

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135313.7 - phase: 0
(199 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
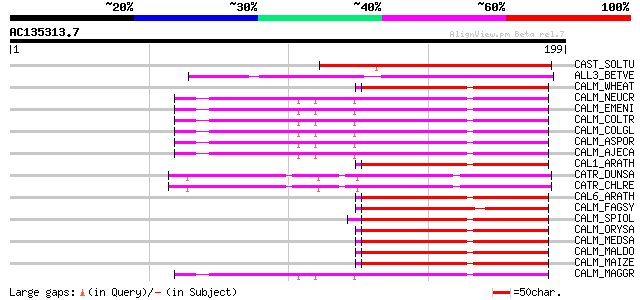
Score E
Sequences producing significant alignments: (bits) Value
CAST_SOLTU (Q09011) Calcium-binding protein CAST 80 4e-15
ALL3_BETVE (P43187) Calcium-binding allergen Bet v 3 (Bet v III) 72 6e-13
CALM_WHEAT (P04464) Calmodulin (CaM) 56 5e-08
CALM_NEUCR (P61859) Calmodulin (CaM) 56 5e-08
CALM_EMENI (P60204) Calmodulin (CaM) 56 5e-08
CALM_COLTR (P61860) Calmodulin (CaM) 56 5e-08
CALM_COLGL (P61861) Calmodulin (CaM) 56 5e-08
CALM_ASPOR (P60205) Calmodulin (CaM) 56 5e-08
CALM_AJECA (P60206) Calmodulin (CaM) 56 5e-08
CAL1_ARATH (P25854) Calmodulin 1/4 (CaM 1/4) 55 8e-08
CATR_DUNSA (P54213) Caltractin (Centrin) 55 1e-07
CATR_CHLRE (P05434) Caltractin (Centrin) (20 kDa calcium-binding... 55 1e-07
CAL6_ARATH (Q03509) Calmodulin 6 (CaM 6) 55 1e-07
CALM_FAGSY (Q39752) Calmodulin (CaM) 55 1e-07
CALM_SPIOL (P04353) Calmodulin (CaM) 54 2e-07
CALM_ORYSA (P29612) Calmodulin (CaM) 54 2e-07
CALM_MEDSA (P17928) Calmodulin (CaM) 54 2e-07
CALM_MALDO (P48976) Calmodulin (CaM) 54 2e-07
CALM_MAIZE (P41040) Calmodulin (CaM) 54 2e-07
CALM_MAGGR (Q9UWF0) Calmodulin (CaM) 54 2e-07
>CAST_SOLTU (Q09011) Calcium-binding protein CAST
Length = 199
Score = 79.7 bits (195), Expect = 4e-15
Identities = 42/85 (49%), Positives = 60/85 (70%), Gaps = 2/85 (2%)
Query: 112 YGSKELCEVFEENEPSLEE--LKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENC 169
+GSK ++ +P+ +E LK+AFDVFDEN DGFI AKELQ VL LGL +GSE +
Sbjct: 109 FGSKCEDKLGLNPDPAQDESDLKEAFDVFDENGDGFISAKELQVVLEKLGLPEGSEIDRV 168
Query: 170 QKMITIFDENQDGRIDFIEFVNIMK 194
+ MI+ +++ DGR+DF EF ++M+
Sbjct: 169 EMMISSVEQDHDGRVDFFEFKDMMR 193
Score = 37.0 bits (84), Expect = 0.030
Identities = 22/77 (28%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query: 121 FEENEPSLEE--LKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDE 178
F PSL L++ FDVFD N D I +EL + L +LGL ++ + M+ + +
Sbjct: 29 FRLRSPSLNSIRLRRIFDVFDRNHDCLISVEELSQALNLLGL--DADLSEIESMVKLHIK 86
Query: 179 NQDGRIDFIEFVNIMKN 195
++ + F +F + ++
Sbjct: 87 PENTGLRFEDFETLHRS 103
>ALL3_BETVE (P43187) Calcium-binding allergen Bet v 3 (Bet v III)
Length = 205
Score = 72.4 bits (176), Expect = 6e-13
Identities = 47/131 (35%), Positives = 65/131 (48%), Gaps = 9/131 (6%)
Query: 65 ESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCSSESEELEEKYGSKELCEVFEEN 124
ES S E NIG D I + + F E E+ E+ K + E +
Sbjct: 78 ESTVKSFTREGNIGLQFEDFIS---LHQSLNDSYFAYGGEDEDDNEEDMRKSILSQEEAD 134
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
F VFDE+ DG+I A+ELQ VL LG +GSE + +KMI D N+DGR+
Sbjct: 135 SFG------GFKVFDEDGDGYISARELQMVLGKLGFSEGSEIDRVEKMIVSVDSNRDGRV 188
Query: 185 DFIEFVNIMKN 195
DF EF ++M++
Sbjct: 189 DFFEFKDMMRS 199
Score = 40.0 bits (92), Expect = 0.004
Identities = 25/65 (38%), Positives = 38/65 (58%), Gaps = 8/65 (12%)
Query: 131 LKQAFDVFDENKDGFIDAKELQRVLVILGLKQG-SEFENCQKMITIFDENQDGRI--DFI 187
L++ FD+FD+N DG I EL R L +LGL+ SE E+ K T ++G I F
Sbjct: 41 LRRIFDLFDKNSDGIITVDELSRALNLLGLETDLSELESTVKSFT-----REGNIGLQFE 95
Query: 188 EFVNI 192
+F+++
Sbjct: 96 DFISL 100
