

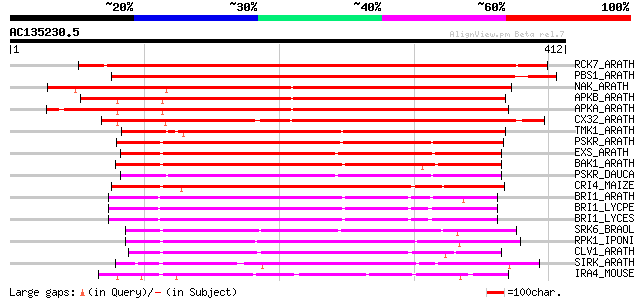
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135230.5 + phase: 0
(412 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 558 e-158
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 461 e-129
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 316 8e-86
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 311 2e-84
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 303 4e-82
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 276 7e-74
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 227 4e-59
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 225 2e-58
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 224 3e-58
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 222 1e-57
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 220 6e-57
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 218 2e-56
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 218 2e-56
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 210 5e-54
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 209 8e-54
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 192 2e-48
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 186 9e-47
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 185 2e-46
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 184 5e-46
IRA4_MOUSE (Q8R4K2) Interleukin-1 receptor-associated kinase-4 (... 182 1e-45
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 558 bits (1437), Expect = e-158
Identities = 278/348 (79%), Positives = 303/348 (86%), Gaps = 3/348 (0%)
Query: 52 KHDQLSLDVKNLNLNDGVSPDGKVAQTFTFAELAAATENFRADCFVGEGGFGKVYKGYLE 111
K DQLSLDVK LNLND V+ GK AQTFTF ELA AT NFR+DCF+GEGGFGKV+KG +E
Sbjct: 66 KEDQLSLDVKGLNLNDQVT--GKKAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIE 123
Query: 112 KINQVVAIKQLDRNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLG 171
K++QVVAIKQLDRNGVQGIREFVVEV+TL LADHPNLVKL+GFCAEG+QRLLVYEYMP G
Sbjct: 124 KLDQVVAIKQLDRNGVQGIREFVVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQG 183
Query: 172 SLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYH 231
SLE+HLH L G+KPLDWNTRMKIAAGAARGLEYLHD+M PPVIYRDLKCSNILLGEDY
Sbjct: 184 SLEDHLHVLPSGKKPLDWNTRMKIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQ 243
Query: 232 SKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITG 291
KLSDFGLAKVGP GDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGV LLELITG
Sbjct: 244 PKLSDFGLAKVGPSGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITG 303
Query: 292 RKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQ 351
RKAID+ K K+QNLV WARPLF+DRR F +M+DPLL+GQYPVRGLYQALAI+AMCVQEQ
Sbjct: 304 RKAIDNTKTRKDQNLVGWARPLFKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQ 363
Query: 352 PNMRPVIADVVTALNYLASQKYDPQIHHIQGSRKGSSSPRSRSERHRR 399
P MRPV++DVV ALN+LAS KYDP S K S R R + +R
Sbjct: 364 PTMRPVVSDVVLALNFLASSKYDPN-SPSSSSGKNPSFHRDRDDEEKR 410
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 461 bits (1187), Expect = e-129
Identities = 225/331 (67%), Positives = 266/331 (79%), Gaps = 9/331 (2%)
Query: 76 AQTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVV 135
A TF F ELAAAT NF D F+GEGGFG+VYKG L+ QVVA+KQLDRNG+QG REF+V
Sbjct: 71 AHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLV 130
Query: 136 EVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKI 195
EV+ L L HPNLV L+G+CA+G+QRLLVYE+MPLGSLE+HLHDL P ++ LDWN RMKI
