

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135100.7 + phase: 0 /pseudo
(855 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
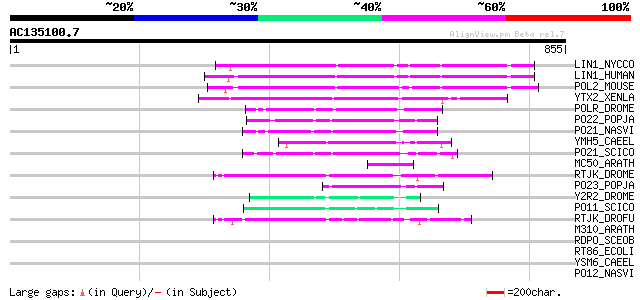
Score E
Sequences producing significant alignments: (bits) Value
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 169 4e-41
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 166 2e-40
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 166 3e-40
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 151 6e-36
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 111 9e-24
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 78 1e-13
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 73 3e-12
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 70 2e-11
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 64 2e-09
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 63 3e-09
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 60 3e-08
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 59 7e-08
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 55 1e-06
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 54 2e-06
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 52 7e-06
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 44 0.002
RDPO_SCEOB (P19593) Probable reverse transcriptase (EC 2.7.7.49) 39 0.058
RT86_ECOLI (P23070) RNA-directed DNA polymerase from retron EC86... 36 0.49
YSM6_CAEEL (Q10126) Hypothetical protein F52C9.6 in chromosome III 35 0.65
PO12_NASVI (Q03270) Retrovirus-related Pol polyprotein from type... 34 1.4
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 169 bits (427), Expect = 4e-41
Identities = 129/500 (25%), Positives = 232/500 (45%), Gaps = 22/500 (4%)
Query: 318 TYDTQLQKTAISFYQNLFQKD----RTVNPHSLRVPLIPTLCALGIRSLSSPVLKEEVKD 373
T +++QK +Y+ L+ + ++ + L +P L + L+ P+ E+
Sbjct: 401 TDPSEIQKILNEYYKKLYSHKYENLKEIDQY-LEACHLPRLSQKEVEMLNRPISSSEIAS 459
Query: 374 ALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFIDEAIVETLLVLIPKE 433
+ ++ K+PGPDGF F++ F E+ L L + G + E + LIPK
Sbjct: 460 TIQNLPKKKSPGPDGFTSEFYQTFKEELVPILLNLFQNIEKEGILPNTFYEANITLIPKP 519
Query: 434 N-NPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDELVSPFQSNFIPRRGTTDNAIIAQ 492
+PT +N+RPISL N+ K++ K+L NRI+ + +++ Q FIP N +
Sbjct: 520 GKDPTRKENYRPISLMNIDAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQGWFNIRKSI 579
Query: 493 EVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELTLHDFGFPHHIIGLIMSCVKASDL 552
V+ +++ ++ K + L I EKA+D + F+ TL G + LI +
Sbjct: 580 NVIQHINKLKN-KDHMILSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKLIEAIYSKPTA 638
Query: 553 AILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RPIHISKDGPG 612
I+ NG K F G RQG PLSP LF + ME LA+ I+ E+++ + IHI +
Sbjct: 639 NIILNGVKLKSFPLRSGTRQGCPLSPLLFNIVMEVLAIAIR---EEKAIKGIHIGSEEIK 695
Query: 613 LSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSKRVPERTKRDI 672
LS FADD++++ E +++ + E+ + SG K+N KS + + ++ +
Sbjct: 696 LS--LFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKSVAFIYTN-NNQAEKTV 752
Query: 673 QGISSIRLVSNLGHYLGFPLVQG--RVTNETYKHIIEKMHHRLASWKGKLLNKSGRVCLE 730
+ +V YLG L + + E Y+ + +++ + WK + GR+ +
Sbjct: 753 KDSIPFTVVPKKMKYLGVYLTKDVKDLYKENYETLRKEIAEDVNKWKNIPCSWLGRINIV 812