>CALM_WHEAT (P04464) Calmodulin (CaM)
Length = 148
Score = 56.2 bits (134), Expect = 5e-08
Identities = 32/67 (47%), Positives = 44/67 (64%), Gaps = 2/67 (2%)
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD+++DGFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 81 SEEELKEAFRVFDKDQDGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 138
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 139 EEFVKVM 145
Score = 48.5 bits (114), Expect = 1e-05
Identities = 28/69 (40%), Positives = 39/69 (55%), Gaps = 2/69 (2%)
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 6 DEQIAEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 63
Query: 185 DFIEFVNIM 193
DF EF+N+M
Sbjct: 64 DFPEFLNLM 72
>CALM_NEUCR (P61859) Calmodulin (CaM)
Length = 148
Score = 56.2 bits (134), Expect = 5e-08
Identities = 47/146 (32%), Positives = 74/146 (50%), Gaps = 18/146 (12%)
Query: 60 EKKVSESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCS-SESEEL-----EEKYG 113
E++VSE K E S +D I E+ VM +G S SE +++ + G
Sbjct: 6 EEQVSEFK----EAFSLFDKDGDGQITTKELGTVMRSLGQNPSESELQDMINEVDADNNG 61
Query: 114 SKELCEVFE------ENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFE 167
+ + E ++ S EE+++AF VFD + +GFI A EL+ V+ +G K +
Sbjct: 62 TIDFPEFLTMMARKMKDTDSEEEIREAFKVFDRDNNGFISAAELRHVMTSIGEKLTD--D 119
Query: 168 NCQKMITIFDENQDGRIDFIEFVNIM 193
+MI D++ DGRID+ EFV +M
Sbjct: 120 EVDEMIREADQDGDGRIDYNEFVQLM 145
>CALM_EMENI (P60204) Calmodulin (CaM)
Length = 148
Score = 56.2 bits (134), Expect = 5e-08
Identities = 47/146 (32%), Positives = 74/146 (50%), Gaps = 18/146 (12%)
Query: 60 EKKVSESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCS-SESEEL-----EEKYG 113
E++VSE K E S +D I E+ VM +G S SE +++ + G
Sbjct: 6 EEQVSEYK----EAFSLFDKDGDGQITTKELGTVMRSLGQNPSESELQDMINEVDADNNG 61
Query: 114 SKELCEVFE------ENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFE 167
+ + E ++ S EE+++AF VFD + +GFI A EL+ V+ +G K +
Sbjct: 62 TIDFPEFLTMMARKMKDTDSEEEIREAFKVFDRDNNGFISAAELRHVMTSIGEKLTD--D 119
Query: 168 NCQKMITIFDENQDGRIDFIEFVNIM 193
+MI D++ DGRID+ EFV +M
Sbjct: 120 EVDEMIREADQDGDGRIDYNEFVQLM 145
>CALM_COLTR (P61860) Calmodulin (CaM)
Length = 148
Score = 56.2 bits (134), Expect = 5e-08
Identities = 47/146 (32%), Positives = 74/146 (50%), Gaps = 18/146 (12%)
Query: 60 EKKVSESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCS-SESEEL-----EEKYG 113
E++VSE K E S +D I E+ VM +G S SE +++ + G
Sbjct: 6 EEQVSEFK----EAFSLFDKDGDGQITTKELGTVMRSLGQNPSESELQDMINEVDADNNG 61
Query: 114 SKELCEVFE------ENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFE 167
+ + E ++ S EE+++AF VFD + +GFI A EL+ V+ +G K +
Sbjct: 62 TIDFPEFLTMMARKMKDTDSEEEIREAFKVFDRDNNGFISAAELRHVMTSIGEKLTD--D 119
Query: 168 NCQKMITIFDENQDGRIDFIEFVNIM 193
+MI D++ DGRID+ EFV +M
Sbjct: 120 EVDEMIREADQDGDGRIDYNEFVQLM 145
>CALM_COLGL (P61861) Calmodulin (CaM)
Length = 148
Score = 56.2 bits (134), Expect = 5e-08
Identities = 47/146 (32%), Positives = 74/146 (50%), Gaps = 18/146 (12%)
Query: 60 EKKVSESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCS-SESEEL-----EEKYG 113
E++VSE K E S +D I E+ VM +G S SE +++ + G
Sbjct: 6 EEQVSEFK----EAFSLFDKDGDGQITTKELGTVMRSLGQNPSESELQDMINEVDADNNG 61
