

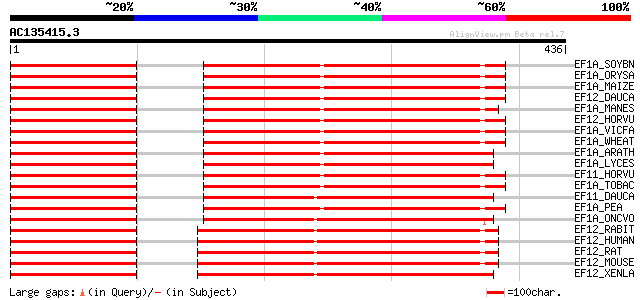
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135415.3 + phase: 0 /pseudo
(436 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
EF1A_SOYBN (P25698) Elongation factor 1-alpha (EF-1-alpha) 309 8e-84
EF1A_ORYSA (O64937) Elongation factor 1-alpha (EF-1-alpha) 308 2e-83
EF1A_MAIZE (Q41803) Elongation factor 1-alpha (EF-1-alpha) 304 3e-82
EF12_DAUCA (P34823) Elongation factor 1-alpha (EF-1-alpha) 304 3e-82
EF1A_MANES (O49169) Elongation factor 1-alpha (EF-1-alpha) 303 6e-82
EF12_HORVU (Q40034) Elongation factor 1-alpha (EF-1-alpha) 302 1e-81
EF1A_VICFA (O24534) Elongation factor 1-alpha (EF-1-alpha) 301 2e-81
EF1A_WHEAT (Q03033) Elongation factor 1-alpha (EF-1-alpha) 301 2e-81
EF1A_ARATH (P13905) Elongation factor 1-alpha (EF-1-alpha) 301 2e-81
EF1A_LYCES (P17786) Elongation factor 1-alpha (EF-1-alpha) 301 3e-81
EF11_HORVU (P34824) Elongation factor 1-alpha (EF-1-alpha) 300 4e-81
EF1A_TOBAC (P43643) Elongation factor 1-alpha (EF-1-alpha) (Vitr... 298 1e-80
EF11_DAUCA (P29521) Elongation factor 1-alpha (EF-1-alpha) 297 4e-80
EF1A_PEA (Q41011) Elongation factor 1-alpha (EF-1-alpha) 296 7e-80
EF1A_ONCVO (P27592) Elongation factor 1-alpha (EF-1-alpha) 275 2e-73
EF12_RABIT (Q71V39) Elongation factor 1-alpha 2 (EF-1-alpha-2) (... 274 3e-73
EF12_HUMAN (Q05639) Elongation factor 1-alpha 2 (EF-1-alpha-2) (... 274 3e-73
EF12_RAT (P62632) Elongation factor 1-alpha 2 (EF-1-alpha-2) (El... 273 5e-73
EF12_MOUSE (P62631) Elongation factor 1-alpha 2 (EF-1-alpha-2) (... 273 5e-73
EF12_XENLA (P17507) Elongation factor 1-alpha, oocyte form (EF-1... 273 6e-73
>EF1A_SOYBN (P25698) Elongation factor 1-alpha (EF-1-alpha)
Length = 447
Score = 309 bits (792), Expect = 8e-84
Identities = 160/237 (67%), Positives = 185/237 (77%), Gaps = 5/237 (2%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L A+DQISEPKRP KPLRLPLQ VYKIGGIGTVPVGRVETGVLKPGMV+TFAPT L +
Sbjct: 216 LLDALDQISEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGVLKPGMVVTFAPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E++ EA P D VGF + N + D L RGYVASNS+D+PA EAA FT++VI
Sbjct: 276 EVKSVEMHHESLTEAHPGDNVGFNVKNVAVKD--LKRGYVASNSKDDPAKEAANFTAQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
IINHP I GY P+LDCHTSH+AVKFA+L TK DR +G ELEK+PK LKNGDAG VKMI
Sbjct: 334 IINHPGQIGNGYAPVLDCHTSHIAVKFAELMTKIDRRSGKELEKEPKFLKNGDAGFVKMI 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE +EYP LGRFAVRDMRQTVAVGVI +V+K K+ K+T + +K
Sbjct: 394 PTKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKNVEK---KDPTGAKVTKAAQKKK 447
Score = 154 bits (390), Expect = 3e-37
Identities = 76/99 (76%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +I+IV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKVHISIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>EF1A_ORYSA (O64937) Elongation factor 1-alpha (EF-1-alpha)
Length = 447
Score = 308 bits (789), Expect = 2e-83
Identities = 158/237 (66%), Positives = 186/237 (77%), Gaps = 5/237 (2%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETGVLKPGMV+TF P+ L +
Sbjct: 216 LLEALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGVLKPGMVVTFGPSGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D VGF + N + D L RGYVASNS+D+PA EAA FTS+VI
Sbjct: 276 EVKSVEMHHEALQEALPGDNVGFNVKNVAVKD--LKRGYVASNSKDDPAKEAASFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA+L TK DR +G ELEK+PK LKNGDAG+VKMI
