

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135319.2 - phase: 0
(323 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
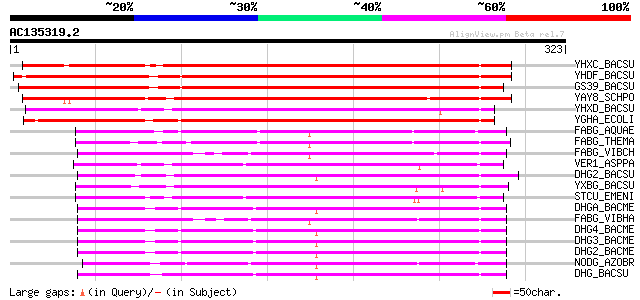
Score E
Sequences producing significant alignments: (bits) Value
YHXC_BACSU (P40397) Hypothetical oxidoreductase yhxC (EC 1.-.-.-... 294 2e-79
YHDF_BACSU (O07575) Hypothetical oxidoreductase yhdF (EC 1.-.-.-) 288 1e-77
GS39_BACSU (P80873) General stress protein 39 (GSP39) (EC 1.-.-.-) 280 3e-75
YAY8_SCHPO (Q10216) Hypothetical oxidoreductase C4H3.08 in chrom... 224 2e-58
YHXD_BACSU (P40398) Hypothetical oxidoreductase yhxD (EC 1.-.-.-... 198 2e-50
YGHA_ECOLI (P25887) Hypothetical oxidoreductase yghA (EC 1.-.-.-) 193 6e-49
FABG_AQUAE (O67610) 3-oxoacyl-[acyl-carrier-protein] reductase (... 132 2e-30
FABG_THEMA (Q9X248) 3-oxoacyl-[acyl-carrier-protein] reductase (... 131 3e-30
FABG_VIBCH (Q9KQH7) 3-oxoacyl-[acyl-carrier-protein] reductase (... 129 8e-30
VER1_ASPPA (P50161) Versicolorin reductase (EC 1.1.-.-) (VER-1) 128 2e-29
DHG2_BACSU (P80869) Glucose 1-dehydrogenase II (EC 1.1.1.47) (GL... 128 2e-29
YXBG_BACSU (P46331) Hypothetical oxidoreductase yxbG (EC 1.-.-.-) 127 4e-29
STCU_EMENI (Q00791) Versicolorin reductase (EC 1.1.-.-) 127 4e-29
DHGA_BACME (P10528) Glucose 1-dehydrogenase A (EC 1.1.1.47) 126 8e-29
FABG_VIBHA (P55336) 3-oxoacyl-[acyl-carrier-protein] reductase (... 125 1e-28
DHG4_BACME (P39485) Glucose 1-dehydrogenase IV (EC 1.1.1.47) (GL... 124 4e-28
DHG3_BACME (P39484) Glucose 1-dehydrogenase III (EC 1.1.1.47) (G... 124 4e-28
DHG2_BACME (P39483) Glucose 1-dehydrogenase II (EC 1.1.1.47) (GL... 123 7e-28
NODG_AZOBR (P17611) Nodulation protein G 122 1e-27
DHG_BACSU (P12310) Glucose 1-dehydrogenase (EC 1.1.1.47) 122 1e-27
>YHXC_BACSU (P40397) Hypothetical oxidoreductase yhxC (EC 1.-.-.-)
(ORFX)
Length = 285
Score = 294 bits (753), Expect = 2e-79
Identities = 155/285 (54%), Positives = 199/285 (69%), Gaps = 9/285 (3%)
Query: 8 IPPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALEGA 67
+PPQ Q+ QPG E+ M+P P F P K A KL+GK A++TGGDSGIGRAV LFA EGA
Sbjct: 9 LPPQHQNQQPGFEYLMDPRPVFDKP--KKAKKLEGKTAIITGGDSGIGRAVSVLFAKEGA 66
Query: 68 TVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEIINAYGRI 127
V+ Y+ H+D A +T K + I D+G + C V+ + + I
Sbjct: 67 NVVIVYLNEHQD--AEET----KQYVEKEGVKCLLIAGDVGDEAFCNDVVGQASQVFPSI 120
Query: 128 DILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNIINTTSVNA 187
DILVNNAAEQ+ S+E+I +L R F+TNIFS F++T+ L H+K+GS+IINT S+ A
Sbjct: 121 DILVNNAAEQHVQPSIEKITSHQLIRTFQTNIFSMFYLTKAVLPHLKKGSSIINTASITA 180
Query: 188 YKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQF 247
YKGN TLIDY++TKGAIV FTR+LS LV +GIRVN VAPGPIWTPLIPASF + F
Sbjct: 181 YKGNKTLIDYSATKGAIVTFTRSLSQSLVQQGIRVNAVAPGPIWTPLIPASFAAKDVEVF 240
Query: 248 GSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNGDMLIN 292
GSDVPM+R GQPVEVAPS+++LAS+ S+Y+TGQ +H NG ++N
Sbjct: 241 GSDVPMERPGQPVEVAPSYLYLASDD-STYVTGQTIHVNGGTIVN 284
>YHDF_BACSU (O07575) Hypothetical oxidoreductase yhdF (EC 1.-.-.-)
Length = 289
Score = 288 bits (738), Expect = 1e-77