Query: 114 SKELCEVFE------ENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFE 167
+ + E ++ S EE+++AF VFD + +GFI A EL+ V+ +G K +
Sbjct: 62 TIDFPEFLTMMARKMKDTDSEEEIREAFKVFDRDNNGFISAAELRHVMTSIGEKLTD--D 119
Query: 168 NCQKMITIFDENQDGRIDFIEFVNIM 193
+MI D++ DGRID+ EFV +M
Sbjct: 120 EVDEMIREADQDGDGRIDYNEFVQLM 145
>CALM_ASPOR (P60205) Calmodulin (CaM)
Length = 148
Score = 56.2 bits (134), Expect = 5e-08
Identities = 47/146 (32%), Positives = 74/146 (50%), Gaps = 18/146 (12%)
Query: 60 EKKVSESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCS-SESEEL-----EEKYG 113
E++VSE K E S +D I E+ VM +G S SE +++ + G
Sbjct: 6 EEQVSEYK----EAFSLFDKDGDGQITTKELGTVMRSLGQNPSESELQDMINEVDADNNG 61
Query: 114 SKELCEVFE------ENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFE 167
+ + E ++ S EE+++AF VFD + +GFI A EL+ V+ +G K +
Sbjct: 62 TIDFPEFLTMMARKMKDTDSEEEIREAFKVFDRDNNGFISAAELRHVMTSIGEKLTD--D 119
Query: 168 NCQKMITIFDENQDGRIDFIEFVNIM 193
+MI D++ DGRID+ EFV +M
Sbjct: 120 EVDEMIREADQDGDGRIDYNEFVQLM 145
>CALM_AJECA (P60206) Calmodulin (CaM)
Length = 148
Score = 56.2 bits (134), Expect = 5e-08
Identities = 47/146 (32%), Positives = 74/146 (50%), Gaps = 18/146 (12%)
Query: 60 EKKVSESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCS-SESEEL-----EEKYG 113
E++VSE K E S +D I E+ VM +G S SE +++ + G
Sbjct: 6 EEQVSEYK----EAFSLFDKDGDGQITTKELGTVMRSLGQNPSESELQDMINEVDADNNG 61
Query: 114 SKELCEVFE------ENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFE 167
+ + E ++ S EE+++AF VFD + +GFI A EL+ V+ +G K +
Sbjct: 62 TIDFPEFLTMMARKMKDTDSEEEIREAFKVFDRDNNGFISAAELRHVMTSIGEKLTD--D 119
Query: 168 NCQKMITIFDENQDGRIDFIEFVNIM 193
+MI D++ DGRID+ EFV +M
Sbjct: 120 EVDEMIREADQDGDGRIDYNEFVQLM 145
>CAL1_ARATH (P25854) Calmodulin 1/4 (CaM 1/4)
Length = 148
Score = 55.5 bits (132), Expect = 8e-08
Identities = 32/67 (47%), Positives = 45/67 (66%), Gaps = 2/67 (2%)
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E ++MI D + DG+I++
Sbjct: 81 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTD--EEVEEMIREADVDGDGQINY 138
Query: 187 IEFVNIM 193
EFV IM
Sbjct: 139 EEFVKIM 145
Score = 48.9 bits (115), Expect = 8e-06
Identities = 28/69 (40%), Positives = 39/69 (55%), Gaps = 2/69 (2%)
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 6 DEQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 63
Query: 185 DFIEFVNIM 193
DF EF+N+M
Sbjct: 64 DFPEFLNLM 72
>CATR_DUNSA (P54213) Caltractin (Centrin)
Length = 169
Score = 55.1 bits (131), Expect = 1e-07
Identities = 51/159 (32%), Positives = 72/159 (45%), Gaps = 28/159 (17%)
Query: 58 REEKK------VSESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCSSESEELE-- 109
R++KK E K +E D I+ E+K+ M +GF + EE++
Sbjct: 12 RDQKKGRVGGLTEEQKQEIREAFDLFDTDGSGTIDAKELKVAMRALGF--EPKKEEIKKM 69
Query: 110 ----EKYGSKELCEVFEE----------NEPSLEELKQAFDVFDENKDGFIDAKELQRVL 155
+K GS + FEE S EE+ +AF +FD++ GFI K L+RV
Sbjct: 70 IADIDKAGSGTID--FEEFLQMMTSKMGERDSREEIIKAFKLFDDDNTGFITLKNLKRVA 127
Query: 156 VILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIMK 194
LG + E Q+M D N DG+ID EF IMK
Sbjct: 128 KELG--ENLTDEELQEMTDEADRNGDGQIDEDEFYRIMK 164
>CATR_CHLRE (P05434) Caltractin (Centrin) (20 kDa calcium-binding
protein)
Length = 169
Score = 55.1 bits (131), Expect = 1e-07
