

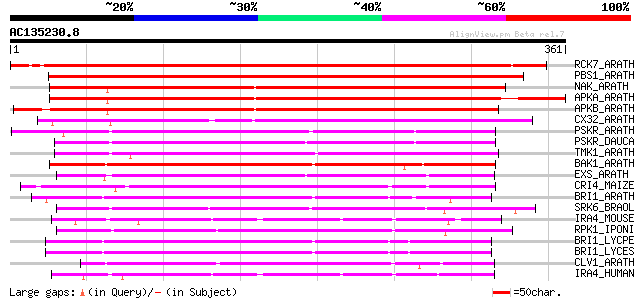
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135230.8 + phase: 1 /partial
(361 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 488 e-137
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 428 e-119
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 281 1e-75
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 276 6e-74
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 275 1e-73
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 246 8e-65
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 219 8e-57
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 218 2e-56
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 215 1e-55
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 215 2e-55
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 214 2e-55
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 201 2e-51
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 193 5e-49
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 188 2e-47
IRA4_MOUSE (Q8R4K2) Interleukin-1 receptor-associated kinase-4 (... 182 1e-45
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 181 2e-45
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 181 2e-45
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 181 3e-45
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 179 1e-44
IRA4_HUMAN (Q9NWZ3) Interleukin-1 receptor-associated kinase-4 (... 175 1e-43
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 488 bits (1255), Expect = e-137
Identities = 248/349 (71%), Positives = 282/349 (80%), Gaps = 4/349 (1%)
Query: 1 KEVDLSVNEKTEDELKDGVSTNGKLAKTFTLNELAAATGNFSANCFVGEGGFGKVYKGYL 60
KE LS++ K + L D V+ GK A+TFT ELA ATGNF ++CF+GEGGFGKV+KG +
Sbjct: 66 KEDQLSLDVKGLN-LNDQVT--GKKAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTI 122
Query: 61 QKTNQFVAIKQLDPKGIQGTREFVVEVLTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPL 120
+K +Q VAIKQLD G+QG REFVVEVLTLSLA+H NLVKL+GF AEGDQRLLVYEYMP
Sbjct: 123 EKLDQVVAIKQLDRNGVQGIREFVVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQ 182
Query: 121 GSLESHLHDLPPGKNPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDY 180
GSLE HLH LP GK PLDWNTRM+IAAG A+GLEYLHD M PPVIYRDLKCSNILLG DY
Sbjct: 183 GSLEDHLHVLPSGKKPLDWNTRMKIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDY 242
Query: 181 HPKLSDFGLAKIGPMGDQTHVSTRVMGTHGYCAPDYGMTGQLTFKSDIYSFGVALLELIT 240
PKLSDFGLAK+GP GD+THVSTRVMGT+GYCAPDY MTGQLTFKSDIYSFGV LLELIT
Sbjct: 243 QPKLSDFGLAKVGPSGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELIT 302
Query: 241 GRKAYDESKPSKKRHLVKWATPLFRDQKNFSKMVDPLLKGQYPARGLYQALAIASMCVVE 300
GRKA D +K K ++LV WA PLF+D++NF KMVDPLL+GQYP RGLYQALAI++MCV E
Sbjct: 303 GRKAIDNTKTRKDQNLVGWARPLFKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQE 362
Query: 301 QTNMRPVIADVVSALDFLASQKYEPRVHPIQRSRYGSSSSRSRAKGHRR 349
Q MRPV++DVV AL+FLAS KY+P P S S R R +R
Sbjct: 363 QPTMRPVVSDVVLALNFLASSKYDPN-SPSSSSGKNPSFHRDRDDEEKR 410
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 428 bits (1100), Expect = e-119
Identities = 206/309 (66%), Positives = 243/309 (77%)
Query: 26 AKTFTLNELAAATGNFSANCFVGEGGFGKVYKGYLQKTNQFVAIKQLDPKGIQGTREFVV 85
A TF ELAAAT NF + F+GEGGFG+VYKG L T Q VA+KQLD G+QG REF+V
Sbjct: 71 AHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLV 130
Query: 86 EVLTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPPGKNPLDWNTRMRI 145
EVL LSL H NLV L+G+ A+GDQRLLVYE+MPLGSLE HLHDLPP K LDWN RM+I