Identities = 150/290 (51%), Positives = 200/290 (68%), Gaps = 9/290 (3%)
Query: 3 TGGQKIPPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCNLF 62
T GQ+ PQ QD QPG E MNP P DY+ + KL+GK+A++TGGDSGIGRA F
Sbjct: 8 TEGQE--PQHQDRQPGIESKMNPLPLSEDEDYRGSGKLKGKVAIITGGDSGIGRAAAIAF 65
Query: 63 ALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEIIN 122
A EGA + Y+ H D + + K + N + + IP D+G + +C++ + + ++
Sbjct: 66 AKEGADISILYLDEHSDAE-----ETRKRIEKENVRC-LLIPGDVGDENHCEQAVQQTVD 119
Query: 123 AYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNIINT 182
+G++DILVNNAAEQ+ S+ I +LE+ FRTNIFS F MT+ AL H++EG IINT
Sbjct: 120 HFGKLDILVNNAAEQHPQDSILNISTEQLEKTFRTNIFSMFHMTKKALPHLQEGCAIINT 179
Query: 183 TSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEE 242
TS+ AY+G++ LIDY+STKGAIV+FTR+++ L KGIRVN VAPGPIWTPLIPA+F EE
Sbjct: 180 TSITAYEGDTALIDYSSTKGAIVSFTRSMAKSLADKGIRVNAVAPGPIWTPLIPATFPEE 239
Query: 243 KTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNGDMLIN 292
K Q G D PM R GQPVE A ++V LAS++ SSY+TGQ +H NG I+
Sbjct: 240 KVKQHGLDTPMGRPGQPVEHAGAYVLLASDE-SSYMTGQTIHVNGGRFIS 288
>GS39_BACSU (P80873) General stress protein 39 (GSP39) (EC 1.-.-.-)
Length = 285
Score = 280 bits (717), Expect = 3e-75
Identities = 145/282 (51%), Positives = 190/282 (66%), Gaps = 7/282 (2%)
Query: 6 QKIPPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALE 65
+++P Q Q QPG E MNP+P + DYK A+KL+GK+A++TGGDSGIGRAV +A E
Sbjct: 5 KELPAQTQSRQPGIESEMNPSPVYEYEDYKGADKLKGKVALITGGDSGIGRAVSVAYAKE 64
Query: 66 GATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEIINAYG 125
GA + Y HED + + K + K + I D+G +E C +++ + G
Sbjct: 65 GADIAIVYKDEHEDAE-----ETKKRVEQEGVKC-LLIAGDVGEEEFCNEAVEKTVKELG 118
Query: 126 RIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNIINTTSV 185
+DILVNNA EQ+ S+++I +L R F+TN +S F++T+ A+ ++K GS IINTTS+
Sbjct: 119 GLDILVNNAGEQHPKESIKDITSEQLHRTFKTNFYSQFYLTKKAIDYLKPGSAIINTTSI 178
Query: 186 NAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTA 245
N Y GN TLIDYT+TKGAI AFTR ++ LV GIRVN VAPGPIWTPLIPA+F EE A
Sbjct: 179 NPYVGNPTLIDYTATKGAINAFTRTMAQALVKDGIRVNAVAPGPIWTPLIPATFPEETVA 238
Query: 246 QFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNG 287
QFG D PM R GQPVE +V LAS++ SSY+TGQ LH NG
Sbjct: 239 QFGQDTPMGRPGQPVEHVGCYVLLASDE-SSYMTGQTLHVNG 279
>YAY8_SCHPO (Q10216) Hypothetical oxidoreductase C4H3.08 in
chromosome I (EC 1.-.-.-)
Length = 286
Score = 224 bits (572), Expect = 2e-58
Identities = 124/289 (42%), Positives = 178/289 (60%), Gaps = 11/289 (3%)
Query: 8 IPPQKQDTQPGKEHAMNPTPQFT--CPD--YKPANKLQGKIAVVTGGDSGIGRAVCNLFA 63
+P + PGK ++P P C + + KL K ++TGGDSGIG+A +FA
Sbjct: 4 VPSTQTQKWPGKHADLDPEPSLLRYCDGRVHVGSGKLAEKKTLLTGGDSGIGKAAAVMFA 63
Query: 64 LEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEIINA 123
EG+ ++ + + D DA T D+++ ++ L +NC+ ++D +
Sbjct: 64 REGSDLVISCLPEERD-DAEVTRDLIER----EGRNCWIWEGKLDKSDNCRDLVDFALKK 118
Query: 124 YGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNIINTT 183
G ID+LVNN A Q S+E+ID+ + + F+TNIFS+F++T+ A+ HMK GS+I+N +
Sbjct: 119 LGWIDVLVNNIAYQQVAQSIEDIDDEQWDLTFKTNIFSFFWVTKAAISHMKSGSSIVNCS 178