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFAELVTKIDRRSGKELEKEPKFLKNGDAGMVKMI 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE +EYP LGRFAVRDMRQTVAVGVI +V+K K+ K+T + +K
Sbjct: 394 PTKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKNVEK---KDPTGAKVTKAARKKK 447
Score = 157 bits (396), Expect = 7e-38
Identities = 77/99 (77%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKTHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>EF1A_MAIZE (Q41803) Elongation factor 1-alpha (EF-1-alpha)
Length = 447
Score = 304 bits (778), Expect = 3e-82
Identities = 155/237 (65%), Positives = 184/237 (77%), Gaps = 5/237 (2%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+D I+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETGV+KPGMV+TF PT L +
Sbjct: 216 LLEALDLINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGVIKPGMVVTFGPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D VGF + N + D L RGYVAS S+D+PA EAA FTS+VI
Sbjct: 276 EVKSVEMHHEALQEALPGDNVGFNVKNVAVKD--LKRGYVASGSKDDPAKEAASFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA+L TK DR +G ELEK+PK LKNGDAG+VKM+
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFAELVTKIDRRSGKELEKEPKFLKNGDAGMVKMV 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE ++YP LGRFAVRDMRQTVAVGVI SV+K K+ K+T + +K
Sbjct: 394 PTKPMVVETFSQYPPLGRFAVRDMRQTVAVGVIKSVEK---KDPTGAKVTKAAAKKK 447
Score = 157 bits (396), Expect = 7e-38
Identities = 77/99 (77%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKTHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>EF12_DAUCA (P34823) Elongation factor 1-alpha (EF-1-alpha)
Length = 447
Score = 304 bits (778), Expect = 3e-82
Identities = 156/237 (65%), Positives = 183/237 (76%), Gaps = 5/237 (2%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQISEPKRP KPLRLPLQ VYKIGGIGTVPVGRVETGV+KPGMV+TF P+ L +
Sbjct: 216 LLEALDQISEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGVIKPGMVVTFGPSGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D VGF + N + D L RGYVASNS+D+PA EAA FT++VI
Sbjct: 276 EVKSVEMHHEALQEALPGDNVGFNVKNVAVKD--LKRGYVASNSKDDPAKEAANFTAQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA++ TK DR +G ELEK+PK LKNGDAG VKMI
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFAEIQTKIDRRSGKELEKEPKFLKNGDAGFVKMI 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE YP LGRFAVRDMRQTVAVGVI SV+K K+ K+T + +K
Sbjct: 394 PTKPMVVETFMSYPPLGRFAVRDMRQTVAVGVIKSVEK---KDPTGAKVTKAAIKKK 447
Score = 154 bits (389), Expect = 4e-37
Identities = 76/99 (76%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +I+IV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKIHISIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>EF1A_MANES (O49169) Elongation factor 1-alpha (EF-1-alpha)
Length = 449
Score = 303 bits (776), Expect = 6e-82
Identities = 156/232 (67%), Positives = 181/232 (77%), Gaps = 5/232 (2%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETG+LKPGMV+TF PT L +
Sbjct: 216 LLEALDQIQEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGILKPGMVVTFGPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D VGF + N + D L RG VASNS+D+PA EAA FTS+VI
Sbjct: 276 EVKSVEMHHEALQEALPGDNVGFNVKNVAVKD--LKRGIVASNSKDDPAKEAANFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA++ TK DR +G ELEK+PK LKNGDAG VKMI