Query: 731 KSVTTSMPVYSMQINLLRKELC--DGIDKITKSFIWGGNGVT*KWNMVKWTTVTSPRKFG 788
K +Y+ ++ L ++KI FIW K + T +++ K G
Sbjct: 813 KMSILPKAIYNFNAIPIKAPLSYFKDLEKIILHFIWNQ-----KKPQIAKTLLSNKNKAG 867
Query: 789 GLAIRDARNTNLALLGKLVW 808
G+ + D R +++ K W
Sbjct: 868 GITLPDLRLYYKSIVIKTAW 887
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 166 bits (420), Expect = 2e-40
Identities = 131/522 (25%), Positives = 241/522 (46%), Gaps = 30/522 (5%)
Query: 300 RRKHNKIHGLHLDDGTWCTYDTQLQKTAISFYQNLF--------QKDRTVNPHSLRVPLI 351
+R+ N+I + D G T T++Q T +Y++L+ + D+ ++ ++L
Sbjct: 383 KREKNQIDTIKNDRGDITTDPTEIQTTIREYYKHLYANKLENLEEMDKFLDTYTL----- 437
Query: 352 PTLCALGIRSLSSPVLKEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDI 411
P L + SL+ P+ E++ + S+ + K+PGP+GF F++ + E+ L +L
Sbjct: 438 PRLNQEEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQRYKEELVPFLLKLFQS 497
Query: 412 SFRRGFIDEAIVETLLVLIPKENNPTHLK-NFRPISLCNVIFKVITKVLVNRIRPFLDEL 470
+ G + + E ++LIPK T K NFRPISL N+ K++ K+L N+I+ + +L
Sbjct: 498 IEKEGILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQQHIKKL 557
Query: 471 VSPFQSNFIPRRGTTDNAIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELTL 530
+ Q FIP N + ++ +++ ++ + + I EKA+D++ F+ L
Sbjct: 558 IHHDQVGFIPAMQGWFNIRKSINIIQHINRTK-DTNHMIISIDAEKAFDKIQQPFMLKPL 616
Query: 531 HDFGFPHHIIGLIMSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAM 590
+ G + +I + I+ NG K G RQG PLSP L + +E LA
Sbjct: 617 NKLGIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGCPLSPLLPNIVLEVLAR 676
Query: 591 HIQGKVEDRS*RPIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLK 650
I+ +++ + I + K+ LS FADD++++ E Q ++ ++ F SG K
Sbjct: 677 AIR---QEKEIKGIQLGKEEVKLS--LFADDMIVYLENPIVSAQNLLKLISNFSKVSGYK 731
Query: 651 VNVTKSKSMCSKRVPERTKRDIQGISSIRLVSNLGHYLGFPLVQG--RVTNETYKHIIEK 708
+NV KS++ +T+ I + S YLG L + + E YK ++ +
Sbjct: 732 INVQKSQAFLYTN-NRQTESQIMSELPFTIASKRIKYLGIQLTRDVKDLFKENYKPLLNE 790
Query: 709 MHHRLASWKGKLLNKSGRVCLEKSVTTSMPVYSMQINLLRKELC--DGIDKITKSFIWGG 766
+ WK + GR+ + K +Y ++ + ++K T FIW
Sbjct: 791 IKEDTNKWKNIPCSWVGRINIVKMAILPKVIYRFNAIPIKLPMTFFTELEKTTLKFIWNQ 850
Query: 767 NGVT*KWNMVKWTTVTSPRKFGGLAIRDARNTNLALLGKLVW 808
K + +T++ K GG+ + D + A + K W
Sbjct: 851 -----KRAHIAKSTLSQKNKAGGITLPDFKLYYKATVTKTAW 887
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 166 bits (419), Expect = 3e-40
Identities = 131/522 (25%), Positives = 243/522 (46%), Gaps = 30/522 (5%)
Query: 306 IHGLHLDDGTWCTYDTQLQKTAISFY--------QNLFQKDRTVNPHSLRVPLIPTLCAL 357
I+ + + G T ++Q T SFY +NL + D+ ++ + +P L
Sbjct: 416 INKIRNEKGDITTDPEEIQNTIRSFYKRLYSTKLENLDEMDKFLDRYQ-----VPKLNQD 470
Query: 358 GIRSLSSPVLKEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGF 417
+ L+SP+ +E++ + S+ + K+PGPDGF F++ F E L +L G
Sbjct: 471 QVDHLNSPISPKEIEAVINSLPTKKSPGPDGFSAEFYQTFKEDLIPILHKLFHKIEVEGT 530
Query: 418 IDEAIVETLLVLIPK-ENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDELVSPFQS 476
+ + E + LIPK + +PT ++NFRPISL N+ K++ K+L NRI+ + ++ P Q
Sbjct: 531 LPNSFYEATITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQV 590
Query: 477 NFIPRRGTTDNAIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELTLHDFGFP 536
FIP N + V+HY++ + K + + + EKA+D++ F+ L G
Sbjct: 591 GFIPGMQGWFNIRKSINVIHYINKLK-DKNHMIISLDAEKAFDKIQHPFMIKVLERSGIQ 649
