPVHHYSPPHQPYLYKSP 368
Score = 35.8 bits (81), Expect = 0.10
Identities = 29/119 (24%), Positives = 44/119 (36%), Gaps = 20/119 (16%)
Query: 10 FLLLQITSFVVFAEELETLHHKPTTPLHPPTKSPVHKPLAKPPTH---------APHHHH 60
FL+L + V + P P+ + PV+K P H P
Sbjct: 4 FLVLAFSLAFVSQTTANYFYSSPPPPVKHYSPPPVYKSPPPPVKHYSPPPVYKSPPPPVK 63
Query: 61 HHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
H+SP PP P K ++SP P P P+ + + PP + H+ PP +
Sbjct: 64 HYSPPPVYKSPPPPVK----------YYSPPPVYKSPPPPVYK-SPPPPVKHYSPPPVY 111
Score = 33.1 bits (74), Expect = 0.67
Identities = 22/75 (29%), Positives = 31/75 (41%), Gaps = 10/75 (13%)
Query: 28 LHHKPTTPLHPPTKSPVHKPLAKPPTH----APHHHHHHSPS---HAPLPPPHPAKPLTH 80
++H P P+H V+ P H +P H+SP H+P PP H P
Sbjct: 302 VYHSPPPPVHYSPPPVVYHSPPPPKKHYEYKSPPPPVHYSPPTVYHSPPPPVHHYSP--- 358
Query: 81 HHHQHQHHSPAPSPY 95
H + + SP P Y
Sbjct: 359 PHQPYLYKSPPPPHY 373
>APG_ARATH (P40602) Anter-specific proline-rich protein APG
precursor
Length = 534
Score = 52.4 bits (124), Expect = 1e-06
Identities = 40/134 (29%), Positives = 51/134 (37%), Gaps = 17/134 (12%)
Query: 5 ILALIFLLLQITSFVVFAEELETLHHK------------PTTPLHPPTKSPVHKPLAKP- 51
I IF LL F + + +H + P++PPT P KP+A P
Sbjct: 15 IFRSIFCLLSFCIFFLTTTNAQVMHRRLWPWPLWPRPYPQPWPMNPPTPDPSPKPVAPPG 74
Query: 52 PTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLH 111
P+ P SP +P P P P P P+P P P P+ PA PPT
Sbjct: 75 PSSKPVAPPGPSPCPSPPPKPQPKPPPA----PSPSPCPSPPPKPQPKPVPPPACPPTPP 130
Query: 112 HHQHPPAHAPTHMP 125
Q PA P P
Sbjct: 131 KPQPKPAPPPEPKP 144
Score = 47.4 bits (111), Expect = 3e-05
Identities = 36/99 (36%), Positives = 38/99 (38%), Gaps = 17/99 (17%)
Query: 32 PTTPLHPPTKSPVHKPLAKP--PTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHS 89
P P P SP KP KP P P P AP P P PA P
Sbjct: 100 PPAPSPSPCPSPPPKPQPKPVPPPACPPTPPKPQPKPAPPPEPKPAPP------------ 147
Query: 90 PAPSPYHVPTPLQRPA---KPPTLHHHQHPPAHAPTHMP 125
PAP P P+P + PA KP H PA APT P
Sbjct: 148 PAPKPVPCPSPPKPPAPTPKPVPPHGPPPKPAPAPTPAP 186
Score = 42.0 bits (97), Expect = 0.001
Identities = 27/76 (35%), Positives = 31/76 (40%), Gaps = 5/76 (6%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
PT P P +P +P PP AP SP P P P P P H +PA
Sbjct: 127 PTPPKPQPKPAPPPEPKPAPPP-APKPVPCPSPPKPPAPTPKPVPP----HGPPPKPAPA 181
Query: 92 PSPYHVPTPLQRPAKP 107
P+P P P P KP
Sbjct: 182 PTPAPSPKPAPSPPKP 197
Score = 31.2 bits (69), Expect = 2.5
Identities = 24/72 (33%), Positives = 26/72 (35%), Gaps = 20/72 (27%)
Query: 31 KPTTPLHP-PTKSPVHKPLAKP-------PTHAPHHHHHHSPSHAPLPPPHPAKPLTHHH 82
KP P P P P KP+ P PT P H P AP P P P+
Sbjct: 135 KPAPPPEPKPAPPPAPKPVPCPSPPKPPAPTPKPVPPHGPPPKPAPAPTPAPSP------ 188
Query: 83 HQHQHHSPAPSP 94
PAPSP
Sbjct: 189 ------KPAPSP 194
>FXL2_HUMAN (P58012) Forkhead box protein L2
Length = 376
Score = 52.0 bits (123), Expect = 1e-06
Identities = 39/112 (34%), Positives = 52/112 (45%), Gaps = 11/112 (9%)
Query: 48 LAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYH----VPTPLQ- 102
+A PP ++ P AP PPPHP P H HH H +P P+P H P P Q
Sbjct: 264 MALPPGVVNSYNGLGGPPAAPPPPPHP-HPHPHAHHLHAAAAPPPAPPHHGAAAPPPGQL 322
Query: 103 RPAKPPTLHHHQHPPAHAPTHMPRVSRSSIAVEGVVYVKSCHHAGFDTLKGA 154
PA P T PPA APT P + + + A + + + C + D+ GA
Sbjct: 323 SPASPAT----AAPPAPAPTSAPGL-QFACARQPELAMMHCSYWDHDSKTGA 369