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFAEILTKIDRRSGKELEKEPKFLKNGDAGFVKMI 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGS 384
P KPMVVE + YP LGRFAVRDMRQTVAVGVI SV+K K+ + K+T S
Sbjct: 394 PTKPMVVETFSAYPPLGRFAVRDMRQTVAVGVIKSVEK---KDPSGAKVTKS 442
Score = 156 bits (395), Expect = 9e-38
Identities = 77/99 (77%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKVHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>EF12_HORVU (Q40034) Elongation factor 1-alpha (EF-1-alpha)
Length = 447
Score = 302 bits (774), Expect = 1e-81
Identities = 155/237 (65%), Positives = 183/237 (76%), Gaps = 5/237 (2%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETGV+KPGMV+TF PT L +
Sbjct: 216 LLEALDQINEPKRPSEKPLRLPLQDVYKIGGIGTVPVGRVETGVIKPGMVVTFGPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E++ EALP D VGF + N + D L RGYVASNS+D+PA EAA FT++VI
Sbjct: 276 EVKSVEMHHESLLEALPGDNVGFNVKNVAVKD--LKRGYVASNSKDDPAKEAANFTAQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I+ GY P+LDCHTSH+AVKFA++ TK DR +G ELE PK LKNGDAG VKMI
Sbjct: 334 IMNHPGQISNGYAPVLDCHTSHIAVKFAEIQTKIDRRSGKELEAAPKFLKNGDAGFVKMI 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE +YP LGRFAVRDMRQTVAVGVI SV+K K K+T + +K
Sbjct: 394 PTKPMVVETFAQYPPLGRFAVRDMRQTVAVGVIKSVEK---KEPTGAKVTKAAIKKK 447
Score = 155 bits (392), Expect = 2e-37
Identities = 76/99 (76%), Positives = 84/99 (84%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +I+IV IGHV+SGKSTT GHL+YKLGGI K IE+ KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKIHISIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIESFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYSCTVIDAPGHRDFI 99
>EF1A_VICFA (O24534) Elongation factor 1-alpha (EF-1-alpha)
Length = 447
Score = 301 bits (772), Expect = 2e-81
Identities = 155/237 (65%), Positives = 183/237 (76%), Gaps = 5/237 (2%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L A+D I+EPKRP KPLRLPLQ VYKIGGIG VPVGRVETGV+KPGM++TFAPT L +
Sbjct: 216 LLDALDNINEPKRPSDKPLRLPLQDVYKIGGIGIVPVGRVETGVVKPGMLVTFAPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D VGF + N + D L RG+VASNS+D+PA EAA FTS+VI
Sbjct: 276 EVKSVEMHHEALTEALPGDNVGFNVKNVAVKD--LKRGFVASNSKDDPAKEAANFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA+L TK DR +G E+EK+PK LKNGDAG+VKMI
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFAELITKIDRRSGKEIEKEPKFLKNGDAGMVKMI 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE EYP LGRFAVRDMRQTVAVGVI SV+K K+ K+T + +K
Sbjct: 394 PTKPMVVETFAEYPPLGRFAVRDMRQTVAVGVIKSVEK---KDPTGAKVTKAAAKKK 447
Score = 155 bits (391), Expect = 3e-37
Identities = 76/99 (76%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKVHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFET+KY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETSKYYCTVIDAPGHRDFI 99
>EF1A_WHEAT (Q03033) Elongation factor 1-alpha (EF-1-alpha)
Length = 447
Score = 301 bits (771), Expect = 2e-81
Identities = 156/237 (65%), Positives = 182/237 (75%), Gaps = 5/237 (2%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETGV+KPGMV+TF PT L +
Sbjct: 216 LLEALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGVIKPGMVVTFGPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E++ EALP D VGF + N + D L RG+VASNS+D+PA EAA FTS+VI
Sbjct: 276 EVKSVEMHHESLLEALPGDNVGFNVKNVAVKD--LKRGFVASNSKDDPAKEAANFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA+L TK DR +G ELE PK LKNGDAGIVKMI