Sbjct: 131 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKI 190
Query: 146 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGDQTHVSTRV 205
AAG AKGLE+LHD+ PPVIYRD K SNILL +HPKLSDFGLAK+GP GD++HVSTRV
Sbjct: 191 AAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRV 250
Query: 206 MGTHGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESKPSKKRHLVKWATPLFR 265
MGT+GYCAP+Y MTGQLT KSD+YSFGV LELITGRKA D P +++LV WA PLF
Sbjct: 251 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFN 310
Query: 266 DQKNFSKMVDPLLKGQYPARGLYQALAIASMCVVEQTNMRPVIADVVSALDFLASQKYEP 325
D++ F K+ DP LKG++P R LYQALA+ASMC+ EQ RP+IADVV+AL +LA+Q Y+P
Sbjct: 311 DRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTALSYLANQAYDP 370
Query: 326 RVHPIQRSR 334
+R+R
Sbjct: 371 SKDDSRRNR 379
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 281 bits (720), Expect = 1e-75
Identities = 144/305 (47%), Positives = 203/305 (66%), Gaps = 10/305 (3%)
Query: 27 KTFTLNELAAATGNFSANCFVGEGGFGKVYKGYLQK---------TNQFVAIKQLDPKGI 77
K F+L+EL +AT NF + VGEGGFG V+KG++ + T +A+K+L+ +G
Sbjct: 54 KNFSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEGF 113
Query: 78 QGTREFVVEVLTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPPGKNPL 137
QG RE++ E+ L +H NLVKL+G+ E + RLLVYE+M GSLE+HL PL
Sbjct: 114 QGHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTFYQPL 173
Query: 138 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGD 197
WNTR+R+A G A+GL +LH+ +P VIYRD K SNILL ++Y+ KLSDFGLA+ GPMGD
Sbjct: 174 SWNTRVRMALGAARGLAFLHNA-QPQVIYRDFKASNILLDSNYNAKLSDFGLARDGPMGD 232
Query: 198 QTHVSTRVMGTHGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESKPSKKRHLV 257
+HVSTRVMGT GY AP+Y TG L+ KSD+YSFGV LLEL++GR+A D+++P + +LV
Sbjct: 233 NSHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVGEHNLV 292
Query: 258 KWATPLFRDQKNFSKMVDPLLKGQYPARGLYQALAIASMCVVEQTNMRPVIADVVSALDF 317
WA P +++ +++DP L+GQY + +A C+ RP + ++V ++
Sbjct: 293 DWARPYLTNKRRLLRVMDPRLQGQYSLTRALKIAVLALDCISIDAKSRPTMNEIVKTMEE 352
Query: 318 LASQK 322
L QK
Sbjct: 353 LHIQK 357
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 276 bits (706), Expect = 6e-74
Identities = 149/344 (43%), Positives = 217/344 (62%), Gaps = 20/344 (5%)
Query: 27 KTFTLNELAAATGNFSANCFVGEGGFGKVYKGYLQK---------TNQFVAIKQLDPKGI 77
K+F+ EL +AT NF + +GEGGFG V+KG++ + T +A+K+L+ G
Sbjct: 54 KSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGW 113
Query: 78 QGTREFVVEVLTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPPGKNPL 137
QG +E++ EV L H +LVKL+G+ E + RLLVYE+MP GSLE+HL PL
Sbjct: 114 QGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQPL 173
Query: 138 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGD 197
W R+++A G AKGL +LH + VIYRD K SNILL ++Y+ KLSDFGLAK GP+GD
Sbjct: 174 SWKLRLKVALGAAKGLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGD 232
Query: 198 QTHVSTRVMGTHGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESKPSKKRHLV 257
++HVSTRVMGTHGY AP+Y TG LT KSD+YSFGV LLEL++GR+A D+++PS +R+LV
Sbjct: 233 KSHVSTRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLV 292
Query: 258 KWATPLFRDQKNFSKMVDPLLKGQYPARGLYQALAIASMCVVEQTNMRPVIADVVSALDF 317
+WA P +++ +++D L+ QY + ++ C+ + +RP +++VVS L+
Sbjct: 293 EWAKPYLVNKRKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHLEH 352
Query: 318 LASQKYEPRVHPIQRSRYGSSSSRSRAKGHRRVTSNVSEKDKLG 361
+ S + G + ++ + RR S VS+K G
Sbjct: 353 IQS----------LNAAIGGNMDKTDRRMRRRSDSVVSKKVNAG 386
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 275 bits (703), Expect = 1e-73