Identities = 50/158 (31%), Positives = 72/158 (44%), Gaps = 27/158 (17%)
Query: 58 REEKK-----VSESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCSSESEELE--- 109
R++KK E K +E D I+ E+K+ M +GF + EE++
Sbjct: 13 RDQKKGRVGLTEEQKQEIREAFDLFDTDGSGTIDAKELKVAMRALGF--EPKKEEIKKMI 70
Query: 110 ---EKYGSKELCEVFEE----------NEPSLEELKQAFDVFDENKDGFIDAKELQRVLV 156
+K GS + FEE S EE+ +AF +FD++ G I K+L+RV
Sbjct: 71 SEIDKDGSGTID--FEEFLTMMTAKMGERDSREEILKAFRLFDDDNSGTITIKDLRRVAK 128
Query: 157 ILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIMK 194
LG + E Q+MI D N D ID EF+ IMK
Sbjct: 129 ELG--ENLTEEELQEMIAEADRNDDNEIDEDEFIRIMK 164
>CAL6_ARATH (Q03509) Calmodulin 6 (CaM 6)
Length = 148
Score = 55.1 bits (131), Expect = 1e-07
Identities = 31/67 (46%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 81 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLSD--EEVDEMIREADVDGDGQINY 138
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 139 EEFVKVM 145
Score = 48.9 bits (115), Expect = 8e-06
Identities = 28/69 (40%), Positives = 39/69 (55%), Gaps = 2/69 (2%)
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 6 DDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 63
Query: 185 DFIEFVNIM 193
DF EF+N+M
Sbjct: 64 DFPEFLNLM 72
>CALM_FAGSY (Q39752) Calmodulin (CaM)
Length = 147
Score = 54.7 bits (130), Expect = 1e-07
Identities = 31/67 (46%), Positives = 45/67 (66%), Gaps = 3/67 (4%)
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E + +MI D + DG+I++
Sbjct: 81 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEVD---EMIREADVDGDGQINY 137
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 138 EEFVKVM 144
Score = 49.3 bits (116), Expect = 6e-06
Identities = 28/69 (40%), Positives = 39/69 (55%), Gaps = 2/69 (2%)
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 6 DDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDRDGNGTI 63
Query: 185 DFIEFVNIM 193
DF EF+N+M
Sbjct: 64 DFPEFLNLM 72
>CALM_SPIOL (P04353) Calmodulin (CaM)
Length = 148
Score = 54.3 bits (129), Expect = 2e-07
Identities = 31/67 (46%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 81 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 138
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 139 EEFVKVM 145
Score = 48.9 bits (115), Expect = 8e-06
Identities = 28/72 (38%), Positives = 40/72 (54%), Gaps = 2/72 (2%)
Query: 122 EENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQD 181
Z + + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +
Sbjct: 3 ZLTDEQIAEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGN 60
Query: 182 GRIDFIEFVNIM 193
G IDF EF+N+M
Sbjct: 61 GTIDFPEFLNLM 72
>CALM_ORYSA (P29612) Calmodulin (CaM)
Length = 148
Score = 54.3 bits (129), Expect = 2e-07
Identities = 31/67 (46%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 81 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 138
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 139 EEFVKVM 145
Score = 48.5 bits (114), Expect = 1e-05
Identities = 28/69 (40%), Positives = 39/69 (55%), Gaps = 2/69 (2%)
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 6 DDQIAEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 63
Query: 185 DFIEFVNIM 193
DF EF+N+M