Query: 537 HHIIGLIMSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKV 596
+ +I + I NG K G RQG PLSPYLF + +E LA I+ +
Sbjct: 650 GPYLNMIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVLEVLARAIRQQK 709
Query: 597 EDRS*RPIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKS 656
E + I I K+ +S L ADD++++ + +++ +N F G K+N KS
Sbjct: 710 EIKG---IQIGKEEVKISLL--ADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKS 764
Query: 657 KSMCSKRVPERTKRDIQGISSIRLVSNLGHYLGFPLVQ--GRVTNETYKHIIEKMHHRLA 714
+ + ++ +++I+ + +V+N YLG L + + ++ +K + +++ L
Sbjct: 765 MAFLYTK-NKQAEKEIRETTPFSIVTNNIKYLGVTLTKEVKDLYDKNFKSLKKEIKEDLR 823
Query: 715 SWKGKLLNKSGRVCLEKSVTTSMPVYSMQINLLR--KELCDGIDKITKSFIWGGNGVT*K 772
WK + GR+ + K +Y ++ + + ++ F+W K
Sbjct: 824 RWKDLPCSWIGRINIVKMAILPKAIYRFNAIPIKIPTQFFNELEGAICKFVWNN-----K 878
Query: 773 WNMVKWTTVTSPRKFGGLAIRDARNTNLALLGKLVWSLLHDK 814
+ + + R GG+ + D + A++ K W D+
Sbjct: 879 KPRIAKSLLKDKRTSGGITMPDLKLYYRAIVIKTAWYWYRDR 920
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 151 bits (382), Expect = 6e-36
Identities = 119/483 (24%), Positives = 225/483 (45%), Gaps = 22/483 (4%)
Query: 292 FFHTQTIVRRKHNKIHGLHLDDGTWCTYDTQLQKTAISFYQNLFQKDRTVNPHSLRVPL- 350
FF+ + +I L +DGT ++ A SFYQNLF D ++P +
Sbjct: 374 FFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARSFYQNLFSPD-PISPDACEELWD 432
Query: 351 -IPTLCALGIRSLSSPVLKEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELV 409
+P + L +P+ +E+ AL M +K+PG DG FF+ FW+ G ++
Sbjct: 433 GLPVVSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEFFQFFWDTLGPDFHRVL 492
Query: 410 DISFRRGFIDEAIVETLLVLIPKENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDE 469
+F++G + + +L L+PK+ + +KN+RP+SL + +K++ K + R++ L E
Sbjct: 493 TEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYKIVAKAISLRLKSVLAE 552
Query: 470 LVSPFQSNFIPRRGTTDNAIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELT 529
++ P QS +P R DN + ++++H+ R+ L + EKA+DR+D +L T
Sbjct: 553 VIHPDQSYTVPGRTIFDNVFLIRDLLHF--ARRTGLSLAFLSLDQEKAFDRVDHQYLIGT 610
Query: 530 LHDFGFPHHIIGLIMSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLA 589
L + F +G + + +++ + N + T +G+RQG PLS L+ L +E
Sbjct: 611 LQAYSFGPQFVGYLKTMYASAECLVKINWSLTAPLAFGRGVRQGCPLSGQLYSLAIEPFL 670
Query: 590 MHIQGKVEDRS*RPIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGL 649
++ ++ + + + + +ADDV+L + + ++ + + +AS
Sbjct: 671 CLLRKRLTG-----LVLKEPDMRVVLSAYADDVILVAQ-DLVDLERAQECQEVYAAASSA 724
Query: 650 KVNVTKSKSMCSKRVP----ERTKRDIQGISSIRLVSNLGHYLGFPLVQGRVTNETYKHI 705
++N +KS + + RDI S +++ LG YL + ++ + +
Sbjct: 725 RINWSKSSGLLEGSLKVDFLPPAFRDISWES--KIIKYLGVYLS---AEEYPVSQNFIEL 779
Query: 706 IEKMHHRLASWKG--KLLNKSGRVCLEKSVTTSMPVYSMQINLLRKELCDGIDKITKSFI 763
E + RL WKG K+L+ GR + + S Y + +E I + F+
Sbjct: 780 EECVLTRLGKWKGFAKVLSMRGRALVINQLVASQIWYRLICLSPTQEFIAKIQRRLLDFL 839
Query: 764 WGG 766
W G
Sbjct: 840 WIG 842
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 111 bits (277), Expect = 9e-24
Identities = 80/305 (26%), Positives = 151/305 (49%), Gaps = 20/305 (6%)
Query: 364 SPVLKEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFIDEAIV 423
S + +++++ + S+ S +PGPDG P K E + ++++ G + +I
Sbjct: 342 SAITEQDLRASRVSLSS--SPGPDGITP---KSAREVPSGIMLRIMNLILWCGNLPHSIR 396