Identities = 144/325 (44%), Positives = 209/325 (64%), Gaps = 15/325 (4%)
Query: 3 VDLSVNEKTEDELKDGVSTNGKLAKTFTLNELAAATGNFSANCFVGEGGFGKVYKGYLQK 62
V + N +TE E+ + K+FT EL AAT NF + +GEGGFG V+KG++ +
Sbjct: 36 VSIRTNPRTEGEILQSPNL-----KSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDE 90
Query: 63 ---------TNQFVAIKQLDPKGIQGTREFVVEVLTLSLAEHTNLVKLLGFGAEGDQRLL 113
T +A+K+L+ G QG +E++ EV L H NLVKL+G+ E + RLL
Sbjct: 91 QTLTASKPGTGVVIAVKKLNQDGWQGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLL 150
Query: 114 VYEYMPLGSLESHLHDLPPGKNPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSN 173
VYE+MP GSLE+HL PL W R+++A G AKGL +LH+ + VIYRD K SN
Sbjct: 151 VYEFMPRGSLENHLFRRGSYFQPLSWTLRLKVALGAAKGLAFLHNA-ETSVIYRDFKTSN 209
Query: 174 ILLGNDYHPKLSDFGLAKIGPMGDQTHVSTRVMGTHGYCAPDYGMTGQLTFKSDIYSFGV 233
ILL ++Y+ KLSDFGLAK GP GD++HVSTR+MGT+GY AP+Y TG LT KSD+YS+GV
Sbjct: 210 ILLDSEYNAKLSDFGLAKDGPTGDKSHVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGV 269
Query: 234 ALLELITGRKAYDESKPSKKRHLVKWATPLFRDQKNFSKMVDPLLKGQYPARGLYQALAI 293
LLE+++GR+A D+++P ++ LV+WA PL +++ +++D L+ QY + +
Sbjct: 270 VLLEVLSGRRAVDKNRPPGEQKLVEWARPLLANKRKLFRVIDNRLQDQYSMEEACKVATL 329
Query: 294 ASMCVVEQTNMRPVIADVVSALDFL 318
A C+ + +RP + +VVS L+ +
Sbjct: 330 ALRCLTFEIKLRPNMNEVVSHLEHI 354
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 246 bits (627), Expect = 8e-65
Identities = 135/336 (40%), Positives = 197/336 (58%), Gaps = 18/336 (5%)
Query: 19 VSTNGKLA-----KTFTLNELAAATGNFSANCFVGEGGFGKVYKGYLQKTN--------- 64
+S +GKL K + +L AT NF + +G+GGFGKVY+G++ T
Sbjct: 59 ISDSGKLLESPNLKVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSG 118
Query: 65 QFVAIKQLDPKGIQGTREFVVEVLTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLE 124
VAIK+L+ + +QG E+ EV L + H NLVKLLG+ E + LLVYE+MP GSLE
Sbjct: 119 MIVAIKRLNSESVQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLE 178
Query: 125 SHLHDLPPGKNPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKL 184
SHL +P W+ R++I G A+GL +LH ++ VIYRD K SNILL ++Y KL
Sbjct: 179 SHLFRR---NDPFPWDLRIKIVIGAARGLAFLHS-LQREVIYRDFKASNILLDSNYDAKL 234
Query: 185 SDFGLAKIGPMGDQTHVSTRVMGTHGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKA 244
SDFGLAK+GP +++HV+TR+MGT+GY AP+Y TG L KSD+++FGV LLE++TG A
Sbjct: 235 SDFGLAKLGPADEKSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTA 294
Query: 245 YDESKPSKKRHLVKWATPLFRDQKNFSKMVDPLLKGQYPARGLYQALAIASMCVVEQTNM 304
++ +P + LV W P ++ +++D +KGQY + + I C+
Sbjct: 295 HNTKRPRGQESLVDWLRPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKN 354
Query: 305 RPVIADVVSALDFLASQKYEPRVHPIQRSRYGSSSS 340
RP + +VV L+ + P +++ SS S
Sbjct: 355 RPHMKEVVEVLEHIQGLNVVPNRSSTKQAVANSSRS 390
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 219 bits (558), Expect = 8e-57
Identities = 132/323 (40%), Positives = 181/323 (55%), Gaps = 12/323 (3%)
Query: 2 EVDLSVNEKTEDELKDGVSTNGKLAKTFTLNE-------LAAATGNFSANCFVGEGGFGK 54
EVD + E K+ KL F N+ L +T +F +G GGFG
Sbjct: 688 EVDPEIEESESMNRKELGEIGSKLVVLFQSNDKELSYDDLLDSTNSFDQANIIGCGGFGM 747
Query: 55 VYKGYLQKTNQFVAIKQLDPKGIQGTREFVVEVLTLSLAEHTNLVKLLGFGAEGDQRLLV 114
VYK L + VAIK+L Q REF EV TLS A+H NLV L GF + RLL+
Sbjct: 748 VYKATLPDGKK-VAIKKLSGDCGQIEREFEAEVETLSRAQHPNLVLLRGFCFYKNDRLLI 806
Query: 115 YEYMPLGSLESHLHDLPPGKNPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNI 174
Y YM GSL+ LH+ G L W TR+RIA G AKGL YLH+ P +++RD+K SNI
Sbjct: 807 YSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGAAKGLLYLHEGCDPHILHRDIKSSNI 866
Query: 175 LLGNDYHPKLSDFGLAKI-GPMGDQTHVSTRVMGTHGYCAPDYGMTGQLTFKSDIYSFGV 233