Query: 184 SVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEK 243
S+NAY G L+DYTSTKGAI AFTR LS Q GIRVN VAPGPI+TPL+ ++F +EK
Sbjct: 179 SINAYVGRPDLLDYTSTKGAITAFTRGLSNQYAQHGIRVNAVAPGPIYTPLVSSTFPKEK 238
Query: 244 TAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNGDMLIN 292
+ VP+ R GQPVEVA ++FLA + Y+TGQ LHPNG +IN
Sbjct: 239 -IELSDQVPLGRMGQPVEVASCYLFLACSD-GGYMTGQTLHPNGGTVIN 285
>YHXD_BACSU (P40398) Hypothetical oxidoreductase yhxD (EC 1.-.-.-)
(ORFY)
Length = 299
Score = 198 bits (503), Expect = 2e-50
Identities = 112/277 (40%), Positives = 161/277 (57%), Gaps = 10/277 (3%)
Query: 10 PQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALEGATV 69
P++ PG + M P P YK + KL G+ A+VTGGDSGIGRA +A EGA V
Sbjct: 19 PEQYQEPPGLQKNMKPVPDCGEKSYKGSGKLTGRKALVTGGDSGIGRAAAIAYAREGADV 78
Query: 70 IFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEIINAYGRIDI 129
Y+ E DA + ++++ A + + IP DL + C+ ++ + + G +D+
Sbjct: 79 AINYLP-EEQPDAEEVKELIE----AEGRKAVLIPGDLSDESFCQDLVKQSHHELGGLDV 133
Query: 130 LVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNIINTTSVNAYK 189
L A +Q ++E++ ++ + F N+FS +++ + AL ++ EG++II TTSV Y
Sbjct: 134 LALVAGKQQAVENIEDLPTEQIYKTFEVNVFSLYWVVKAALPYLPEGASIITTTSVEGYN 193
Query: 190 GNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPL-IPASFNEEKTAQFG 248
+ L+DY +TK AI+ FT L QL SKGIRVN VAPGPIWTPL I E +FG
Sbjct: 194 PSPMLLDYAATKNAIIGFTVGLGKQLASKGIRVNSVAPGPIWTPLQISGGQPTENIPKFG 253
Query: 249 S---DVPMKRAGQPVEVAPSFVFLASNQCSSYITGQV 282
P+ RAGQPVE+A +VFLAS SSY+T QV
Sbjct: 254 QGTPPAPLNRAGQPVELADVYVFLASEN-SSYVTSQV 289
>YGHA_ECOLI (P25887) Hypothetical oxidoreductase yghA (EC 1.-.-.-)
Length = 294
Score = 193 bits (490), Expect = 6e-49
Identities = 108/275 (39%), Positives = 166/275 (60%), Gaps = 8/275 (2%)
Query: 9 PPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALEGAT 68
P QKQ T PG + M P P Y + +L+ + A+VTGGDSGIGRA +A EGA
Sbjct: 17 PKQKQPT-PGIQAKMTPVPDCGEKTYVGSGRLKDRKALVTGGDSGIGRAAAIAYAREGAD 75
Query: 69 VIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEIINAYGRID 128
V +Y+ E+ D D+ K+ + K + +P DL ++ + ++ E A G +D
Sbjct: 76 VAISYLPVEEE----DAQDVKKIIEECGRK-AVLLPGDLSDEKFARSLVHEAHKALGGLD 130
Query: 129 ILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNIINTTSVNAY 188
I+ A +Q + ++ + ++ F N+F+ F++T+ A+ + +G++II T+S+ AY
Sbjct: 131 IMALVAGKQVAIPDIADLTSEQFQKTFAINVFALFWLTQEAIPLLPKGASIITTSSIQAY 190
Query: 189 KGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPL-IPASFNEEKTAQF 247
+ + L+DY +TK AI+ ++R L+ Q+ KGIRVN VAPGPIWT L I ++K QF
Sbjct: 191 QPSPHLLDYAATKAAILNYSRGLAKQVAEKGIRVNIVAPGPIWTALQISGGQTQDKIPQF 250
Query: 248 GSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQV 282
G PMKRAGQP E+AP +V+LAS + SSY+T +V
Sbjct: 251 GQQTPMKRAGQPAELAPVYVYLASQE-SSYVTAEV 284
>FABG_AQUAE (O67610) 3-oxoacyl-[acyl-carrier-protein] reductase (EC
1.1.1.100) (3-ketoacyl-acyl carrier protein reductase)
Length = 248
Score = 132 bits (331), Expect = 2e-30
Identities = 87/253 (34%), Positives = 138/253 (54%), Gaps = 11/253 (4%)
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