Query: 424 ETLLVLIPKENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDELVSPFQSNFIPRRG 483
V IPK ++FRPIS+ +V+ + + +L R+ ++ P Q F+P G
Sbjct: 397 LARTVFIPKTVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSINW--DPRQRGFLPTDG 454
Query: 484 TTDNAIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELTLHDFGFPHHIIGLI 543
DNA I V+ + H + + + + KA+D L + + TL +G P + +
Sbjct: 455 CADNATIVDLVLRHSH--KHFRSCYIANLDVSKAFDSLSHASIYDTLRAYGAPKGFVDYV 512
Query: 544 MSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RP 603
+ + ++ +G + +F P +G++QGDPLSP LF L M++L + ++
Sbjct: 513 QNTYEGGGTSLNGDGWSSEEFVPARGVKQGDPLSPILFNLVMDRLLRTLPSEIG------ 566
Query: 604 IHISKDGPGLSH-LTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSK 662
+K G +++ FADD++LF E +Q+++D +F S GLK+N K ++ K
Sbjct: 567 ---AKVGNAITNAAAFADDLVLFAETR-MGLQVLLDKTLDFLSIVGLKLNADKCFTVGIK 622
Query: 663 RVPER 667
P++
Sbjct: 623 GQPKQ 627
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 77.8 bits (190), Expect = 1e-13
Identities = 73/297 (24%), Positives = 132/297 (43%), Gaps = 20/297 (6%)
Query: 365 PVLKEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFIDEAIVE 424
P+ +EE++ A+ + APG DG V + RG +
Sbjct: 5 PIAREEIQCAIKGWKPS-APGSDGLTVQAITRTRLPRN-----FVQLHLLRGHVPTPWTA 58
Query: 425 TLLVLIPKENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDELVSPFQSNFIPRRGT 484
LIPK+ + + N+RPI++ + + +++ ++L R+ ++ + P Q + GT
Sbjct: 59 MRTTLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVE--LHPAQKGYARIDGT 116
Query: 485 TDNAIIAQEVVHYMHTSRSQKGTL-ALKIYLEKAYDRLDWSFLELTLHDFGFPHHIIGLI 543
N+++ Y+ + R Q+ T + + + KA+D + S + L G I
Sbjct: 117 LVNSLLLDT---YISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYI 173
Query: 544 MSCVKASDLAI-LWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*R 602
+ S I + G++T K +G++QGDPLSP+LF +++L +Q S
Sbjct: 174 TGSLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLFNAVLDELLCSLQ------STP 227
Query: 603 PIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSM 659
I + + L FADD+LL E N + + T+ F G+ +N KS S+
Sbjct: 228 GIGGTIGEEKIPVLAFADDLLLL-EDNDVLLPTTLATVANFFRLRGMSLNAKKSVSI 283
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 73.2 bits (178), Expect = 3e-12
Identities = 70/301 (23%), Positives = 122/301 (40%), Gaps = 19/301 (6%)
Query: 359 IRSLSSPVLKEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFI 418
I++L P+ +E+K+ ++ A GPDG W E + L ++ G
Sbjct: 312 IKNLWRPISNDEIKEVEACKRT--AAGPDGMTTTA----WNSIDECIKSLFNMIMYHGQC 365
Query: 419 DEAIVETLLVLIPKENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDELVSPFQSNF 478
+++ VLIPKE FRP+S+ +V + ++L NRI L+ Q F
Sbjct: 366 PRRYLDSRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANRIGE--HGLLDTRQRAF 423
Query: 479 IPRRGTTDNAIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELTLHDFGFPHH 538
I G +N + ++ KG + ++KA+D ++ + L P
Sbjct: 424 IVADGVAENTSLLSAMI--KEARMKIKGLYIAILDVKKAFDSVEHRSILDALRRKKLPLE 481
Query: 539 IIGLIMSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVED 598
+ IM + S + K +P +G+RQGDPLSP LF M+ + +
Sbjct: 482 MRNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLFNCVMDAVLRRLPENT-- 539
Query: 599 RS*RPIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKS 658
+ L FADD++L E + +Q + + GL++ K +