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFAELVTKIDRRSGKELEALPKFLKNGDAGIVKMI 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE YP LGRFAVRDMRQTVAVGVI V+K K+ K+T + +K
Sbjct: 394 PTKPMVVETFATYPPLGRFAVRDMRQTVAVGVIKGVEK---KDPTGAKVTKAAIKKK 447
Score = 157 bits (396), Expect = 7e-38
Identities = 77/99 (77%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKTHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>EF1A_ARATH (P13905) Elongation factor 1-alpha (EF-1-alpha)
Length = 449
Score = 301 bits (771), Expect = 2e-81
Identities = 152/228 (66%), Positives = 179/228 (77%), Gaps = 2/228 (0%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETG++KPGMV+TFAPT L +
Sbjct: 216 LLEALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGMIKPGMVVTFAPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E++ EALP D VGF + N + D L RGYVASNS+D+PA AA FTS+VI
Sbjct: 276 EVKSVEMHHESLLEALPGDNVGFNVKNVAVKD--LKRGYVASNSKDDPAKGAANFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKF+++ TK DR +G E+EK+PK LKNGDAG+VKM
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFSEILTKIDRRSGKEIEKEPKFLKNGDAGMVKMT 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATK 380
P KPMVVE +EYP LGRFAVRDMRQTVAVGVI SV K TK
Sbjct: 394 PTKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKSVDKKDPTGAKVTK 441
Score = 155 bits (393), Expect = 1e-37
Identities = 77/99 (77%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKFHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>EF1A_LYCES (P17786) Elongation factor 1-alpha (EF-1-alpha)
Length = 448
Score = 301 bits (770), Expect = 3e-81
Identities = 152/228 (66%), Positives = 177/228 (76%), Gaps = 2/228 (0%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETGV+KPGMV+TF PT L +
Sbjct: 216 LLEALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGVIKPGMVVTFGPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D VGF + N + D L RGYVASNS+D+PA AA FT++VI
Sbjct: 276 EVKSVEMHHEALQEALPGDNVGFNVKNVAVKD--LKRGYVASNSKDDPAKGAASFTAQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA++ TK DR +G ELEK+PK LKNGDAG+VKMI
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFAEILTKIDRRSGKELEKEPKFLKNGDAGMVKMI 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATK 380
P KPMVVE EYP LGRFAVRDMRQTVAVGV+ +V K TK
Sbjct: 394 PTKPMVVETFAEYPPLGRFAVRDMRQTVAVGVVKNVDKKDPTGAKVTK 441
Score = 154 bits (389), Expect = 4e-37
Identities = 76/99 (76%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +I+IV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKIHISIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>EF11_HORVU (P34824) Elongation factor 1-alpha (EF-1-alpha)
Length = 447
Score = 300 bits (769), Expect = 4e-81
Identities = 154/237 (64%), Positives = 182/237 (75%), Gaps = 5/237 (2%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPL LPLQ VYKIGGIGTVPVGRVETGV+KPGMV+TF PT L +
Sbjct: 216 LLEALDQINEPKRPSDKPLHLPLQDVYKIGGIGTVPVGRVETGVIKPGMVVTFGPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E++ EALP D VGF + N + D L RGYVASNS+D+PA EAA FT++VI
Sbjct: 276 EVKSVEVHHESLLEALPGDNVGFNVKNVAVKD--LKRGYVASNSKDDPAKEAANFTAQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I+ GY P+LDCHTSH+AVKFA++ TK DR +G ELE PK LKNGDAG VKMI