Sbjct: 131 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKI 190
Query: 196 AAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRV 255
AAGAA+GLE+LHDK PPVIYRD K SNILL E +H KLSDFGLAK+GP GDK+HVSTRV
Sbjct: 191 AAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRV 250
Query: 256 MGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFR 315
MGTYGYCAP+YAMTGQLT KSD+YSFGV LELITGRKAID + P EQNLVAWARPLF
Sbjct: 251 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFN 310
Query: 316 DRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYLASQKYDP 375
DRR+F ++ DP L+G++P R LYQALA+A+MC+QEQ RP+IADVVTAL+YLA+Q YDP
Sbjct: 311 DRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTALSYLANQAYDP 370
Query: 376 QIHHIQGSRKGSSSPRSRSERHRRVTSKDSE 406
S R+R ER R+ +++ +
Sbjct: 371 ---------SKDDSRRNRDERGARLITRNDD 392
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 316 bits (809), Expect = 8e-86
Identities = 164/355 (46%), Positives = 225/355 (63%), Gaps = 12/355 (3%)
Query: 29 CFVDKVKVDLNLSELKDKK--EDDSKHDQLSLDVKNLNLNDGVSPDGKVAQTFTFAELAA 86
CF +++K D+ S K D + + +G + F+ +EL +
Sbjct: 4 CFSNRIKTDIASSTWLSSKFLSRDGSKGSSTASFSYMPRTEGEILQNANLKNFSLSELKS 63
Query: 87 ATENFRADCFVGEGGFGKVYKGYLEKINQ---------VVAIKQLDRNGVQGIREFVVEV 137
AT NFR D VGEGGFG V+KG++++ + V+A+K+L++ G QG RE++ E+
Sbjct: 64 ATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEGFQGHREWLAEI 123
Query: 138 ITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAA 197
LG DHPNLVKL+G+C E E RLLVYE+M GSLENHL +PL WNTR+++A
Sbjct: 124 NYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTFYQPLSWNTRVRMAL 183
Query: 198 GAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMG 257
GAARGL +LH+ +P VIYRD K SNILL +Y++KLSDFGLA+ GP+GD +HVSTRVMG
Sbjct: 184 GAARGLAFLHNA-QPQVIYRDFKASNILLDSNYNAKLSDFGLARDGPMGDNSHVSTRVMG 242
Query: 258 TYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDR 317
T GY AP+Y TG L+ KSD+YSFGV LLEL++GR+AID +P E NLV WARP ++
Sbjct: 243 TQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVGEHNLVDWARPYLTNK 302
Query: 318 RRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYLASQK 372
RR ++DP L+GQY + + +A C+ RP + ++V + L QK
Sbjct: 303 RRLLRVMDPRLQGQYSLTRALKIAVLALDCISIDAKSRPTMNEIVKTMEELHIQK 357
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 311 bits (797), Expect = 2e-84
Identities = 158/330 (47%), Positives = 222/330 (66%), Gaps = 15/330 (4%)
Query: 53 HDQLSLDVKNLNLNDGVSPDGKVAQT-----FTFAELAAATENFRADCFVGEGGFGKVYK 107
+D L ++++ +G++ Q+ FTFAEL AAT NFR D +GEGGFG V+K
Sbjct: 26 NDSLGSKSSSVSIRTNPRTEGEILQSPNLKSFTFAELKAATRNFRPDSVLGEGGFGSVFK 85
Query: 108 GYLEK---------INQVVAIKQLDRNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEG 158
G++++ V+A+K+L+++G QG +E++ EV LG HPNLVKL+G+C E
Sbjct: 86 GWIDEQTLTASKPGTGVVIAVKKLNQDGWQGHQEWLAEVNYLGQFSHPNLVKLIGYCLED 145
Query: 159 EQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRD 218
E RLLVYE+MP GSLENHL +PL W R+K+A GAA+GL +LH+ + VIYRD
Sbjct: 146 EHRLLVYEFMPRGSLENHLFRRGSYFQPLSWTLRLKVALGAAKGLAFLHNA-ETSVIYRD 204
Query: 219 LKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDI 278
K SNILL +Y++KLSDFGLAK GP GDK+HVSTR+MGTYGY AP+Y TG LT KSD+
Sbjct: 205 FKTSNILLDSEYNAKLSDFGLAKDGPTGDKSHVSTRIMGTYGYAAPEYLATGHLTTKSDV 264
Query: 279 YSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRGLY 338
YS+GV LLE+++GR+A+D +P EQ LV WARPL ++R+ +ID L+ QY +
Sbjct: 265 YSYGVVLLEVLSGRRAVDKNRPPGEQKLVEWARPLLANKRKLFRVIDNRLQDQYSMEEAC 324
Query: 339 QALAIAAMCVQEQPNMRPVIADVVTALNYL 368