Sbjct: 540 ------GFLMGAEKIGALVFADDLVLLAETR-EGLQASLSRIEAGLQEQGLEMMPRKCHT 592
Query: 659 M 659
+
Sbjct: 593 L 593
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 70.5 bits (171), Expect = 2e-11
Identities = 68/285 (23%), Positives = 123/285 (42%), Gaps = 39/285 (13%)
Query: 415 RGFIDEAIVE----TLLVLIPKENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDEL 470
+ F D AI +++ IPK+ NP+ N+RPISL + +++ +++ +RIR L
Sbjct: 654 QSFSDSAIPNRWKHAVIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHL 713
Query: 471 VSPFQSNFIPRRGTTDNAIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELTL 530
+SP Q F+ R + + + + Y +++K L KA+D++ L L
Sbjct: 714 LSPHQHGFLNFRSCPSS--LVRSISLYHSILKNEKSLDILFFDFAKAFDKVSHPILLKKL 771
Query: 531 HDFGFPHHIIGLIMSCVKASDLAILWNG-AKTNKFKPTKGLRQGDPLSPYLFVLCMEKLA 589
FG + ++ N +N + + G+ QG P LF+L + L
Sbjct: 772 ALFGLDKLTCSWFKEFLHLRTFSVKINKFVSSNAYPISSGVPQGSVSGPLLFILFINDLL 831
Query: 590 MHIQGKVEDRS*RPIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGL 649
+ ++ IH+S FADD+ +F N +Q IDT+ ++ + L
Sbjct: 832 IDLEPN--------IHVS---------CFADDIKIF-HHNPSTLQNSIDTIVKWSKKNKL 873
Query: 650 KVNVTKSK--SMCSKR------------VPERTKRDIQGISSIRL 680
+ KS S+ S+ +P T RD+ I+ ++L
Sbjct: 874 PLAPAKSSVLSLGSRNSNHTYRVDNVPILPSSTVRDLGLITDLKL 918
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 63.9 bits (154), Expect = 2e-09
Identities = 84/344 (24%), Positives = 143/344 (41%), Gaps = 36/344 (10%)
Query: 359 IRSLSSPVLKEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFI 418
+ +L PV E+K A S ++K GPDG P + ++ L+ +I G +
Sbjct: 152 METLWDPVSLIEIKSARAS--NEKGAGPDGVTPRSWNALDDRYKRLLY---NIFVFYGRV 206
Query: 419 DEAIVETLLVLIPKENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDELVSPFQSNF 478
I + V PK FRP+S+C+VI + K+L R Q+ +
Sbjct: 207 PSPIKGSRTVFTPKIEGGPDPGVFRPLSICSVILREFNKILARRFVSCY--TYDERQTAY 264
Query: 479 IPRRGTTDNAIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELTLHDFGFPHH 538
+P G N + ++ R +K + L KA++ + S L + + G P
Sbjct: 265 LPIDGVCINVSMLTAII--AEAKRLRKELHIAILDLVKAFNSVYHSALIDAITEAGCPPG 322
Query: 539 IIGLIMSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKL--AMHIQGKV 596
++ I + + G K G+ QGDPLS LF L EK A++ +G+
Sbjct: 323 VVDYIADMYNNVITEMQFEG-KCELASILAGVYQGDPLSGPLFTLAYEKALRALNNEGRF 381
Query: 597 EDRS*RPIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKS 656
+ R ++ ++DD LL +Q +D E + GL++N KS
Sbjct: 382 DIADVR----------VNASAYSDDGLLLA-MTVIGLQHNLDKFGETLAKIGLRINSRKS 430
Query: 657 KSMCSKRVPERTKRDIQGISSIRL-----------VSNLGHYLG 689
K++ VP ++ ++ +S+ RL +S+L YLG
Sbjct: 431 KTV--SLVPSGREKKMKIVSNRRLLLKASELKPLTISDLWKYLG 472
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 63.2 bits (152), Expect = 3e-09
Identities = 32/70 (45%), Positives = 42/70 (59%)
Query: 552 LAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RPIHISKDGP 611
L + NGA P++GLRQGDPLSPYLF+LC E L+ + E I +S + P
Sbjct: 10 LLFIINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSP 69
Query: 612 GLSHLTFADD 621
++HL FADD
Sbjct: 70 RINHLLFADD 79
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 59.7 bits (143), Expect = 3e-08
Identities = 100/451 (22%), Positives = 187/451 (41%), Gaps = 42/451 (9%)
Query: 314 GTWCTYDTQLQKTAISFYQNLFQKDRTVN--PHSLRVPLIPTLCALGIRSLS-SPVLKEE 370
G WC T L+KT + F NL Q+ N P SL + L + SL S V EE