KLQGK+++VTG GIGRA+ A G+TVI T G K + ++A K
Sbjct: 4 KLQGKVSLVTGSTRGIGRAIAEKLASAGSTVIITGTSGERAKAVAE-----EIANKYGVK 58
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
+ +L +E+ + +EI N IDILVNNA + + + E V + N
Sbjct: 59 -AHGVEMNLLSEESINKAFEEIYNLVDGIDILVNNAGITRDKLFLR-MSLLDWEEVLKVN 116
Query: 159 IFSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLV 216
+ F +T+++L+ M + I+N +SV + GN ++Y++TK ++ FT++L+ +L
Sbjct: 117 LTGTFLVTQNSLRKMIKQRWGRIVNISSVVGFTGNVGQVNYSTTKAGLIGFTKSLAKELA 176
Query: 217 SKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSS 276
+ + VN VAPG I T + A +EE ++ +P+ R G P EVA +FL S + +S
Sbjct: 177 PRNVLVNAVAPGFIETDM-TAVLSEEIKQKYKEQIPLGRFGSPEEVANVVLFLCS-ELAS 234
Query: 277 YITGQVLHPNGDM 289
YITG+V+H NG M
Sbjct: 235 YITGEVIHVNGGM 247
>FABG_THEMA (Q9X248) 3-oxoacyl-[acyl-carrier-protein] reductase (EC
1.1.1.100) (3-ketoacyl-acyl carrier protein reductase)
Length = 246
Score = 131 bits (329), Expect = 3e-30
Identities = 90/256 (35%), Positives = 144/256 (56%), Gaps = 14/256 (5%)
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+L+GK+ ++TG SGIG+A LFA EGATVI G K+ D+L +K A+ K
Sbjct: 2 RLEGKVCLITGAASGIGKATTLLFAQEGATVI----AGDISKENLDSL--VKEAEGLPGK 55
Query: 99 -DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRT 157
DP + ++ + K V+++++ YGRID+LVNNA + V + E + V
Sbjct: 56 VDPYVL--NVTDRDQIKEVVEKVVQKYGRIDVLVNNAGITRDALLVR-MKEEDWDAVINV 112
Query: 158 NIFSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQL 215
N+ F +T+ + +M + +I+N +SV GN +Y ++K ++ T+ + +L
Sbjct: 113 NLKGVFNVTQMVVPYMIKQRNGSIVNVSSVVGIYGNPGQTNYAASKAGVIGMTKTWAKEL 172
Query: 216 VSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCS 275
+ IRVN VAPG I TP+ E+ S +P+ R G+P EVA +FLAS++ S
Sbjct: 173 AGRNIRVNAVAPGFIETPM-TEKLPEKARETALSRIPLGRFGKPEEVAQVILFLASDE-S 230
Query: 276 SYITGQVLHPNGDMLI 291
SY+TGQV+ +G ++I
Sbjct: 231 SYVTGQVIGIDGGLVI 246
>FABG_VIBCH (Q9KQH7) 3-oxoacyl-[acyl-carrier-protein] reductase (EC
1.1.1.100) (3-ketoacyl-acyl carrier protein reductase)
Length = 244
Score = 129 bits (325), Expect = 8e-30
Identities = 89/252 (35%), Positives = 133/252 (52%), Gaps = 15/252 (5%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L+GK+A+VTG GIG+A+ L A GA VI T + D L N +
Sbjct: 3 LEGKVALVTGASRGIGKAIAELLAERGAKVIGTATSESGAQAISDYLGDNGKGMALNVTN 62
Query: 100 PMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNI 159
P +I A L K + DE +G +DILVNNA + + + E + TN+
Sbjct: 63 PESIEAVL------KAITDE----FGGVDILVNNAGITRD-NLLMRMKEEEWSDIMETNL 111
Query: 160 FSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVS 217
S F +++ L+ M K IIN SV GN+ +Y + K ++ FT++++ ++ S
Sbjct: 112 TSIFRLSKAVLRGMMKKRQGRIINVGSVVGTMGNAGQANYAAAKAGVIGFTKSMAREVAS 171
Query: 218 KGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSY 277
+G+ VN VAPG I T + A +E++TA + VP R G P E+A + FLAS + ++Y
Sbjct: 172 RGVTVNTVAPGFIETDMTKALNDEQRTATL-AQVPAGRLGDPREIASAVAFLASPE-AAY 229
Query: 278 ITGQVLHPNGDM 289
ITG+ LH NG M
Sbjct: 230 ITGETLHVNGGM 241
>VER1_ASPPA (P50161) Versicolorin reductase (EC 1.1.-.-) (VER-1)
Length = 262
Score = 128 bits (322), Expect = 2e-29