Sbjct: 334 IMNHPGQISNGYAPVLDCHTSHIAVKFAEIQTKIDRRSGKELEAAPKFLKNGDAGFVKMI 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE +YP LGRFAVRDMRQTVAVGVI SV+K K K+T + +K
Sbjct: 394 PTKPMVVETFAQYPPLGRFAVRDMRQTVAVGVIKSVEK---KEPTGAKVTKAAIKKK 447
Score = 154 bits (390), Expect = 3e-37
Identities = 76/99 (76%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +I+IV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKIHISIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYSCTVIDAPGHRDFI 99
>EF1A_TOBAC (P43643) Elongation factor 1-alpha (EF-1-alpha)
(Vitronectin-like adhesion protein 1) (PVN1)
Length = 447
Score = 298 bits (764), Expect = 1e-80
Identities = 153/237 (64%), Positives = 183/237 (76%), Gaps = 5/237 (2%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI++ KRP KPLRLPLQ VYKIGGIGTVPVGRVETGVLKPGMV+TF PT L +
Sbjct: 216 LLEALDQINDAKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGVLKPGMVVTFGPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D VGF + N + D L RG+VASNS+D+PA AA FTS+VI
Sbjct: 276 EVKSVEMHHEALQEALPGDNVGFNVKNVAVKD--LKRGFVASNSKDDPAKGAASFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA++ TK DR +G E+EK+PK LKNGDAG+VKMI
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFAEILTKIDRRSGKEIEKEPKFLKNGDAGMVKMI 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE +EYP LGRFAVRDMRQTVAVGVI +V K K+ K+T + +K
Sbjct: 394 PTKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKNVDK---KDPTGAKVTKAAQKKK 447
Score = 155 bits (393), Expect = 1e-37
Identities = 77/99 (77%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKFHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>EF11_DAUCA (P29521) Elongation factor 1-alpha (EF-1-alpha)
Length = 449
Score = 297 bits (760), Expect = 4e-80
Identities = 152/228 (66%), Positives = 178/228 (77%), Gaps = 2/228 (0%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETG +KPGMV+TF P+ L +
Sbjct: 216 LLDALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGTIKPGMVVTFGPSGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E++ EALP D VGF + N +S K+L RGYVASNS+D+PA AA FTS+VI
Sbjct: 276 EVKSVEMHHESLLEALPGDNVGFNVKN--VSVKDLKRGYVASNSKDDPAKGAASFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA+L TK DR +G ELEK+PK LKNGDAG+VKM+
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFAELLTKIDRRSGKELEKEPKFLKNGDAGMVKML 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATK 380
P KPMVVE EYP LGRFAVR MRQTVAVGVI +V+K TK
Sbjct: 394 PTKPMVVETFAEYPPLGRFAVRVMRQTVAVGVIKAVEKKDPTGAKVTK 441
Score = 154 bits (390), Expect = 3e-37
Identities = 76/99 (76%), Positives = 82/99 (82%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKVHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFET KY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETNKYYCTVIDAPGHRDFI 99
>EF1A_PEA (Q41011) Elongation factor 1-alpha (EF-1-alpha)
Length = 447
Score = 296 bits (758), Expect = 7e-80
Identities = 153/237 (64%), Positives = 181/237 (75%), Gaps = 5/237 (2%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L A+D I+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETGV+KPGM++TFAPT L +
Sbjct: 216 LLDALDNINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGVVKPGMLVTFAPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D V F + N + D L G VASNS+D+PA +AA FTS+VI
Sbjct: 276 EVKSVEMHHEALTEALPGDNVRFNVKNVAVKD--LKHGLVASNSKDDPAKDAANFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA+L TK DR +G E+EK+PK LKNGDAG+VKMI