Sbjct: 386 GGWCR--TSLEKTEV-FANNLEQRFTPYNYAPESLCRQVEEYLESPFQMSLPLSAVTLEE 442
Query: 371 VKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFIDEAIVETLLVLI 430
VK+ + + KAPG D + ++ + L + + G+ +A +++I
Sbjct: 443 VKNLIAKLPLKKAPGEDLLDNRTIRLLPDQALQFLALIFNSVLDVGYFPKAWKSASIIMI 502
Query: 431 PKENN-PTHLKNFRPISLCNVIFKVITKVLVNRIRPFLD--ELVSPFQSNFIPRRGTTDN 487
K PT + ++RP SL + K++ ++++NR+ D + + FQ F + GT +
Sbjct: 503 HKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRLLTCKDVTKAIPKFQFGFRLQHGTPEQ 562
Query: 488 AIIAQEVVHY-MHTSRSQKGTLALKIYLEKAYDRLDWSFLELTLHDFGFPHHIIGLIMSC 546
VV++ + +++ + + +++A+DR+ W L FP + ++ S
Sbjct: 563 ---LHRVVNFALEAMENKEYAVGAFLDIQQAFDRV-WHPGLLYKAKRLFPPQLYLVVKSF 618
Query: 547 VKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RPIHI 606
++ + +G K++ G+ QG L P L+ + + H P+
Sbjct: 619 LEERTFHVSVDGYKSSIKPIAAGVPQGSVLGPTLYSVFASDMPTH----------TPV-T 667
Query: 607 SKDGPGLSHLTFADDVLLFCE-----ANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCS 661
D + T+ADD + + A T +Q +D ++ +++N K C+
Sbjct: 668 EVDEEDVLIATYADDTAVLTKSKSILAATSGLQEYLDAFQQWAENWNVRINAEK----CA 723
Query: 662 KRVPERTKRDIQGIS-SIRLVSN--LGHYLGFPLVQGRVTNETYKHIIEKMHHRLA--SW 716
G+S + RL+ + YLG L + + +I + ++A SW
Sbjct: 724 NVTFANRTGSCPGVSLNGRLIRHHQAYKYLGITLDRKLTFSRHITNIQQAFRTKVARMSW 783
Query: 717 KGKLLNKSGRVC---LEKSVTTSMPVYSMQI 744
NK C + KS+ Y +Q+
Sbjct: 784 LIAPRNKLSLGCKVNIYKSILAPCLFYGLQV 814
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 58.5 bits (140), Expect = 7e-08
Identities = 50/187 (26%), Positives = 83/187 (43%), Gaps = 11/187 (5%)
Query: 483 GTTDNAIIAQEVVHYMHTSRSQKGTL-ALKIYLEKAYDRLDWSFLELTLHDFGFPHHIIG 541
GT N I+ + HY+ R + T + + + KA+D + + + FG +
Sbjct: 1 GTLANIIMLE---HYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQD 57
Query: 542 LIMSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS* 601
IMS + + I+ G TNK G++QGDPLSP LF + +++L + + S
Sbjct: 58 FIMSTITDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQPGASM 117
Query: 602 RPIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCS 661
P ++ L FADD+LL E V + T + G+ +N K S+ +
Sbjct: 118 TP------ACKIASLAFADDLLLL-EDRDIDVPNSLATTCAYFRTRGMTLNPEKCASISA 170
Query: 662 KRVPERT 668
V R+
Sbjct: 171 ATVSGRS 177
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 54.7 bits (130), Expect = 1e-06
Identities = 63/266 (23%), Positives = 107/266 (39%), Gaps = 27/266 (10%)
Query: 370 EVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFI--DEAIVETLL 427
EV + ++S ++PG DG K W E L L R G+ + +
Sbjct: 438 EVDTCVARLKSRRSPGLDGINGTICKAVWRAIPEHLASLFSRCIRLGYFPAEWKCPRVVS 497
Query: 428 VLIPKENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDELVSPFQSNFIPRRGTTDN 487
+L + + ++R I L V KV+ ++VNR+R L E +Q F R D
Sbjct: 498 LLKGPDKDKCEPSSYRGICLLPVFGKVLEAIMVNRVREVLPE-GCRWQFGFRQGRCVED- 555
Query: 488 AIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELTLHDFGFPHHIIGLIMSCV 547
A V + + + L + + A+D ++WS L D G +GL S
Sbjct: 556 ---AWRHVKSSVGASAAQYVLGTFVDFKGAFDNVEWSAALSRLADLGCRE--MGLWQSFF 610
Query: 548 KASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RPIHIS 607
+ A++ + + T + T+G QG P+++ + M+ L +Q
Sbjct: 611 -SGRRAVIRSSSGTVEVPVTRGCPQGSISGPFIWDILMDVLLQRLQ-------------- 655
Query: 608 KDGPGLSHLTFADDVLLFCEANTQQV 633