Identities = 87/262 (33%), Positives = 139/262 (52%), Gaps = 20/262 (7%)
Query: 38 NKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANA 97
++L GK+A+VTG GIG A+ GA V+ Y H + A ++ +K AN
Sbjct: 5 HRLDGKVALVTGAGRGIGAAIAVALGERGAKVVVNYA--HSREAAEKVVEQIK----ANG 58
Query: 98 KDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRT 157
D +AI AD+G E +++ E + +G +DI+ +NA G ++++ +RVFR
Sbjct: 59 TDAIAIQADVGDPEATAKLMAETVRHFGYLDIVSSNAGI-VSFGHLKDVTPEEFDRVFRV 117
Query: 158 NIFSYFFMTRHALKHMKEGSNIINTTSVNA-YKGNSTLIDYTSTKGAIVAFTRALSLQLV 216
N FF+ R A +HM+EG II T+S A KG Y+ +KGAI F R +++
Sbjct: 118 NTRGQFFVAREAYRHMREGGRIILTSSNTACVKGVPKHAVYSGSKGAIDTFVRCMAIDCG 177
Query: 217 SKGIRVNGVAPGPIWTPLIPA----------SFNEEKTAQFGSDV-PMKRAGQPVEVAPS 265
K I VN VAPG I T + A +F +E+ + + + P+ R G PV+VA
Sbjct: 178 DKKITVNAVAPGAIKTDMFLAVSREYIPNGETFTDEQVDECAAWLSPLNRVGLPVDVARV 237
Query: 266 FVFLASNQCSSYITGQVLHPNG 287
FLAS+ + +++G+++ +G
Sbjct: 238 VSFLASD-TAEWVSGKIIGVDG 258
>DHG2_BACSU (P80869) Glucose 1-dehydrogenase II (EC 1.1.1.47)
(GLCDH-II) (GDH-II) (General stress protein 74) (GSP74)
Length = 258
Score = 128 bits (322), Expect = 2e-29
Identities = 82/261 (31%), Positives = 131/261 (49%), Gaps = 12/261 (4%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARD-TLDMLKMAKTANAK 98
L GK A+VTG GIG+A+ F E V+ Y H D D TL+++K N
Sbjct: 5 LTGKTAIVTGSSKGIGKAIAERFGKEKMNVVVNY---HSDPSGADETLEIIKQ----NGG 57
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
+++ AD+ +E + ++D + +G +D++VNN+ E+ +RV N
Sbjct: 58 KAVSVEADVSKEEGIQALLDTALEHFGTLDVMVNNSGFNGVEAMPHEMSLEDWQRVIDVN 117
Query: 159 IFSYFFMTRHALKHMKEGS---NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQL 215
+ F + AL HM + + N++N +SV+ + Y+++KG I T L+L
Sbjct: 118 VTGTFLGAKAALNHMMKNNIKGNVLNISSVHQQIPRPVNVQYSTSKGGIKMMTETLALNY 177
Query: 216 VSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCS 275
KGIRVN +APG I T + EE + +PMK G+P EVA + +L S + +
Sbjct: 178 ADKGIRVNAIAPGTIATESNVDTKKEESRQKQLKKIPMKAFGKPEEVAAAAAWLVSEE-A 236
Query: 276 SYITGQVLHPNGDMLINASNV 296
SY+TG L +G M + S +
Sbjct: 237 SYVTGATLFVDGGMTLYPSQL 257
>YXBG_BACSU (P46331) Hypothetical oxidoreductase yxbG (EC 1.-.-.-)
Length = 273
Score = 127 bits (319), Expect = 4e-29
Identities = 88/261 (33%), Positives = 137/261 (51%), Gaps = 18/261 (6%)
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDA-RDTLDMLKMAKTANA 97
+L+ K AV+TG +GIG+A +FA EGA VI G +KD +T+D ++ N
Sbjct: 3 RLENKTAVITGAATGIGQATAEVFANEGARVII----GDINKDQMEETVDAIRK----NG 54
Query: 98 KDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRT 157
+ D+ + + K D+I +A G IDIL NNA E G V E +R+
Sbjct: 55 GQAESFHLDVSDENSVKAFADQIKDACGTIDILFNNAGVDQEGGKVHEYPVDLFDRIIAV 114
Query: 158 NIFSYFFMTRHALKHMKE-GSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLV 216
++ F +++ + M E G +IINT+S++ + Y + KG I T+A+++
Sbjct: 115 DLRGTFLCSKYLIPLMLENGGSIINTSSMSGRAADLDRSGYNAAKGGITNLTKAMAIDYA 174
Query: 217 SKGIRVNGVAPGPIWTPLI---PASFNEEKTAQFGSD----VPMKRAGQPVEVAPSFVFL 269
GIRVN ++PG I TPLI + +E QF P+ R GQP E+A +FL
Sbjct: 175 RNGIRVNSISPGTIETPLIDKLAGTKEQEMGEQFREANKWITPLGRLGQPKEMATVALFL 234
Query: 270 ASNQCSSYITGQVLHPNGDML 290