>FXGB_MOUSE (Q60987) Forkhead box protein G1B (Forkhead-related
protein FKHL1) (Transcription factor BF-1) (Brain factor
1) (BF1)
Length = 481
Score = 52.0 bits (123), Expect = 1e-06
Identities = 27/69 (39%), Positives = 29/69 (41%), Gaps = 21/69 (30%)
Query: 54 HAPH-HHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHH 112
HA H HH+ H P H HHHH H HH P P+P Q P PP
Sbjct: 34 HASHGHHNSHHPQH-------------HHHHHHHHHPPPPAP-------QPPPPPPQQQQ 73
Query: 113 HQHPPAHAP 121
Q PPA P
Sbjct: 74 QQPPPAPQP 82
Score = 46.6 bits (109), Expect = 6e-05
Identities = 21/44 (47%), Positives = 22/44 (49%), Gaps = 6/44 (13%)
Query: 51 PPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSP 94
P H HHHHHH P AP PPP P + Q Q PAP P
Sbjct: 45 PQHHHHHHHHHHPPPPAPQPPPPPPQ------QQQQQPPPAPQP 82
Score = 38.5 bits (88), Expect = 0.016
Identities = 23/84 (27%), Positives = 31/84 (36%), Gaps = 16/84 (19%)
Query: 52 PTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLH 111
P + +HH S H H+ H QHH +H P P +P PP
Sbjct: 25 PEAVQNDNHHASHGH-------------HNSHHPQHHHHHHHHHHPPPPAPQPPPPPPQQ 71
Query: 112 HHQHPPAHAPTHMPRVSRSSIAVE 135
Q PP P P +R + A +
Sbjct: 72 QQQQPP---PAPQPPQARGAPAAD 92
>EXTN_SORBI (P24152) Extensin precursor (Proline-rich glycoprotein)
Length = 283
Score = 52.0 bits (123), Expect = 1e-06
Identities = 41/136 (30%), Positives = 46/136 (33%), Gaps = 22/136 (16%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHS-- 89
P P P PV+ P KPP P P + P P P KP TH S
Sbjct: 134 PKPPATKPPTPPVYTPSPKPPVTKPPTPKPTPPVYTPNPKPPVTKPPTHTPSPKPPTSKP 193
Query: 90 -----------PAPS-PYHVPTPLQRPAKPPT--------LHHHQHPPAHAPTHMPRVSR 129
P PS P + PTP KPPT H +P AH PT+ P
Sbjct: 194 TPPVYTPSPKPPKPSPPTYTPTPKPPATKPPTSTPTHPKPTPHTPYPQAHPPTYKPAPKP 253
Query: 130 SSIAVEGVVYVKSCHH 145
S A Y H
Sbjct: 254 SPPAPTPPTYTPPVSH 269
Score = 50.4 bits (119), Expect = 4e-06
Identities = 34/116 (29%), Positives = 46/116 (39%), Gaps = 20/116 (17%)
Query: 30 HKPTTPLH----------------PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPH 73
HKPT P + PPT +P KP K P + P P++ P P P
Sbjct: 67 HKPTPPTYTPSPKPTPPPATPKPTPPTYTPSPKP--KSPVYPPPPKASTPPTYTPSPKPP 124
Query: 74 PAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSR 129
KP T+ + P P + P+P KPPT PP + P P V++
Sbjct: 125 ATKPPTYPTPKPPATKPPTPPVYTPSPKPPVTKPPT--PKPTPPVYTPNPKPPVTK 178
Score = 48.1 bits (113), Expect = 2e-05
Identities = 35/96 (36%), Positives = 41/96 (42%), Gaps = 8/96 (8%)
Query: 31 KPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSP 90
K P PPTK HKP P H P ++PS P PPP KP + SP
Sbjct: 44 KGPKPEKPPTKGHGHKPEKPPKEHKP-TPPTYTPSPKPTPPPATPKPTPPTYTP----SP 98
Query: 91 AP-SPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
P SP + P P + + PPT PPA P P
Sbjct: 99 KPKSPVYPPPP--KASTPPTYTPSPKPPATKPPTYP 132
Score = 47.0 bits (110), Expect = 4e-05
Identities = 31/91 (34%), Positives = 34/91 (37%), Gaps = 17/91 (18%)
Query: 31 KPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSP 90
KP+ P + PT P P KPPT P H P P PH P H + P
Sbjct: 206 KPSPPTYTPTPKP---PATKPPTSTPTH---------PKPTPHTPYPQAH----PPTYKP 249
Query: 91 APSPYHVPTPLQRPAKPPTLHHHQHPPAHAP 121
AP P P P PP H PP P
Sbjct: 250 APKP-SPPAPTPPTYTPPVSHTPSSPPPPPP 279
Score = 39.7 bits (91), Expect = 0.007
Identities = 21/48 (43%), Positives = 23/48 (47%), Gaps = 5/48 (10%)