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFAELITKIDRRSGKEIEKEPKFLKNGDAGMVKMI 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE EYP LGRFAVRDMRQTVAVGVI SV+K K+ K+T + +K
Sbjct: 394 PTKPMVVETFAEYPPLGRFAVRDMRQTVAVGVIKSVEK---KDPTGAKVTKAAAKKK 447
Score = 144 bits (362), Expect = 6e-34
Identities = 71/99 (71%), Positives = 80/99 (80%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT H++YKLGGI K IE KEA EM RSFKYAW+L
Sbjct: 1 MGKEKVHINIVVIGHVDSGKSTTTVHVIYKLGGIDKRVIERFEKEADEMNKRSFKYAWLL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY T++DAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALLKFETTKYYSTVMDAPGHRDFI 99
>EF1A_ONCVO (P27592) Elongation factor 1-alpha (EF-1-alpha)
Length = 464
Score = 275 bits (702), Expect = 2e-73
Identities = 139/231 (60%), Positives = 175/231 (75%), Gaps = 5/231 (2%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+D + P+RP KPLRLPLQ VYKIGGIGTVPVGRVETG+LKPGM++TFAP L +
Sbjct: 228 LLEALDSVVPPQRPTDKPLRLPLQDVYKIGGIGTVPVGRVETGILKPGMIVTFAPQNLTT 287
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D VGF + N +S K++ RG VAS+S+++PA E FT++VI
Sbjct: 288 EVKSVEMHHEALQEALPGDNVGFNVKN--ISIKDIRRGSVASDSKNDPAKETKMFTAQVI 345
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I+ GYTP+LDCHT+H+A KFA+L K DR +G ++E PKSLK+GDAGI+ +I
Sbjct: 346 IMNHPGQISAGYTPVLDCHTAHIACKFAELKEKVDRRSGKKVEDNPKSLKSGDAGIIDLI 405
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKN---GYKNKAATK 380
P KP+ VE EYP LGRFAVRDMRQTVAVGVI +V K+ G KAA K
Sbjct: 406 PTKPLCVETFTEYPPLGRFAVRDMRQTVAVGVIKNVDKSEGVGKVQKAAQK 456
Score = 141 bits (356), Expect = 3e-33
Identities = 71/99 (71%), Positives = 77/99 (77%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YK GGI K IE KEA EM SFKYAWVL
Sbjct: 1 MGKEKTHINIVVIGHVDSGKSTTTGHLIYKCGGIDKRTIEKFEKEAQEMGKGSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGI IDI++ KFET KY T+IDAPGHRDFI
Sbjct: 61 DKLKAERERGIQIDIALWKFETPKYYITIIDAPGHRDFI 99
>EF12_RABIT (Q71V39) Elongation factor 1-alpha 2 (EF-1-alpha-2)
(Elongation factor 1 A-2) (eEF1A-2) (Statin S1)
Length = 463
Score = 274 bits (701), Expect = 3e-73
Identities = 139/237 (58%), Positives = 180/237 (75%), Gaps = 5/237 (2%)
Query: 148 ANDLLLQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAP 207
A+ + L +A+D I P RP KPLRLPLQ VYKIGGIGTVPVGRVETG+L+PGMV+TFAP
Sbjct: 223 ASGVSLLEALDTILPPTRPTDKPLRLPLQDVYKIGGIGTVPVGRVETGILRPGMVVTFAP 282
Query: 208 TKLQSGVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKF 267
+ + VKS++ E ++EALP D VGF + N +S K++ RG V +S+ +P EAA+F
Sbjct: 283 VNITTEVKSVEMHHEALSEALPGDNVGFNVKN--VSVKDIRRGNVCGDSKSDPPQEAAQF 340
Query: 268 TSRVIIINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAG 327
TS+VII+NHP I+ GY+P++DCHT+H+A KFA+L K DR +G +LE PKSLK+GDA
Sbjct: 341 TSQVIILNHPGQISAGYSPVIDCHTAHIACKFAELKEKIDRRSGKKLEDNPKSLKSGDAA 400
Query: 328 IVKMIPMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGS 384
IV+M+P KPM VE ++YP LGRFAVRDMRQTVAVGVI +V+K K+ A K+T S
Sbjct: 401 IVEMVPGKPMCVESFSQYPPLGRFAVRDMRQTVAVGVIKNVEK---KSGGAGKVTKS 454
Score = 149 bits (376), Expect = 1e-35
Identities = 75/99 (75%), Positives = 80/99 (80%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YK GGI K IE KEAAEM SFKYAWVL
Sbjct: 1 MGKEKTHINIVVIGHVDSGKSTTTGHLIYKCGGIDKRTIEKFEKEAAEMGKGSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDIS+ KFETTKY T+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDISLWKFETTKYYITIIDAPGHRDFI 99