LL +++ L+DFGLA++ P +THVST ++GT GY P+YG T+K D+YSFGV
Sbjct: 867 LLDENFNSHLADFGLARLMSPY--ETHVSTDLVGTLGYIPPEYGQASVATYKGDVYSFGV 924
Query: 234 ALLELITGRKAYDESKPSKKRHLVKWATPLFRDQKNFSKMVDPLLKGQYPARGLYQALAI 293
LLEL+T ++ D KP R L+ W + + + S++ DPL+ + + +++ L I
Sbjct: 925 VLLELLTDKRPVDMCKPKGCRDLISWVVKM-KHESRASEVFDPLIYSKENDKEMFRVLEI 983
Query: 294 ASMCVVEQTNMRPVIADVVSALD 316
A +C+ E RP +VS LD
Sbjct: 984 ACLCLSENPKQRPTTQQLVSWLD 1006
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 218 bits (554), Expect = 2e-56
Identities = 122/287 (42%), Positives = 172/287 (59%), Gaps = 3/287 (1%)
Query: 30 TLNELAAATGNFSANCFVGEGGFGKVYKGYLQKTNQFVAIKQLDPKGIQGTREFVVEVLT 89
+L+++ +T +F+ +G GGFG VYK L + VAIK+L Q REF EV T
Sbjct: 732 SLDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTK-VAIKRLSGDTGQMDREFQAEVET 790
Query: 90 LSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPPGKNPLDWNTRMRIAAGV 149
LS A+H NLV LLG+ + +LL+Y YM GSL+ LH+ G LDW TR+RIA G
Sbjct: 791 LSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGA 850
Query: 150 AKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGDQTHVSTRVMGTH 209
A+GL YLH +P +++RD+K SNILL + + L+DFGLA++ + THV+T ++GT
Sbjct: 851 AEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARL-ILPYDTHVTTDLVGTL 909
Query: 210 GYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESKPSKKRHLVKWATPLFRDQKN 269
GY P+YG T+K D+YSFGV LLEL+TGR+ D KP R L+ W + + +K
Sbjct: 910 GYIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQM-KTEKR 968
Query: 270 FSKMVDPLLKGQYPARGLYQALAIASMCVVEQTNMRPVIADVVSALD 316
S++ DP + + A + L IA C+ E RP +VS L+
Sbjct: 969 ESEIFDPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLE 1015
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 215 bits (548), Expect = 1e-55
Identities = 119/294 (40%), Positives = 177/294 (59%), Gaps = 7/294 (2%)
Query: 30 TLNELAAATGNFSANCFVGEGGFGKVYKGYLQKTNQFVAIKQLDPKGI--QGTREFVVEV 87
++ L + T NFS++ +G GGFG VYKG L + +A+K+++ I +G EF E+
Sbjct: 577 SIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTK-IAVKRMENGVIAGKGFAEFKSEI 635
Query: 88 LTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPP-GKNPLDWNTRMRIA 146
L+ H +LV LLG+ +G+++LLVYEYMP G+L HL + G PL W R+ +A
Sbjct: 636 AVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLA 695
Query: 147 AGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGDQTHVSTRVM 206
VA+G+EYLH I+RDLK SNILLG+D K++DFGL ++ P G + + TR+
Sbjct: 696 LDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGS-IETRIA 754
Query: 207 GTHGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESKPSKKRHLVKWATPLF-R 265
GT GY AP+Y +TG++T K D+YSFGV L+ELITGRK+ DES+P + HLV W ++
Sbjct: 755 GTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMYIN 814
Query: 266 DQKNFSKMVDPLLK-GQYPARGLYQALAIASMCVVEQTNMRPVIADVVSALDFL 318
+ +F K +D + + ++ +A C + RP + V+ L L
Sbjct: 815 KEASFKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGHAVNILSSL 868
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 215 bits (547), Expect = 2e-55
Identities = 124/293 (42%), Positives = 177/293 (60%), Gaps = 6/293 (2%)
Query: 27 KTFTLNELAAATGNFSANCFVGEGGFGKVYKGYLQKTNQFVAIKQLDPKGIQGTR-EFVV 85
K F+L EL A+ NFS +G GGFGKVYKG L VA+K+L + QG +F
Sbjct: 275 KRFSLRELQVASDNFSNKNILGRGGFGKVYKGRLAD-GTLVAVKRLKEERTQGGELQFQT 333
Query: 86 EVLTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPPGKNPLDWNTRMRI 145
EV +S+A H NL++L GF +RLLVY YM GS+ S L + P + PLDW R RI
Sbjct: 334 EVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRI 393