Sbjct: 64 DFPEFLNLM 72
>CALM_MEDSA (P17928) Calmodulin (CaM)
Length = 148
Score = 54.3 bits (129), Expect = 2e-07
Identities = 31/67 (46%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 81 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 138
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 139 EEFVKVM 145
Score = 48.9 bits (115), Expect = 8e-06
Identities = 28/69 (40%), Positives = 39/69 (55%), Gaps = 2/69 (2%)
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 6 DEQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 63
Query: 185 DFIEFVNIM 193
DF EF+N+M
Sbjct: 64 DFPEFLNLM 72
>CALM_MALDO (P48976) Calmodulin (CaM)
Length = 148
Score = 54.3 bits (129), Expect = 2e-07
Identities = 31/67 (46%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 81 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 138
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 139 EEFVKVM 145
Score = 45.1 bits (105), Expect = 1e-04
Identities = 27/69 (39%), Positives = 38/69 (54%), Gaps = 2/69 (2%)
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 6 DDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 63
Query: 185 DFIEFVNIM 193
DF E +N+M
Sbjct: 64 DFPEPLNLM 72
>CALM_MAIZE (P41040) Calmodulin (CaM)
Length = 148
Score = 54.3 bits (129), Expect = 2e-07
Identities = 31/67 (46%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 81 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 138
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 139 EEFVKVM 145
Score = 46.2 bits (108), Expect = 5e-05
Identities = 27/69 (39%), Positives = 38/69 (54%), Gaps = 2/69 (2%)
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 6 DEQIAEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 63
Query: 185 DFIEFVNIM 193
DF E +N+M
Sbjct: 64 DFPELLNLM 72
>CALM_MAGGR (Q9UWF0) Calmodulin (CaM)
Length = 148
Score = 54.3 bits (129), Expect = 2e-07
Identities = 46/146 (31%), Positives = 73/146 (49%), Gaps = 18/146 (12%)
Query: 60 EKKVSESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCS-SESEEL-----EEKYG 113
E++VSE K E S +D I E+ VM +G S SE +++ + G
Sbjct: 6 EEQVSEFK----EAFSLFDKDGDGQITTKELGTVMRSLGQNPSESELQDMINEVDADNNG 61
Query: 114 SKELCEVFE------ENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFE 167
+ + E ++ S EE+++AF FD + +GFI A EL+ V+ +G K +
Sbjct: 62 TIDFPEFLTMMARKMKDTDSEEEIREAFKFFDRDNNGFISAAELRHVMTSIGEKLTD--D 119
Query: 168 NCQKMITIFDENQDGRIDFIEFVNIM 193
+MI D++ DGRID+ EFV +M
Sbjct: 120 EVDEMIREADQDGDGRIDYNEFVQLM 145
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,259,480
Number of Sequences: 164201
Number of extensions: 988622
Number of successful extensions: 5336
Number of sequences better than 10.0: 552
Number of HSP's better than 10.0 without gapping: 325
Number of HSP's successfully gapped in prelim test: 231
Number of HSP's that attempted gapping in prelim test: 4111
Number of HSP's gapped (non-prelim): 1001
length of query: 199
length of database: 59,974,054
effective HSP length: 105
effective length of query: 94
effective length of database: 42,732,949
effective search space: 4016897206
effective search space used: 4016897206
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC135313.7