>EF12_HUMAN (Q05639) Elongation factor 1-alpha 2 (EF-1-alpha-2)
(Elongation factor 1 A-2) (eEF1A-2) (Statin S1)
Length = 463
Score = 274 bits (701), Expect = 3e-73
Identities = 139/237 (58%), Positives = 180/237 (75%), Gaps = 5/237 (2%)
Query: 148 ANDLLLQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAP 207
A+ + L +A+D I P RP KPLRLPLQ VYKIGGIGTVPVGRVETG+L+PGMV+TFAP
Sbjct: 223 ASGVSLLEALDTILPPTRPTDKPLRLPLQDVYKIGGIGTVPVGRVETGILRPGMVVTFAP 282
Query: 208 TKLQSGVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKF 267
+ + VKS++ E ++EALP D VGF + N +S K++ RG V +S+ +P EAA+F
Sbjct: 283 VNITTEVKSVEMHHEALSEALPGDNVGFNVKN--VSVKDIRRGNVCGDSKSDPPQEAAQF 340
Query: 268 TSRVIIINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAG 327
TS+VII+NHP I+ GY+P++DCHT+H+A KFA+L K DR +G +LE PKSLK+GDA
Sbjct: 341 TSQVIILNHPGQISAGYSPVIDCHTAHIACKFAELKEKIDRRSGKKLEDNPKSLKSGDAA 400
Query: 328 IVKMIPMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGS 384
IV+M+P KPM VE ++YP LGRFAVRDMRQTVAVGVI +V+K K+ A K+T S
Sbjct: 401 IVEMVPGKPMCVESFSQYPPLGRFAVRDMRQTVAVGVIKNVEK---KSGGAGKVTKS 454
Score = 149 bits (376), Expect = 1e-35
Identities = 75/99 (75%), Positives = 80/99 (80%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YK GGI K IE KEAAEM SFKYAWVL
Sbjct: 1 MGKEKTHINIVVIGHVDSGKSTTTGHLIYKCGGIDKRTIEKFEKEAAEMGKGSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDIS+ KFETTKY T+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDISLWKFETTKYYITIIDAPGHRDFI 99
>EF12_RAT (P62632) Elongation factor 1-alpha 2 (EF-1-alpha-2)
(Elongation factor 1 A-2) (eEF1A-2) (Statin S1)
Length = 463
Score = 273 bits (699), Expect = 5e-73
Identities = 139/237 (58%), Positives = 180/237 (75%), Gaps = 5/237 (2%)
Query: 148 ANDLLLQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAP 207
A+ + L +A+D I P RP KPLRLPLQ VYKIGGIGTVPVGRVETG+L+PGMV+TFAP
Sbjct: 223 ASGVSLLEALDTILPPTRPTDKPLRLPLQDVYKIGGIGTVPVGRVETGILRPGMVVTFAP 282
Query: 208 TKLQSGVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKF 267
+ + VKS++ E ++EALP D VGF + N +S K++ RG V +S+ +P EAA+F
Sbjct: 283 VNITTEVKSVEMHHEALSEALPGDNVGFNVKN--VSVKDIRRGNVCGDSKADPPQEAAQF 340
Query: 268 TSRVIIINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAG 327
TS+VII+NHP I+ GY+P++DCHT+H+A KFA+L K DR +G +LE PKSLK+GDA
Sbjct: 341 TSQVIILNHPGQISAGYSPVIDCHTAHIACKFAELKEKIDRRSGKKLEDNPKSLKSGDAA 400
Query: 328 IVKMIPMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGS 384
IV+M+P KPM VE ++YP LGRFAVRDMRQTVAVGVI +V+K K+ A K+T S
Sbjct: 401 IVEMVPGKPMCVESFSQYPPLGRFAVRDMRQTVAVGVIKNVEK---KSGGAGKVTKS 454
Score = 149 bits (376), Expect = 1e-35
Identities = 75/99 (75%), Positives = 80/99 (80%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YK GGI K IE KEAAEM SFKYAWVL
Sbjct: 1 MGKEKTHINIVVIGHVDSGKSTTTGHLIYKCGGIDKRTIEKFEKEAAEMGKGSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDIS+ KFETTKY T+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDISLWKFETTKYYITIIDAPGHRDFI 99
>EF12_MOUSE (P62631) Elongation factor 1-alpha 2 (EF-1-alpha-2)
(Elongation factor 1 A-2) (eEF1A-2) (Statin S1)
Length = 463
Score = 273 bits (699), Expect = 5e-73
Identities = 139/237 (58%), Positives = 180/237 (75%), Gaps = 5/237 (2%)
Query: 148 ANDLLLQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAP 207