Query: 146 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGDQTHVSTRV 205
A G A+GL YLHD P +I+RD+K +NILL ++ + DFGLAK+ D THV+T V
Sbjct: 394 ALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKD-THVTTAV 452
Query: 206 MGTHGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESKPSKKRH--LVKWATPL 263
GT G+ AP+Y TG+ + K+D++ +GV LLELITG++A+D ++ + L+ W L
Sbjct: 453 RGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGL 512
Query: 264 FRDQKNFSKMVDPLLKGQYPARGLYQALAIASMCVVEQTNMRPVIADVVSALD 316
+++K +VD L+G Y + Q + +A +C RP +++VV L+
Sbjct: 513 LKEKK-LEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLE 564
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 214 bits (546), Expect = 2e-55
Identities = 123/288 (42%), Positives = 174/288 (59%), Gaps = 8/288 (2%)
Query: 31 LNELAAATGNFSANCFVGEGGFGKVYKGYL--QKTNQFVAIKQLDPKGIQGTREFVVEVL 88
L ++ AT +FS +G+GGFG VYK L +KT VA+K+L QG REF+ E+
Sbjct: 907 LGDIVEATDHFSKKNIIGDGGFGTVYKACLPGEKT---VAVKKLSEAKTQGNREFMAEME 963
Query: 89 TLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPPGKNPLDWNTRMRIAAG 148
TL +H NLV LLG+ + +++LLVYEYM GSL+ L + LDW+ R++IA G
Sbjct: 964 TLGKVKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVG 1023
Query: 149 VAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGDQTHVSTRVMGT 208
A+GL +LH P +I+RD+K SNILL D+ PK++DFGLA++ ++HVST + GT
Sbjct: 1024 AARGLAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARL-ISACESHVSTVIAGT 1082
Query: 209 HGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRK-AYDESKPSKKRHLVKWATPLFRDQ 267
GY P+YG + + T K D+YSFGV LLEL+TG++ + K S+ +LV WA
Sbjct: 1083 FGYIPPEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQG 1142
Query: 268 KNFSKMVDPLLKGQYPARGLYQALAIASMCVVEQTNMRPVIADVVSAL 315
K ++DPLL + L IA +C+ E RP + DV+ AL
Sbjct: 1143 KAVD-VIDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKAL 1189
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 201 bits (512), Expect = 2e-51
Identities = 124/313 (39%), Positives = 182/313 (57%), Gaps = 12/313 (3%)
Query: 8 NEKTEDELKDGVSTNGKLAKTFTLNELAAATGNFSANCFVGEGGFGKVYKGYLQKTNQFV 67
N K + +++D + A+ F+ EL ATG FS + VG+G F V+KG L+
Sbjct: 475 NMKIQPDMED---LKIRRAQEFSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVA 531
Query: 68 ---AIKQLDPKGIQGTREFVVEVLTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLE 124
AIK D K + ++EF E+ LS H +L+ LLG+ +G +RLLVYE+M GSL
Sbjct: 532 VKRAIKASDVK--KSSKEFHNELDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLY 589
Query: 125 SHLHDLPPG-KNPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPK 183
HLH P K L+W R+ IA A+G+EYLH PPVI+RD+K SNIL+ D++ +
Sbjct: 590 QHLHGKDPNLKKRLNWARRVTIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNAR 649
Query: 184 LSDFGLAKIGPMGDQTHVSTRVMGTHGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRK 243
++DFGL+ +GP T +S GT GY P+Y LT KSD+YSFGV LLE+++GRK
Sbjct: 650 VADFGLSILGPADSGTPLSELPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRK 709
Query: 244 AYDESKPSKKRHLVKWATPLFRDQKNFSKMVDPLLKGQYPARGLYQALAIASMCVVEQTN 303
A D ++ ++V+WA PL + F+ ++DP+L L + ++A CV +
Sbjct: 710 AID--MQFEEGNIVEWAVPLIKAGDIFA-ILDPVLSPPSDLEALKKIASVACKCVRMRGK 766
Query: 304 MRPVIADVVSALD 316
RP + V +AL+
Sbjct: 767 DRPSMDKVTTALE 779
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 193 bits (491), Expect = 5e-49
Identities = 121/306 (39%), Positives = 173/306 (55%), Gaps = 12/306 (3%)
Query: 15 LKDGVSTN----GKLAKTFTLNELAAATGNFSANCFVGEGGFGKVYKGYLQKTNQFVAIK 70
+K+ +S N K + T +L AT F + +G GGFG VYK L K VAIK