+ +A C+ + +RP + +VV+ L ++
Sbjct: 325 KVATLALRCLTFEIKLRPNMNEVVSHLEHI 354
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 303 bits (777), Expect = 4e-82
Identities = 161/357 (45%), Positives = 231/357 (64%), Gaps = 18/357 (5%)
Query: 28 ICFVDKVKVDLNLSELKDKKEDDSKHDQLSLDVKNLNLNDGVSPDGKVAQT-----FTFA 82
IC +VK + S K D L ++++ +G++ Q+ F+FA
Sbjct: 3 ICLSAQVKAE---SSGASTKYDAKDIGSLGSKASSVSVRPSPRTEGEILQSPNLKSFSFA 59
Query: 83 ELAAATENFRADCFVGEGGFGKVYKGYLEK---------INQVVAIKQLDRNGVQGIREF 133
EL +AT NFR D +GEGGFG V+KG++++ V+A+K+L+++G QG +E+
Sbjct: 60 ELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQGHQEW 119
Query: 134 VVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRM 193
+ EV LG H +LVKL+G+C E E RLLVYE+MP GSLENHL +PL W R+
Sbjct: 120 LAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQPLSWKLRL 179
Query: 194 KIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVST 253
K+A GAA+GL +LH + VIYRD K SNILL +Y++KLSDFGLAK GPIGDK+HVST
Sbjct: 180 KVALGAAKGLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKSHVST 238
Query: 254 RVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPL 313
RVMGT+GY AP+Y TG LT KSD+YSFGV LLEL++GR+A+D +P+ E+NLV WA+P
Sbjct: 239 RVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEWAKPY 298
Query: 314 FRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYLAS 370
++R+ +ID L+ QY + + ++ C+ + +RP +++VV+ L ++ S
Sbjct: 299 LVNKRKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHLEHIQS 355
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 276 bits (706), Expect = 7e-74
Identities = 147/343 (42%), Positives = 213/343 (61%), Gaps = 22/343 (6%)
Query: 69 VSPDGKVAQT-----FTFAELAAATENFRADCFVGEGGFGKVYKGYLEKIN--------- 114
+S GK+ ++ + F +L AT+NF+ D +G+GGFGKVY+G+++
Sbjct: 59 ISDSGKLLESPNLKVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSG 118
Query: 115 QVVAIKQLDRNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLE 174
+VAIK+L+ VQG E+ EV LG+ H NLVKLLG+C E ++ LLVYE+MP GSLE
Sbjct: 119 MIVAIKRLNSESVQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLE 178
Query: 175 NHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKL 234
+HL + P W+ R+KI GAARGL +LH ++ VIYRD K SNILL +Y +KL
Sbjct: 179 SHLFRRND---PFPWDLRIKIVIGAARGLAFLHS-LQREVIYRDFKASNILLDSNYDAKL 234
Query: 235 SDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKA 294
SDFGLAK+GP +K+HV+TR+MGTYGY AP+Y TG L KSD+++FGV LLE++TG A
Sbjct: 235 SDFGLAKLGPADEKSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTA 294
Query: 295 IDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNM 354
+ K+P +++LV W RP ++ R +++D ++GQY + + I C++ P
Sbjct: 295 HNTKRPRGQESLVDWLRPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKN 354
Query: 355 RPVIADVVTALNYLASQKYDPQIHHIQGSRKGSSSPRSRSERH 397
RP + +VV L ++ P + S K + + SRS H
Sbjct: 355 RPHMKEVVEVLEHIQGLNVVPN----RSSTKQAVANSSRSSPH 393
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 227 bits (579), Expect = 4e-59
Identities = 124/291 (42%), Positives = 183/291 (62%), Gaps = 9/291 (3%)
Query: 84 LAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGV---QGIREFVVEVITL 140
L + T NF +D +G GGFG VYKG L ++ A+K+++ NGV +G EF E+ L
Sbjct: 581 LRSVTNNFSSDNILGSGGFGVVYKGELHDGTKI-AVKRME-NGVIAGKGFAEFKSEIAVL 638
Query: 141 GLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSP-GQKPLDWNTRMKIAAGA 199
H +LV LLG+C +G ++LLVYEYMP G+L HL + S G KPL W R+ +A
Sbjct: 639 TKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLALDV 698