A+ + L +A+D I P RP KPLRLPLQ VYKIGGIGTVPVGRVETG+L+PGMV+TFAP
Sbjct: 223 ASGVSLLEALDTILPPTRPTDKPLRLPLQDVYKIGGIGTVPVGRVETGILRPGMVVTFAP 282
Query: 208 TKLQSGVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKF 267
+ + VKS++ E ++EALP D VGF + N +S K++ RG V +S+ +P EAA+F
Sbjct: 283 VNITTEVKSVEMHHEALSEALPGDNVGFNVKN--VSVKDIRRGNVCGDSKADPPQEAAQF 340
Query: 268 TSRVIIINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAG 327
TS+VII+NHP I+ GY+P++DCHT+H+A KFA+L K DR +G +LE PKSLK+GDA
Sbjct: 341 TSQVIILNHPGQISAGYSPVIDCHTAHIACKFAELKEKIDRRSGKKLEDNPKSLKSGDAA 400
Query: 328 IVKMIPMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGS 384
IV+M+P KPM VE ++YP LGRFAVRDMRQTVAVGVI +V+K K+ A K+T S
Sbjct: 401 IVEMVPGKPMCVESFSQYPPLGRFAVRDMRQTVAVGVIKNVEK---KSGGAGKVTKS 454
Score = 149 bits (376), Expect = 1e-35
Identities = 75/99 (75%), Positives = 80/99 (80%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YK GGI K IE KEAAEM SFKYAWVL
Sbjct: 1 MGKEKTHINIVVIGHVDSGKSTTTGHLIYKCGGIDKRTIEKFEKEAAEMGKGSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDIS+ KFETTKY T+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDISLWKFETTKYYITIIDAPGHRDFI 99
>EF12_XENLA (P17507) Elongation factor 1-alpha, oocyte form
(EF-1-alpha-O) (EF-1AO) (42S p48)
Length = 461
Score = 273 bits (698), Expect = 6e-73
Identities = 137/233 (58%), Positives = 174/233 (73%), Gaps = 2/233 (0%)
Query: 148 ANDLLLQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAP 207
A+ + L +A+D I P+RP KPLRLPLQ VYKIGGIGTVPVGRVETGVLKPGM++TFAP
Sbjct: 223 ASGVTLLEALDCIIPPQRPTAKPLRLPLQDVYKIGGIGTVPVGRVETGVLKPGMIVTFAP 282
Query: 208 TKLQSGVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKF 267
+ + + VKS++ E + EALP D VGF + N +S K++ RG VA +S+++P M+A F
Sbjct: 283 SNVTTEVKSVEMHHEALQEALPGDNVGFNVKN--ISVKDIRRGNVAGDSKNDPPMQAGSF 340
Query: 268 TSRVIIINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAG 327
T++VII+NHP I+ GY P+LDCHT+H+A KFA+L K DR +G +LE PK LK+GDA
Sbjct: 341 TAQVIILNHPGQISAGYAPVLDCHTAHIACKFAELKQKIDRRSGKKLEDDPKFLKSGDAA 400
Query: 328 IVKMIPMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATK 380
IV+MIP KPM VE ++YP LGRFAVRDMRQTVAVGVI V K + TK
Sbjct: 401 IVEMIPGKPMCVESFSDYPPLGRFAVRDMRQTVAVGVIKGVDKKAASSGKVTK 453
Score = 144 bits (363), Expect = 4e-34
Identities = 73/99 (73%), Positives = 79/99 (79%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YK GGI K IE KEAAEM SFKYAWVL
Sbjct: 1 MGKEKIHINIVVIGHVDSGKSTTTGHLIYKCGGIDKRTIEKFEKEAAEMGKGSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDIS+ KFET K+ T+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDISLWKFETGKFYITIIDAPGHRDFI 99
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,991,686
Number of Sequences: 164201
Number of extensions: 1997031
Number of successful extensions: 7759
Number of sequences better than 10.0: 756
Number of HSP's better than 10.0 without gapping: 479
Number of HSP's successfully gapped in prelim test: 277
Number of HSP's that attempted gapping in prelim test: 5890
Number of HSP's gapped (non-prelim): 1153
length of query: 436
length of database: 59,974,054
effective HSP length: 113
effective length of query: 323
effective length of database: 41,419,341
effective search space: 13378447143
effective search space used: 13378447143
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC135415.3