Sbjct: 853 VKEALSINLAAFEKPLRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAIL-KDGSAVAIK 911
Query: 71 QLDPKGIQGTREFVVEVLTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDL 130
+L QG REF+ E+ T+ +H NLV LLG+ GD+RLLVYE+M GSLE LHD
Sbjct: 912 KLIHVSGQGDREFMAEMETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDP 971
Query: 131 PPGKNPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLA 190
L+W+TR +IA G A+GL +LH P +I+RD+K SN+LL + ++SDFG+A
Sbjct: 972 KKAGVKLNWSTRRKIAIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMA 1031
Query: 191 KIGPMGDQTHVSTRVM-GTHGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESK 249
++ D TH+S + GT GY P+Y + + + K D+YS+GV LLEL+TG++ D S
Sbjct: 1032 RLMSAMD-THLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTD-SP 1089
Query: 250 PSKKRHLVKWATPLFRDQKNFSKMVDPLLKGQYPAR--GLYQALAIASMCVVEQTNMRPV 307
+LV W + S + DP L + PA L Q L +A C+ ++ RP
Sbjct: 1090 DFGDNNLVGWVKQ--HAKLRISDVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPT 1147
Query: 308 IADVVS 313
+ V++
Sbjct: 1148 MVQVMA 1153
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 188 bits (478), Expect = 2e-47
Identities = 115/322 (35%), Positives = 191/322 (58%), Gaps = 14/322 (4%)
Query: 31 LNELAAATGNFSANCFVGEGGFGKVYKGYLQKTNQFVAIKQLDPKGIQGTREFVVEVLTL 90
+ + AT NFS+ +G+GGFG VYKG L + +A+K+L +QGT EF+ EV +
Sbjct: 518 METVVKATENFSSCNKLGQGGFGIVYKGRLLDGKE-IAVKRLSKTSVQGTDEFMNEVTLI 576
Query: 91 SLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPPGKNPLDWNTRMRIAAGVA 150
+ +H NLV++LG EGD+++L+YEY+ SL+S+L ++ L+WN R I GVA
Sbjct: 577 ARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFG-KTRRSKLNWNERFDITNGVA 635
Query: 151 KGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGDQTHVST-RVMGTH 209
+GL YLH + + +I+RDLK SNILL + PK+SDFG+A+I D+T +T +V+GT+
Sbjct: 636 RGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFER-DETEANTMKVVGTY 694
Query: 210 GYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESKPSKKRHLVKWATPLFRDQKN 269
GY +P+Y M G + KSD++SFGV +LE+++G+K + L+ + +++ +
Sbjct: 695 GYMSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSRWKEGRA 754
Query: 270 FSKMVDPLLKGQ-------YPARGLYQALAIASMCVVEQTNMRPVIADVVSALDFLASQK 322
++VDP++ + + + + + I +CV E RP ++ VV A++
Sbjct: 755 L-EIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVVWMFGSEATEI 813
Query: 323 YEPRV--HPIQRSRYGSSSSRS 342
+P+ + ++RS Y S S
Sbjct: 814 PQPKPPGYCVRRSPYELDPSSS 835
>IRA4_MOUSE (Q8R4K2) Interleukin-1 receptor-associated kinase-4 (EC
2.7.1.37) (IRAK-4)
Length = 459
Score = 182 bits (462), Expect = 1e-45
Identities = 120/305 (39%), Positives = 172/305 (56%), Gaps = 24/305 (7%)
Query: 28 TFTLNELAAATGNF------SANCFVGEGGFGKVYKGYLQKTNQFVAIKQLDPKGIQGTR 81
+F+ +EL + T NF + +GEGGFG VYKG + T VA+K+L T
Sbjct: 167 SFSFHELKSITNNFDEQPASAGGNRMGEGGFGVVYKGCVNNT--IVAVKKLGAMVEISTE 224
Query: 82 E----FVVEVLTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPPGKNPL 137
E F E+ ++ +H NLV+LLGF ++ D LVY YMP GSL L L G PL
Sbjct: 225 ELKQQFDQEIKVMATCQHENLVELLGFSSDSDNLCLVYAYMPNGSLLDRLSCLD-GTPPL 283
Query: 138 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGD 197
W+TR ++A G A G+ +LH+ I+RD+K +NILL D+ K+SDFGLA+
Sbjct: 284 SWHTRCKVAQGTANGIRFLHENHH---IHRDIKSANILLDKDFTAKISDFGLARASARLA 340
Query: 198 QTHVSTRVMGTHGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESKPSKKRHLV 257
QT +++R++GT Y AP+ + G++T KSDIYSFGV LLELITG A DE++ + + L+
Sbjct: 341 QTVMTSRIVGTTAYMAPE-ALRGEITPKSDIYSFGVVLLELITGLAAVDENR--EPQLLL 397
Query: 258 KWATPLFRDQKNFSKMVDPLLKGQYPA--RGLYQALAIASMCVVEQTNMRPVIADVVSAL 315