AS+ SSY+TG+ + +G ++
Sbjct: 235 ASDD-SSYVTGEDITADGGIM 254
>STCU_EMENI (Q00791) Versicolorin reductase (EC 1.1.-.-)
Length = 264
Score = 127 bits (319), Expect = 4e-29
Identities = 88/261 (33%), Positives = 140/261 (52%), Gaps = 20/261 (7%)
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+L GK+A+VTG GIG A+ GA V+ Y E A +D +K +NA+
Sbjct: 8 RLDGKVALVTGAGRGIGAAIAVALGQRGAKVVVNYANSREA--AEKVVDEIK----SNAQ 61
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
++I AD+G + +++D+ + +G +DI+ +NA G V+++ +RVFR N
Sbjct: 62 TAISIQADVGDPDAVTKLMDQAVEHFGYLDIVSSNAGI-VSFGHVKDVTPDEFDRVFRVN 120
Query: 159 IFSYFFMTRHALKHMKEGSNIINTTSVNA-YKGNSTLIDYTSTKGAIVAFTRALSLQLVS 217
FF+ R A +H++EG II T+S A KG Y+ +KGAI F R L++
Sbjct: 121 TRGQFFVAREAYRHLREGGRIILTSSNTASVKGVPRHAVYSGSKGAIDTFVRCLAIDCGD 180
Query: 218 KGIRVNGVAPGPIWTPL--------IP--ASFNEEKTAQFGSDV-PMKRAGQPVEVAPSF 266
K I VN VAPG I T + IP +F +E+ + + + P+ R G PV+VA
Sbjct: 181 KKITVNAVAPGAIKTDMFLSVSREYIPNGETFTDEQVDECAAWLSPLNRVGLPVDVARVV 240
Query: 267 VFLASNQCSSYITGQVLHPNG 287
FLAS+ + +I+G+++ +G
Sbjct: 241 SFLASD-AAEWISGKIIGVDG 260
>DHGA_BACME (P10528) Glucose 1-dehydrogenase A (EC 1.1.1.47)
Length = 261
Score = 126 bits (316), Expect = 8e-29
Identities = 82/254 (32%), Positives = 131/254 (51%), Gaps = 12/254 (4%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L+ K+ V+TGG +G+GRA+ F E A V+ Y E+ LD K + A +
Sbjct: 5 LKDKVVVITGGSTGLGRAMAVRFGQEEAKVVINYYNNEEE-----ALDAKKEVEEAGGQ- 58
Query: 100 PMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNI 159
+ + D+ +E+ ++ I +G +D+++NNA + S E+ +V TN+
Sbjct: 59 AIIVQGDVTKEEDVVNLVQTAIKEFGTLDVMINNAGVENPVPS-HELSLDNWNKVIDTNL 117
Query: 160 FSYFFMTRHALKHMKEGS---NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLV 216
F +R A+K+ E N+IN +SV+ + Y ++KG + T L+L+
Sbjct: 118 TGAFLGSREAIKYFVENDIKGNVINMSSVHEMIPWPLFVHYAASKGGMKLMTETLALEYA 177
Query: 217 SKGIRVNGVAPGPIWTPLIPASF-NEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCS 275
KGIRVN + PG + TP+ F + E+ A S +PM G+P EVA FLAS+Q +
Sbjct: 178 PKGIRVNNIGPGAMNTPINAEKFADPEQRADVESMIPMGYIGKPEEVAAVAAFLASSQ-A 236
Query: 276 SYITGQVLHPNGDM 289
SY+TG L +G M
Sbjct: 237 SYVTGITLFADGGM 250
>FABG_VIBHA (P55336) 3-oxoacyl-[acyl-carrier-protein] reductase (EC
1.1.1.100) (3-ketoacyl-acyl carrier protein reductase)
Length = 244
Score = 125 bits (314), Expect = 1e-28
Identities = 90/252 (35%), Positives = 130/252 (50%), Gaps = 15/252 (5%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L+GKIA+VTG GIGRA+ L GATVI T + L N D
Sbjct: 3 LEGKIALVTGASRGIGRAIAELLVERGATVIGTATSEGGAAAISEYLGENGKGLALNVTD 62
Query: 100 PMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNI 159
+I A L K + DE G IDILVNNA + + + + + TN+
Sbjct: 63 VESIEATL------KTINDEC----GAIDILVNNAGITRD-NLLMRMKDDEWNDIINTNL 111
Query: 160 FSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVS 217
+ M++ L+ M K IIN SV GN+ +Y + K ++ FT++++ ++ S
Sbjct: 112 TPIYRMSKAVLRGMMKKRAGRIINVGSVVGTMGNAGQTNYAAAKAGVIGFTKSMAREVAS 171
Query: 218 KGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSY 277
+G+ VN VAPG I T + A N+++ A S+VP R G P E+A + VFLAS + ++Y