Query: 200 ARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTY 259
ARG+EYLH I+RDLK SNILLG+D +K++DFGL ++ P G K + TR+ GT+
Sbjct: 699 ARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEG-KGSIETRIAGTF 757
Query: 260 GYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRR- 318
GY AP+YA+TG++T K D+YSFGV L+ELITGRK++D +P + +LV+W + ++ ++
Sbjct: 758 GYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMYINKEA 817
Query: 319 RFSEMIDPLLE-GQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYL 368
F + ID ++ + + ++ +A C +P RP + V L+ L
Sbjct: 818 SFKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGHAVNILSSL 868
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 225 bits (573), Expect = 2e-58
Identities = 124/288 (43%), Positives = 179/288 (62%), Gaps = 5/288 (1%)
Query: 80 TFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVIT 139
++ +L +T +F +G GGFG VYK L + VAIK+L + Q REF EV T
Sbjct: 723 SYDDLLDSTNSFDQANIIGCGGFGMVYKATLPD-GKKVAIKKLSGDCGQIEREFEAEVET 781
Query: 140 LGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGA 199
L A HPNLV L GFC RLL+Y YM GSL+ LH+ + G L W TR++IA GA
Sbjct: 782 LSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGA 841
Query: 200 ARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAK-VGPIGDKTHVSTRVMGT 258
A+GL YLH+ P +++RD+K SNILL E+++S L+DFGLA+ + P +THVST ++GT
Sbjct: 842 AKGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPY--ETHVSTDLVGT 899
Query: 259 YGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRR 318
GY P+Y T+K D+YSFGV LLEL+T ++ +D KP ++L++W + +
Sbjct: 900 LGYIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKM-KHES 958
Query: 319 RFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALN 366
R SE+ DPL+ + + +++ L IA +C+ E P RP +V+ L+
Sbjct: 959 RASEVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWLD 1006
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 224 bits (571), Expect = 3e-58
Identities = 125/284 (44%), Positives = 179/284 (63%), Gaps = 4/284 (1%)
Query: 83 ELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVITLGL 142
++ AT++F +G+GGFG VYK L + VA+K+L QG REF+ E+ TLG
Sbjct: 909 DIVEATDHFSKKNIIGDGGFGTVYKACLPG-EKTVAVKKLSEAKTQGNREFMAEMETLGK 967
Query: 143 ADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARG 202
HPNLV LLG+C+ E++LLVYEYM GSL++ L + + + LDW+ R+KIA GAARG
Sbjct: 968 VKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARG 1027
Query: 203 LEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYC 262
L +LH P +I+RD+K SNILL D+ K++DFGLA++ ++HVST + GT+GY
Sbjct: 1028 LAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARL-ISACESHVSTVIAGTFGYI 1086
Query: 263 APDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHK-KPAKEQNLVAWARPLFRDRRRFS 321
P+Y + + T K D+YSFGV LLEL+TG++ K ++ NLV WA ++ +
Sbjct: 1087 PPEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKI-NQGKAV 1145
Query: 322 EMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTAL 365
++IDPLL + L IA +C+ E P RP + DV+ AL
Sbjct: 1146 DVIDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKAL 1189
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 222 bits (566), Expect = 1e-57
Identities = 122/290 (42%), Positives = 179/290 (61%), Gaps = 6/290 (2%)
Query: 79 FTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIR-EFVVEV 137
F+ EL A++NF +G GGFGKVYKG L +VA+K+L QG +F EV
Sbjct: 277 FSLRELQVASDNFSNKNILGRGGFGKVYKGRLAD-GTLVAVKRLKEERTQGGELQFQTEV 335
Query: 138 ITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAA 197