+ ++K D + PA +Y A AS C+ E+ N RP IA V L
Sbjct: 398 DIKEEIEDEEKTIEDYTDEKMSDADPASVEAMYSA---ASQCLHEKKNRRPDIAKVQQLL 454
Query: 316 DFLAS 320
+++
Sbjct: 455 QEMSA 459
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 181 bits (460), Expect = 2e-45
Identities = 112/302 (37%), Positives = 164/302 (54%), Gaps = 8/302 (2%)
Query: 31 LNELAAATGNFSANCFVGEGGFGKVYKGYLQKTNQFVAIKQLDPKGIQ-GTREFVVEVLT 89
LN++ AT N + +G+G G +YK L + A+K+L GI+ G+ V E+ T
Sbjct: 806 LNKVLEATENLNDKYVIGKGAHGTIYKATLSPDKVY-AVKKLVFTGIKNGSVSMVREIET 864
Query: 90 LSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPPGKNPLDWNTRMRIAAGV 149
+ H NL+KL F + L++Y YM GSL LH+ P K PLDW+TR IA G
Sbjct: 865 IGKVRHRNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPPK-PLDWSTRHNIAVGT 923
Query: 150 AKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGDQTHVSTRVMGTH 209
A GL YLH + P +++RD+K NILL +D P +SDFG+AK+ + S V GT
Sbjct: 924 AHGLAYLHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTI 983
Query: 210 GYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESKPSKKRHLVKWATPLFRDQKN 269
GY AP+ T + +SD+YS+GV LLELIT +KA D S + + +V W ++
Sbjct: 984 GYMAPENAFTTVKSRESDVYSYGVVLLELITRKKALDPSF-NGETDIVGWVRSVWTQTGE 1042
Query: 270 FSKMVDPLLKGQY----PARGLYQALAIASMCVVEQTNMRPVIADVVSALDFLASQKYEP 325
K+VDP L + + +AL++A C ++ + RP + DVV L + + Y
Sbjct: 1043 IQKIVDPSLLDELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQLTRWSIRSYSS 1102
Query: 326 RV 327
V
Sbjct: 1103 SV 1104
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 181 bits (460), Expect = 2e-45
Identities = 117/292 (40%), Positives = 160/292 (54%), Gaps = 6/292 (2%)
Query: 24 KLAKTFTLNELAAATGNFSANCFVGEGGFGKVYKGYLQKTNQFVAIKQLDPKGIQGTREF 83
K + T +L AT F + VG GGFG VYK L K VAIK+L QG REF
Sbjct: 871 KPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQL-KDGSVVAIKKLIHVSGQGDREF 929
Query: 84 VVEVLTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPPGKNPLDWNTRM 143
E+ T+ +H NLV LLG+ G++RLLVYEYM GSLE LHD L+W R
Sbjct: 930 TAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARR 989
Query: 144 RIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGDQTHVST 203
+IA G A+GL +LH P +I+RD+K SN+LL + ++SDFG+A++ D TH+S
Sbjct: 990 KIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMD-THLSV 1048
Query: 204 RVM-GTHGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESKPSKKRHLVKWATP 262
+ GT GY P+Y + + + K D+YS+GV LLEL+TG++ D S +LV W
Sbjct: 1049 STLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVGW-VK 1106
Query: 263 LFRDQKNFSKMVDPLLKGQYPAR-GLYQALAIASMCVVEQTNMRPVIADVVS 313
L K LLK L Q L +A C+ ++ RP + V++
Sbjct: 1107 LHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMA 1158
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 181 bits (458), Expect = 3e-45
Identities = 117/292 (40%), Positives = 160/292 (54%), Gaps = 6/292 (2%)
Query: 24 KLAKTFTLNELAAATGNFSANCFVGEGGFGKVYKGYLQKTNQFVAIKQLDPKGIQGTREF 83
K + T +L AT F + VG GGFG VYK L K VAIK+L QG REF
Sbjct: 871 KPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQL-KDGSVVAIKKLIHVSGQGDREF 929
Query: 84 VVEVLTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPPGKNPLDWNTRM 143
E+ T+ +H NLV LLG+ G++RLLVYEYM GSLE LHD L+W R
Sbjct: 930 TAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARR 989
Query: 144 RIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGDQTHVST 203
+IA G A+GL +LH P +I+RD+K SN+LL + ++SDFG+A++ D TH+S
Sbjct: 990 KIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMD-THLSV 1048