Sbjct: 172 RGVTVNTVAPGFIETDMTKA-LNDDQRAATLSNVPAGRLGDPREIASAVVFLASPE-AAY 229
Query: 278 ITGQVLHPNGDM 289
ITG+ LH NG M
Sbjct: 230 ITGETLHVNGGM 241
>DHG4_BACME (P39485) Glucose 1-dehydrogenase IV (EC 1.1.1.47)
(GLCDH-IV)
Length = 261
Score = 124 bits (310), Expect = 4e-28
Identities = 81/254 (31%), Positives = 130/254 (50%), Gaps = 12/254 (4%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L+ K+ V+TGG +G+GRA+ F E A V+ Y E+ LD K + A +
Sbjct: 5 LKDKVVVITGGSTGLGRAMAVRFGQEEAKVVINYYNNEEE-----ALDAKKEVEEAGGQ- 58
Query: 100 PMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNI 159
+ + D+ +E+ ++ I +G +D+++NNA + S E+ +V TN+
Sbjct: 59 AIIVQGDVTKEEDVVNLVQTAIKEFGTLDVMINNAGVENPVPS-HELSLDNWNKVIDTNL 117
Query: 160 FSYFFMTRHALKHMKEGS---NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLV 216
F +R A+K+ E N+IN +SV+ + Y ++KG + T L+L+
Sbjct: 118 TGAFLGSREAIKYFVENDIKGNVINMSSVHEMIPWPLFVHYAASKGGMKLMTETLALEYA 177
Query: 217 SKGIRVNGVAPGPIWTPLIPASFNEE-KTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCS 275
KGIRVN + PG + TP+ F + + A S +PM G+P EVA FLAS+Q +
Sbjct: 178 PKGIRVNNIGPGAMNTPINAEKFADPVQRADVESMIPMGYIGKPEEVAAVAAFLASSQ-A 236
Query: 276 SYITGQVLHPNGDM 289
SY+TG L +G M
Sbjct: 237 SYVTGITLFADGGM 250
>DHG3_BACME (P39484) Glucose 1-dehydrogenase III (EC 1.1.1.47)
(GLCDH-III)
Length = 261
Score = 124 bits (310), Expect = 4e-28
Identities = 81/254 (31%), Positives = 130/254 (50%), Gaps = 12/254 (4%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L+ K+ V+TGG +G+GRA+ F E A V+ Y E+ LD K + A +
Sbjct: 5 LKDKVVVITGGSTGLGRAMAVRFGQEEAKVVINYYNNEEE-----ALDAKKEVEEAGGQ- 58
Query: 100 PMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNI 159
+ + D+ +E+ ++ I +G +D+++NNA + S E+ +V TN+
Sbjct: 59 AIIVQGDVTKEEDVVNLVQTAIKEFGTLDVMINNAGVENPVPS-HELSLDNWNKVIDTNL 117
Query: 160 FSYFFMTRHALKHMKEGS---NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLV 216
F +R A+K+ E N+IN +SV+ + Y ++KG + T L+L+
Sbjct: 118 TGAFLGSREAIKYFVENDIKGNVINMSSVHEMIPWPLFVHYAASKGGMKQMTETLALEYA 177
Query: 217 SKGIRVNGVAPGPIWTPLIPASFNEE-KTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCS 275
KGIRVN + PG + TP+ F + + A S +PM G+P EVA FLAS+Q +
Sbjct: 178 PKGIRVNNIGPGAMNTPINAEKFADPVQRADVESMIPMGYIGKPEEVAAVAAFLASSQ-A 236
Query: 276 SYITGQVLHPNGDM 289
SY+TG L +G M
Sbjct: 237 SYVTGITLFADGGM 250
>DHG2_BACME (P39483) Glucose 1-dehydrogenase II (EC 1.1.1.47)
(GLCDH-II)
Length = 261
Score = 123 bits (308), Expect = 7e-28
Identities = 80/254 (31%), Positives = 132/254 (51%), Gaps = 12/254 (4%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L+ K+ VVTGG G+GRA+ F E + V+ Y E+ L++ K + A +
Sbjct: 5 LKDKVVVVTGGSKGLGRAMAVRFGQEQSKVVVNYRSNEEE-----ALEVKKEIEEAGGQ- 58
Query: 100 PMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNI 159
+ + D+ +E+ +++ + +G +D+++NNA + S E+ +V TN+
Sbjct: 59 AIIVRGDVTKEEDVVNLVETAVKEFGSLDVMINNAGVENPVPS-HELSLENWNQVIDTNL 117
Query: 160 FSYFFMTRHALKHMKEGS---NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLV 216
F +R A+K+ E N+IN +SV+ + Y ++KG + T L+L+
Sbjct: 118 TGAFLGSREAIKYFVENDIKGNVINMSSVHEMIPWPLFVHYAASKGGMKLMTETLALEYA 177