P +ADD+LL E N++ V
Sbjct: 656 ---PYCQLSAYADDLLLLVEGNSRAV 678
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 53.9 bits (128), Expect = 2e-06
Identities = 71/307 (23%), Positives = 121/307 (39%), Gaps = 29/307 (9%)
Query: 360 RSLSSPVLKEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFID 419
R L + +EV D++ + K+PGPDG + W E ++ L +
Sbjct: 399 RPLPPDLEMDEVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQCLLESYFP 458
Query: 420 EAIVETLLVLIPK--ENNPTHLKNFRPISLCNVIFKVITKVLVNRI-RPFLDELVSPFQS 476
+ LV++ K + + ++RPI L + + KV+ ++V R+ + +D VSP+Q
Sbjct: 459 QKWKIASLVILLKLLDRIRSDPGSYRPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQF 518
Query: 477 NFIPRRGTTDNAIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELTLHDFGFP 536
F + T D A V K + L I + A+D L W + L L +
Sbjct: 519 AFTYGKSTED----AWRCVQRHVECSEMKYVIGLNIDFQGAFDNLGWLSMLLKLDE--AQ 572
Query: 537 HHIIGLIMSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKV 596
+ GL MS + + K T+G QG P ++ L M +L + +
Sbjct: 573 SNEFGLWMSYFGGRKVYYVGKTGIVRK-DVTRGCPQGSKSGPAMWKLVMNELLLALV--- 628
Query: 597 EDRS*RPIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVIDT-LNEFCSASGLK--VNV 653
G + FADD + AN++ + T + C G + V V
Sbjct: 629 -------------AAGFFIVAFADDGTIVIGANSRSALEELGTRCLQLCHEWGKRVCVPV 675
Query: 654 TKSKSMC 660
+ K+ C
Sbjct: 676 SAGKTTC 682
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 52.0 bits (123), Expect = 7e-06
Identities = 94/417 (22%), Positives = 175/417 (41%), Gaps = 49/417 (11%)
Query: 314 GTWCTYDTQLQKTAISFYQNLFQKDRTV----NPHSLRVP---LIPTLCALGIRSLSSPV 366
G WC T +KT + F +L Q+ + + HSL V P AL + PV
Sbjct: 386 GGWCR--TSREKTEV-FANHLEQRFKALAFAPESHSLMVAESLQTPFQMALP----ADPV 438
Query: 367 LKEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFIDEAIVETL 426
EEVK+ + ++ KAPG D + ++ L + + R G+ +A
Sbjct: 439 TLEEVKELVSKLKPKKAPGEDLLDNRTIRLLPDQALLYLVLIFNSILRVGYFPKARPTAS 498
Query: 427 LVLIPKENN-PTHLKNFRPISLCNVIFKVITKVLVNRI--RPFLDELVSPFQSNFIPRRG 483
+++I K P + ++RP SL + K++ ++++NRI + + FQ F + G
Sbjct: 499 IIMILKPGKQPLDVDSYRPTSLLPSLGKMLERLILNRILTSEEVTRAIPKFQFGFRLQHG 558
Query: 484 TTDNAIIAQEVVHYMHTSRSQK---GTLALKIYLEKAYDRLDWSFLELTLHDFGFPHHII 540
T + VV++ + +K G+ L I ++A+DR+ L P +
Sbjct: 559 TPEQ---LHRVVNFALEALEKKEYAGSCFLDI--QQAFDRVWHPGLLYKAKSLLSP-QLF 612
Query: 541 GLIMSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS 600
LI S + ++ +G +++ G+ QG L P L+ + M Q V +
Sbjct: 613 QLIKSFWEGRKFSVTADGCRSSVKFIEAGVPQGSVLGPTLYSIFTAD--MPNQNAVTGLA 670
Query: 601 *RPIHISKDGPGLSHLTFADDVLLFCEAN-----TQQVQMVIDTLNEFCSASGLKVNVTK 655
+ I+ T+ADD+ + ++ T +Q +D E+ +K NV+
Sbjct: 671 EGEVLIA---------TYADDIAVLTKSTCIVEATDALQEYLDAFQEW----AVKWNVSI 717
Query: 656 SKSMCSKRVPERTKRDIQGIS-SIRLVSNLGHYLGFPLVQGRVTNETYKHIIEKMHH 711
+ C+ RD G++ + L+S+ Y ++ R + T++ I + H
Sbjct: 718 NAGKCANVTFTNAIRDCPGVTINGSLLSHTHEYKYLGVILDR--SLTFRRHITSLQH 772
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 43.5 bits (101), Expect = 0.002
Identities = 31/107 (28%), Positives = 46/107 (42%), Gaps = 1/107 (0%)
Query: 736 SMPVYSMQINLLRKELCDGIDKITKSFIWGGNGVT*KWNMVKWTTVTSPRKF-GGLAIRD 794
++PVY+M L K LC + F W K + V W + ++ GGL RD
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 795 ARNTNLALLGKLVWSLLHDKNKLWVRLLSGKIY*EWLFMEQRSSEKP 841
N ALL K + ++H + L RLL + + ME +P