Query: 204 RVM-GTHGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESKPSKKRHLVKWATP 262
+ GT GY P+Y + + + K D+YS+GV LLEL+TG++ D S +LV W
Sbjct: 1049 STLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVGW-VK 1106
Query: 263 LFRDQKNFSKMVDPLLKGQYPAR-GLYQALAIASMCVVEQTNMRPVIADVVS 313
L K LLK L Q L +A C+ ++ RP + V++
Sbjct: 1107 LHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMA 1158
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 179 bits (454), Expect = 1e-44
Identities = 111/276 (40%), Positives = 153/276 (55%), Gaps = 13/276 (4%)
Query: 47 VGEGGFGKVYKGYLQKTNQFVAIKQLDPKGI-QGTREFVVEVLTLSLAEHTNLVKLLGFG 105
+G+GG G VY+G + N VAIK+L +G + F E+ TL H ++V+LLG+
Sbjct: 698 IGKGGAGIVYRGSMPN-NVDVAIKRLVGRGTGRSDHGFTAEIQTLGRIRHRHIVRLLGYV 756
Query: 106 AEGDQRLLVYEYMPLGSLESHLHDLPPGKNPLDWNTRMRIAAGVAKGLEYLHDEMKPPVI 165
A D LL+YEYMP GSL LH G L W TR R+A AKGL YLH + P ++
Sbjct: 757 ANKDTNLLLYEYMPNGSLGELLHGSKGGH--LQWETRHRVAVEAAKGLCYLHHDCSPLIL 814
Query: 166 YRDLKCSNILLGNDYHPKLSDFGLAKIGPMGDQTHVSTRVMGTHGYCAPDYGMTGQLTFK 225
+RD+K +NILL +D+ ++DFGLAK G + + + G++GY AP+Y T ++ K
Sbjct: 815 HRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPEYAYTLKVDEK 874
Query: 226 SDIYSFGVALLELITGRKAYDESKPSKKRHLVKWATPLFR------DQKNFSKMVDPLLK 279
SD+YSFGV LLELI G+K E + +V+W D +VDP L
Sbjct: 875 SDVYSFGVVLLELIAGKKPVGEF--GEGVDIVRWVRNTEEEITQPSDAAIVVAIVDPRLT 932
Query: 280 GQYPARGLYQALAIASMCVVEQTNMRPVIADVVSAL 315
G YP + IA MCV E+ RP + +VV L
Sbjct: 933 G-YPLTSVIHVFKIAMMCVEEEAAARPTMREVVHML 967
>IRA4_HUMAN (Q9NWZ3) Interleukin-1 receptor-associated kinase-4 (EC
2.7.1.37) (IRAK-4) (NY-REN-64 antigen)
Length = 460
Score = 175 bits (444), Expect = 1e-43
Identities = 114/298 (38%), Positives = 166/298 (55%), Gaps = 20/298 (6%)
Query: 28 TFTLNELAAATGNFSANCF------VGEGGFGKVYKGYLQKTNQFVAIKQL----DPKGI 77
+F+ EL T NF +GEGGFG VYKGY+ T VA+K+L D
Sbjct: 167 SFSFYELKNVTNNFDERPISVGGNKMGEGGFGVVYKGYVNNTT--VAVKKLAAMVDITTE 224
Query: 78 QGTREFVVEVLTLSLAEHTNLVKLLGFGAEGDQRLLVYEYMPLGSLESHLHDLPPGKNPL 137
+ ++F E+ ++ +H NLV+LLGF ++GD LVY YMP GSL L L G PL
Sbjct: 225 ELKQQFDQEIKVMAKCQHENLVELLGFSSDGDDLCLVYVYMPNGSLLDRLSCLD-GTPPL 283
Query: 138 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGNDYHPKLSDFGLAKIGPMGD 197
W+ R +IA G A G+ +LH+ I+RD+K +NILL + K+SDFGLA+
Sbjct: 284 SWHMRCKIAQGAANGINFLHENHH---IHRDIKSANILLDEAFTAKISDFGLARASEKFA 340
Query: 198 QTHVSTRVMGTHGYCAPDYGMTGQLTFKSDIYSFGVALLELITGRKAYDESKPSKKRHLV 257
QT +++R++GT Y AP+ + G++T KSDIYSFGV LLE+ITG A DE + + + L+
Sbjct: 341 QTVMTSRIVGTTAYMAPE-ALRGEITPKSDIYSFGVVLLEIITGLPAVDEHR--EPQLLL 397
Query: 258 KWATPLFRDQKNFSKMVDPLLKGQYPARGLYQALAIASMCVVEQTNMRPVIADVVSAL 315
+ ++K +D + + + ++AS C+ E+ N RP I V L
Sbjct: 398 DIKEEIEDEEKTIEDYIDKKM-NDADSTSVEAMYSVASQCLHEKKNKRPDIKKVQQLL 454
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,109,910
Number of Sequences: 164201
Number of extensions: 1843685
Number of successful extensions: 7451
Number of sequences better than 10.0: 1562
Number of HSP's better than 10.0 without gapping: 985
Number of HSP's successfully gapped in prelim test: 577
Number of HSP's that attempted gapping in prelim test: 4184
Number of HSP's gapped (non-prelim): 1748
length of query: 361
length of database: 59,974,054
effective HSP length: 111
effective length of query: 250
effective length of database: 41,747,743
effective search space: 10436935750
effective search space used: 10436935750
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 66 (30.0 bits)
Medicago: description of AC135230.8