+ +A H NL++L GFC +RLLVY YM GS+ + L + Q PLDW R +IA
Sbjct: 336 EMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIAL 395
Query: 198 GAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMG 257
G+ARGL YLHD P +I+RD+K +NILL E++ + + DFGLAK+ D THV+T V G
Sbjct: 396 GSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKD-THVTTAVRG 454
Query: 258 TYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQN--LVAWARPLFR 315
T G+ AP+Y TG+ + K+D++ +GV LLELITG++A D + A + + L+ W + L +
Sbjct: 455 TIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLK 514
Query: 316 DRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTAL 365
+ ++ ++D L+G Y + Q + +A +C Q P RP +++VV L
Sbjct: 515 E-KKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRML 563
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 220 bits (560), Expect = 6e-57
Identities = 120/283 (42%), Positives = 170/283 (59%), Gaps = 3/283 (1%)
Query: 83 ELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVITLGL 142
++ +T +F +G GGFG VYK L +V AIK+L + Q REF EV TL
Sbjct: 735 DILKSTSSFNQANIIGCGGFGLVYKATLPDGTKV-AIKRLSGDTGQMDREFQAEVETLSR 793
Query: 143 ADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARG 202
A HPNLV LLG+C +LL+Y YM GSL+ LH+ G LDW TR++IA GAA G
Sbjct: 794 AQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGAAEG 853
Query: 203 LEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYC 262
L YLH +P +++RD+K SNILL + + + L+DFGLA++ + THV+T ++GT GY
Sbjct: 854 LAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARL-ILPYDTHVTTDLVGTLGYI 912
Query: 263 APDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSE 322
P+Y T+K D+YSFGV LLEL+TGR+ +D KP ++L++W + + +R SE
Sbjct: 913 PPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQM-KTEKRESE 971
Query: 323 MIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTAL 365
+ DP + + + L IA C+ E P RP +V+ L
Sbjct: 972 IFDPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWL 1014
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 218 bits (556), Expect = 2e-56
Identities = 127/295 (43%), Positives = 181/295 (61%), Gaps = 7/295 (2%)
Query: 76 AQTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNG--VQGIREF 133
AQ F++ EL AT F D VG+G F V+KG L VVA+K+ + + +EF
Sbjct: 490 AQEFSYEELEQATGGFSEDSQVGKGSFSCVFKGILRD-GTVVAVKRAIKASDVKKSSKEF 548
Query: 134 VVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPG-QKPLDWNTR 192
E+ L +H +L+ LLG+C +G +RLLVYE+M GSL HLH P +K L+W R
Sbjct: 549 HNELDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARR 608
Query: 193 MKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVS 252
+ IA AARG+EYLH PPVI+RD+K SNIL+ ED++++++DFGL+ +GP T +S
Sbjct: 609 VTIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLS 668
Query: 253 TRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARP 312
GT GY P+Y LT KSD+YSFGV LLE+++GRKAID + +E N+V WA P
Sbjct: 669 ELPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDMQ--FEEGNIVEWAVP 726
Query: 313 LFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNY 367
L + F+ ++DP+L + L + ++A CV+ + RP + V TAL +
Sbjct: 727 LIKAGDIFA-ILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEH 780
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 218 bits (555), Expect = 2e-56
Identities = 125/292 (42%), Positives = 175/292 (59%), Gaps = 8/292 (2%)
Query: 74 KVAQTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREF 133
K + TFA+L AT F D +G GGFG VYK L K VAIK+L QG REF