Query: 217 SKGIRVNGVAPGPIWTPLIPASF-NEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCS 275
KGIRVN + PG I TP+ F + E+ A S +PM G+P E+A FLAS+Q +
Sbjct: 178 PKGIRVNNIGPGAIDTPINAEKFADPEQRADVESMIPMGYIGKPEEIASVAAFLASSQ-A 236
Query: 276 SYITGQVLHPNGDM 289
SY+TG L +G M
Sbjct: 237 SYVTGITLFADGGM 250
>NODG_AZOBR (P17611) Nodulation protein G
Length = 246
Score = 122 bits (306), Expect = 1e-27
Identities = 83/249 (33%), Positives = 130/249 (51%), Gaps = 11/249 (4%)
Query: 43 KIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMA 102
KIA+VTG G+G A+C A +G V + E A L + K +A
Sbjct: 4 KIALVTGAMGGLGTAICQALAKDGCIVAANCLPNFEPAAA-----WLGQQEALGFKFYVA 58
Query: 103 IPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSY 162
D+ E+CK ++ +I G +DILVNNA + ++D+ + + V TN+ S
Sbjct: 59 -EGDVSDFESCKAMVAKIEADLGPVDILVNNAGITRD-KFFAKMDKAQWDAVIATNLSSL 116
Query: 163 FFMTRHALKHMKEGS--NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGI 220
F +T+ M E IIN +SVN KG + +Y++ K ++ FT+AL+ +L +KG+
Sbjct: 117 FNVTQQVSPKMAERGWGRIINISSVNGVKGQAGQTNYSAAKAGVIGFTKALAAELATKGV 176
Query: 221 RVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITG 280
VN +APG I T ++ A E+ VPMKR G+P E+ + +LAS + + Y+TG
Sbjct: 177 TVNAIAPGYIGTDMVMA-IREDIRQAITDSVPMKRLGRPDEIGGAVSYLAS-EIAGYVTG 234
Query: 281 QVLHPNGDM 289
L+ NG +
Sbjct: 235 STLNINGGL 243
>DHG_BACSU (P12310) Glucose 1-dehydrogenase (EC 1.1.1.47)
Length = 261
Score = 122 bits (306), Expect = 1e-27
Identities = 79/254 (31%), Positives = 128/254 (50%), Gaps = 12/254 (4%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L+GK+ +TG SG+G+A+ F E A V+ Y +D + +K +
Sbjct: 5 LKGKVVAITGAASGLGKAMAIRFGKEQAKVVINYYSNKQDPNE------VKEEVIKAGGE 58
Query: 100 PMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNI 159
+ + D+ +E+ K ++ I +G +DI++NNA + S E+ ++V TN+
Sbjct: 59 AVVVQGDVTKEEDVKNIVQTAIKEFGTLDIMINNAGLENPVPS-HEMPLKDWDKVIGTNL 117
Query: 160 FSYFFMTRHALKHMKEGS---NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLV 216
F +R A+K+ E N+IN +SV+ + Y ++KG I T L+L+
Sbjct: 118 TGAFLGSREAIKYFVENDIKGNVINMSSVHEVIPWPLFVHYAASKGGIKLMTETLALEYA 177
Query: 217 SKGIRVNGVAPGPIWTPLIPASFNEEK-TAQFGSDVPMKRAGQPVEVAPSFVFLASNQCS 275
KGIRVN + PG I TP+ F + K A S +PM G+P E+A +LAS + +
Sbjct: 178 PKGIRVNNIGPGAINTPINAEKFADPKQKADVESMIPMGYIGEPEEIAAVAAWLASKE-A 236
Query: 276 SYITGQVLHPNGDM 289
SY+TG L +G M
Sbjct: 237 SYVTGITLFADGGM 250
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,178,420
Number of Sequences: 164201
Number of extensions: 1670941
Number of successful extensions: 4868
Number of sequences better than 10.0: 393
Number of HSP's better than 10.0 without gapping: 224
Number of HSP's successfully gapped in prelim test: 169
Number of HSP's that attempted gapping in prelim test: 4048
Number of HSP's gapped (non-prelim): 409
length of query: 323
length of database: 59,974,054
effective HSP length: 110
effective length of query: 213
effective length of database: 41,911,944
effective search space: 8927244072
effective search space used: 8927244072
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC135319.2