Query: 30 HKPTTPLHPPTKSPVHK---PLAKPPTHAPHHHHHHSPSHAPLPPPHP 74
H P HPPT P K P PPT+ P H+PS P PPP P
Sbjct: 236 HTPYPQAHPPTYKPAPKPSPPAPTPPTYTP--PVSHTPSSPPPPPPPP 281
Score = 32.0 bits (71), Expect = 1.5
Identities = 22/71 (30%), Positives = 28/71 (38%), Gaps = 12/71 (16%)
Query: 64 PSHAPLPPP---HPAKPLTHHHHQH-----QHHSPAPSPYHVPTPLQRPAKPPTLHHHQH 115
P+ P PP P KP T H + H P P Y TP +P PP
Sbjct: 35 PTPTPKPPAKGPKPEKPPTKGHGHKPEKPPKEHKPTPPTY---TPSPKPTPPPAT-PKPT 90
Query: 116 PPAHAPTHMPR 126
PP + P+ P+
Sbjct: 91 PPTYTPSPKPK 101
>FXGB_RAT (Q00939) Forkhead box protein G1B (Forkhead-related
protein FKHL1) (Transcription factor BF-1) (Brain factor
1) (BF1)
Length = 480
Score = 51.2 bits (121), Expect = 2e-06
Identities = 23/56 (41%), Positives = 27/56 (48%), Gaps = 14/56 (25%)
Query: 54 HAPH-HHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPP 108
HA H HH+ H P H HHHH H HH P P+P P P Q+ +PP
Sbjct: 34 HASHGHHNSHHPQH-------------HHHHHHHHHPPPPAPQPPPPPPQQQQQPP 76
Score = 46.2 bits (108), Expect = 8e-05
Identities = 21/44 (47%), Positives = 22/44 (49%), Gaps = 7/44 (15%)
Query: 51 PPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSP 94
P H HHHHHH P AP PPP P + Q Q PAP P
Sbjct: 45 PQHHHHHHHHHHPPPPAPQPPPPPPQ-------QQQQPPPAPQP 81
Score = 39.3 bits (90), Expect = 0.009
Identities = 21/70 (30%), Positives = 26/70 (37%), Gaps = 13/70 (18%)
Query: 52 PTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLH 111
P + +HH S H H+ H QHH +H P P +P PP
Sbjct: 25 PEAVQNDNHHASHGH-------------HNSHHPQHHHHHHHHHHPPPPAPQPPPPPPQQ 71
Query: 112 HHQHPPAHAP 121
Q PPA P
Sbjct: 72 QQQPPPAPQP 81
>FXGA_HUMAN (P55316) Forkhead box protein G1A (Forkhead-related
protein FKHL2) (Transcription factor BF-2) (Brain factor
2) (BF2) (HFK2)
Length = 469
Score = 50.8 bits (120), Expect = 3e-06
Identities = 27/84 (32%), Positives = 34/84 (40%), Gaps = 13/84 (15%)
Query: 54 HAPH-HHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHH 112
HA H HH+ H P H HHHH H HH P P+P P P Q+ PP
Sbjct: 34 HASHGHHNSHHPQHH------------HHHHHHHHHPPPPAPQPPPPPQQQQPPPPPRRG 81
Query: 113 HQHPPAHAPTHMPRVSRSSIAVEG 136
+ P+ + + A EG
Sbjct: 82 ARRRRRRGPSSCCSAAHAHGAPEG 105
Score = 37.4 bits (85), Expect = 0.035
Identities = 19/45 (42%), Positives = 19/45 (42%), Gaps = 13/45 (28%)
Query: 52 PTHAPH--HHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSP 94
P H H HHHHH P AP PPP P Q P P P
Sbjct: 45 PQHHHHHHHHHHHPPPPAPQPPPPP-----------QQQQPPPPP 78
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.134 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,080,970
Number of Sequences: 164201
Number of extensions: 1724497
Number of successful extensions: 20006
Number of sequences better than 10.0: 965
Number of HSP's better than 10.0 without gapping: 337
Number of HSP's successfully gapped in prelim test: 661
Number of HSP's that attempted gapping in prelim test: 9761
Number of HSP's gapped (non-prelim): 4464
length of query: 255
length of database: 59,974,054
effective HSP length: 108
effective length of query: 147
effective length of database: 42,240,346
effective search space: 6209330862
effective search space used: 6209330862
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC135464.6