Sbjct: 866 KPLRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAIL-KDGSAVAIKKLIHVSGQGDREF 924
Query: 134 VVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRM 193
+ E+ T+G H NLV LLG+C G++RLLVYE+M GSLE+ LHD L+W+TR
Sbjct: 925 MAEMETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRR 984
Query: 194 KIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVST 253
KIA G+ARGL +LH P +I+RD+K SN+LL E+ +++SDFG+A++ D TH+S
Sbjct: 985 KIAIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMD-THLSV 1043
Query: 254 RVM-GTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARP 312
+ GT GY P+Y + + + K D+YS+GV LLEL+TG++ D + NLV W +
Sbjct: 1044 STLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTD-SPDFGDNNLVGWVKQ 1102
Query: 313 LFRDRRRFSEMIDPLLEGQYPVR--GLYQALAIAAMCVQEQPNMRPVIADVV 362
+ R S++ DP L + P L Q L +A C+ ++ RP + V+
Sbjct: 1103 --HAKLRISDVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVM 1152
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 210 bits (535), Expect = 5e-54
Identities = 126/292 (43%), Positives = 173/292 (59%), Gaps = 8/292 (2%)
Query: 74 KVAQTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREF 133
K + TFA+L AT F D VG GGFG VYK L K VVAIK+L QG REF
Sbjct: 871 KPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQL-KDGSVVAIKKLIHVSGQGDREF 929
Query: 134 VVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRM 193
E+ T+G H NLV LLG+C GE+RLLVYEYM GSLE+ LHD L+W R
Sbjct: 930 TAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARR 989
Query: 194 KIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVST 253
KIA GAARGL +LH P +I+RD+K SN+LL E+ +++SDFG+A++ D TH+S
Sbjct: 990 KIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMD-THLSV 1048
Query: 254 RVM-GTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARP 312
+ GT GY P+Y + + + K D+YS+GV LLEL+TG++ D + NLV W +
Sbjct: 1049 STLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVGWVK- 1106
Query: 313 LFRDRRRFSEMID-PLLEGQYPVR-GLYQALAIAAMCVQEQPNMRPVIADVV 362
+ + +++ D LL+ + L Q L +A C+ ++ RP + V+
Sbjct: 1107 -LHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVM 1157
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 209 bits (533), Expect = 8e-54
Identities = 126/292 (43%), Positives = 173/292 (59%), Gaps = 8/292 (2%)
Query: 74 KVAQTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREF 133
K + TFA+L AT F D VG GGFG VYK L K VVAIK+L QG REF
Sbjct: 871 KPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQL-KDGSVVAIKKLIHVSGQGDREF 929
Query: 134 VVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRM 193
E+ T+G H NLV LLG+C GE+RLLVYEYM GSLE+ LHD L+W R
Sbjct: 930 TAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARR 989
Query: 194 KIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVST 253
KIA GAARGL +LH P +I+RD+K SN+LL E+ +++SDFG+A++ D TH+S
Sbjct: 990 KIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMD-THLSV 1048
Query: 254 RVM-GTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARP 312
+ GT GY P+Y + + + K D+YS+GV LLEL+TG++ D + NLV W +
Sbjct: 1049 STLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVGWVK- 1106
Query: 313 LFRDRRRFSEMID-PLLEGQYPVR-GLYQALAIAAMCVQEQPNMRPVIADVV 362
+ + +++ D LL+ + L Q L +A C+ ++ RP + V+
Sbjct: 1107 -LHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVM 1157
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 192 bits (487), Expect = 2e-48
Identities = 113/298 (37%), Positives = 181/298 (59%), Gaps = 12/298 (4%)