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRP 108
>RDPO_SCEOB (P19593) Probable reverse transcriptase (EC 2.7.7.49)
Length = 608
Score = 38.9 bits (89), Expect = 0.058
Identities = 29/112 (25%), Positives = 54/112 (47%), Gaps = 18/112 (16%)
Query: 554 ILWNGAK-------TNKFKPTK-GLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RPIH 605
ILW K +N +PT G+ QG +SP + L ++ L HI K++
Sbjct: 222 ILWAWLKCGYIERNSNTLQPTTTGVPQGGIISPLIMNLTLDGLEFHIYKKIQKS------ 275
Query: 606 ISKDGPGLSHLTFADDVLLFCEANTQQVQMV-IDTLNEFCSASGLKVNVTKS 656
S G ++ +ADD+++ T++ ++ + + EF + GL+V + K+
Sbjct: 276 -SSQSKGNTYCRYADDMVIL--TTTEETALIALPAVKEFLAVRGLEVKLAKT 324
>RT86_ECOLI (P23070) RNA-directed DNA polymerase from retron EC86
(EC 2.7.7.49) (Reverse transcriptase) (EC86-RT)
Length = 320
Score = 35.8 bits (81), Expect = 0.49
Identities = 40/142 (28%), Positives = 56/142 (39%), Gaps = 31/142 (21%)
Query: 570 LRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RPIHISKDGPGLSHLTFADDVLLFCEAN 629
L QG P SP L L KL IQG R GL + +ADD+ L + +
Sbjct: 159 LPQGAPSSPKLANLICSKLDYRIQGYAGSR------------GLIYTRYADDLTLSAQ-S 205
Query: 630 TQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSKRVPERTKRDIQGISSIRLVSNLGHYLG 689
++V D L + GL +N SK C R++R + G+ + +G
Sbjct: 206 MKKVVKARDFLFSIIPSEGLVIN---SKKTCIS--GPRSQRKVTGLVISQEKVGIG---- 256
Query: 690 FPLVQGRVTNETYKHIIEKMHH 711
E YK I K+HH
Sbjct: 257 ---------REKYKEIRAKIHH 269
>YSM6_CAEEL (Q10126) Hypothetical protein F52C9.6 in chromosome III
Length = 279
Score = 35.4 bits (80), Expect = 0.65
Identities = 35/139 (25%), Positives = 67/139 (48%), Gaps = 16/139 (11%)
Query: 609 DGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSKRVPERT 668
+G L++L FADD++L + ++ L + CS GL++N K+K + R+ + +
Sbjct: 4 NGRNLTNLRFADDIVLIAN-HPNTASKMLQELVQKCSEVGLEINTGKTK-VLRNRLADPS 61
Query: 669 KRDIQGISSIRLVSNLGHYLGFPLVQGRVTNETYKHIIEKMH-HRLASWKGKLLNKSGRV 727
K SS + ++ Y + GR N + +++ ++H R A+W N S
Sbjct: 62 KVYFGSPSSTTQLDDVDEY----IYLGRQIN-AHNNLMPEIHRRRRAAWAA--FNGS--- 111
Query: 728 CLEKSVTTSMPVYSMQINL 746
K+ T S+ +++NL
Sbjct: 112 ---KNTTDSITDKKIRVNL 127
>PO12_NASVI (Q03270) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 410
Score = 34.3 bits (77), Expect = 1.4
Identities = 30/97 (30%), Positives = 46/97 (46%), Gaps = 12/97 (12%)
Query: 610 GPGLSHLTFADDVLLFCEANT-QQVQMVIDTLNEFCSASGLK--VNVTKSKSMC------ 660
G G+S FADDV + E NT ++V+ VI+ E G K V+V++ K++C
Sbjct: 26 GSGVSVCAFADDVAILAEGNTRKEVERVINEKMEIVYKWGEKMGVSVSEEKTVCMLLKGK 85
Query: 661 --SKRVPERTKRDIQGISSIRLVSNLGHYLGFPLVQG 695
+ R + I I+ VS YLG + +G
Sbjct: 86 YVTSNRAVRVSKTINVYKRIKYVSCF-RYLGVNVTEG 121
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.345 0.152 0.514
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 91,374,305
Number of Sequences: 164201
Number of extensions: 3658955
Number of successful extensions: 14562
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 14504
Number of HSP's gapped (non-prelim): 32
length of query: 855
length of database: 59,974,054
effective HSP length: 119
effective length of query: 736
effective length of database: 40,434,135
effective search space: 29759523360
effective search space used: 29759523360
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC135100.7