Query: 87 ATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVITLGLADHP 146
ATENF + +G+GGFG VYKG L + +A+K+L + VQG EF+ EV + H
Sbjct: 524 ATENFSSCNKLGQGGFGIVYKGRLLD-GKEIAVKRLSKTSVQGTDEFMNEVTLIARLQHI 582
Query: 147 NLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYL 206
NLV++LG C EG++++L+YEY+ SL+++L + K L+WN R I G ARGL YL
Sbjct: 583 NLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSK-LNWNERFDITNGVARGLLYL 641
Query: 207 HDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVST-RVMGTYGYCAPD 265
H + +I+RDLK SNILL ++ K+SDFG+A++ D+T +T +V+GTYGY +P+
Sbjct: 642 HQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFE-RDETEANTMKVVGTYGYMSPE 700
Query: 266 YAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMID 325
YAM G + KSD++SFGV +LE+++G+K E +L+++ +++ R E++D
Sbjct: 701 YAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSRWKEGRAL-EIVD 759
Query: 326 PLLEGQ-------YPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYLASQKYDPQ 376
P++ + + + + + I +CVQE RP ++ VV A++ P+
Sbjct: 760 PVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVVWMFGSEATEIPQPK 817
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 186 bits (472), Expect = 9e-47
Identities = 111/298 (37%), Positives = 166/298 (55%), Gaps = 8/298 (2%)
Query: 87 ATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQ-GIREFVVEVITLGLADH 145
ATEN +G+G G +YK L ++V A+K+L G++ G V E+ T+G H
Sbjct: 812 ATENLNDKYVIGKGAHGTIYKATLSP-DKVYAVKKLVFTGIKNGSVSMVREIETIGKVRH 870
Query: 146 PNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEY 205
NL+KL F E L++Y YM GSL + LH+ +P KPLDW+TR IA G A GL Y
Sbjct: 871 RNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNP-PKPLDWSTRHNIAVGTAHGLAY 929
Query: 206 LHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPD 265
LH P +++RD+K NILL D +SDFG+AK+ + S V GT GY AP+
Sbjct: 930 LHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAPE 989
Query: 266 YAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMID 325
A T + +SD+YS+GV LLELIT +KA+D E ++V W R ++ +++D
Sbjct: 990 NAFTTVKSRESDVYSYGVVLLELITRKKALDPSFNG-ETDIVGWVRSVWTQTGEIQKIVD 1048
Query: 326 PLLEGQY----PVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYLASQKYDPQIHH 379
P L + + + +AL++A C +++ + RP + DVV L + + Y + +
Sbjct: 1049 PSLLDELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQLTRWSIRSYSSSVRN 1106
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 185 bits (470), Expect = 2e-46
Identities = 112/284 (39%), Positives = 164/284 (57%), Gaps = 13/284 (4%)
Query: 89 ENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGV-QGIREFVVEVITLGLADHPN 147
E + + +G+GG G VY+G + N VAIK+L G + F E+ TLG H +
Sbjct: 690 ECLKEENIIGKGGAGIVYRGSMPN-NVDVAIKRLVGRGTGRSDHGFTAEIQTLGRIRHRH 748
Query: 148 LVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLH 207
+V+LLG+ A + LL+YEYMP GSL LH G L W TR ++A AA+GL YLH
Sbjct: 749 IVRLLGYVANKDTNLLLYEYMPNGSLGELLHGSKGGH--LQWETRHRVAVEAAKGLCYLH 806
Query: 208 DKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYA 267
P +++RD+K +NILL D+ + ++DFGLAK G + + + G+YGY AP+YA
Sbjct: 807 HDCSPLILHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPEYA 866
Query: 268 MTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSE----- 322
T ++ KSD+YSFGV LLELI G+K + + + ++V W R + + S+
Sbjct: 867 YTLKVDEKSDVYSFGVVLLELIAGKKPVG--EFGEGVDIVRWVRNTEEEITQPSDAAIVV 924
Query: 323 -MIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTAL 365
++DP L G YP+ + IA MCV+E+ RP + +VV L
Sbjct: 925 AIVDPRLTG-YPLTSVIHVFKIAMMCVEEEAAARPTMREVVHML 967
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 184 bits (466), Expect = 5e-46
Identities = 114/323 (35%), Positives = 169/323 (52%), Gaps = 18/323 (5%)
Query: 79 FTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVI 138
F ++E+ T NF +G+GGFGKVY G + + VA+K L QG +EF EV
Sbjct: 564 FKYSEVVNITNNFER--VIGKGGFGKVYHGVIN--GEQVAVKVLSEESAQGYKEFRAEVD 619
Query: 139 TLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKP--LDWNTRMKIA 196
L H NL L+G+C E +L+YEYM +L D G++ L W R+KI+
Sbjct: 620 LLMRVHHTNLTSLVGYCNEINHMVLIYEYMA----NENLGDYLAGKRSFILSWEERLKIS 675
Query: 197 AGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVM 256
AA+GLEYLH+ KPP+++RD+K +NILL E +K++DFGL++ + +ST V
Sbjct: 676 LDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTVVA 735
Query: 257 GTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRD 316
G+ GY P+Y T Q+ KSD+YS GV LLE+ITG+ AI K K ++ R + +
Sbjct: 736 GSIGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKTEK-VHISDHVRSILAN 794
Query: 317 RRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYLA------S 370
++D L +Y V ++ IA C + RP ++ VV L +
Sbjct: 795 -GDIRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMELKQIVYGIVTDQ 853
Query: 371 QKYDPQIHHIQGSRKGSSSPRSR 393
+ YD + + PR+R
Sbjct: 854 ENYDDSTKMLTVNLDTEMVPRAR 876
>IRA4_MOUSE (Q8R4K2) Interleukin-1 receptor-associated kinase-4 (EC
2.7.1.37) (IRAK-4)
Length = 459
Score = 182 bits (463), Expect = 1e-45
Identities = 121/323 (37%), Positives = 182/323 (55%), Gaps = 31/323 (9%)
Query: 67 DGVSPDGKVAQT-------FTFAELAAATENFRADCF------VGEGGFGKVYKGYLEKI 113
D SPD + ++ F+F EL + T NF +GEGGFG VYKG +
Sbjct: 149 DSSSPDNRSVESSDTRFHSFSFHELKSITNNFDEQPASAGGNRMGEGGFGVVYKGCVN-- 206
Query: 114 NQVVAIKQL----DRNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMP 169
N +VA+K+L + + + ++F E+ + H NLV+LLGF ++ + LVY YMP
Sbjct: 207 NTIVAVKKLGAMVEISTEELKQQFDQEIKVMATCQHENLVELLGFSSDSDNLCLVYAYMP 266
Query: 170 LGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGED 229
GSL + L L G PL W+TR K+A G A G+ +LH+ I+RD+K +NILL +D
Sbjct: 267 NGSLLDRLSCLD-GTPPLSWHTRCKVAQGTANGIRFLHENHH---IHRDIKSANILLDKD 322
Query: 230 YHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELI 289
+ +K+SDFGLA+ +T +++R++GT Y AP+ A+ G++T KSDIYSFGV LLELI
Sbjct: 323 FTAKISDFGLARASARLAQTVMTSRIVGTTAYMAPE-ALRGEITPKSDIYSFGVVLLELI 381
Query: 290 TGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYP--VRGLYQALAIAAMC 347
TG A+D + + Q L+ + + + + D + P V +Y A A+ C
Sbjct: 382 TGLAAVDENR--EPQLLLDIKEEIEDEEKTIEDYTDEKMSDADPASVEAMYSA---ASQC 436
Query: 348 VQEQPNMRPVIADVVTALNYLAS 370
+ E+ N RP IA V L +++
Sbjct: 437 LHEKKNRRPDIAKVQQLLQEMSA 459
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.139 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 49,291,563
Number of Sequences: 164201
Number of extensions: 2113180
Number of successful extensions: 8755
Number of sequences better than 10.0: 1566
Number of HSP's better than 10.0 without gapping: 1052
Number of HSP's successfully gapped in prelim test: 514
Number of HSP's that attempted gapping in prelim test: 5245
Number of HSP's gapped (non-prelim): 1761
length of query: 412
length of database: 59,974,054
effective HSP length: 113
effective length of query: 299
effective length of database: 41,419,341
effective search space: 12384382959
effective search space used: 12384382959
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC135230.5


