

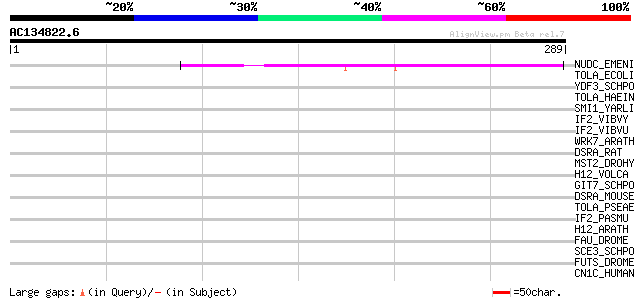
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.6 + phase: 0
(289 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NUDC_EMENI (P17624) Nuclear movement protein nudC 144 2e-34
TOLA_ECOLI (P19934) TolA protein 40 0.005
YDF3_SCHPO (Q10475) Probable eukaryotic initiation factor C17C9.03 40 0.009
TOLA_HAEIN (P44678) TolA protein 39 0.011
SMI1_YARLI (Q6CDX0) KNR4/SMI1 homolog 36 0.096
IF2_VIBVY (Q7MI09) Translation initiation factor IF-2 36 0.096
IF2_VIBVU (Q8DBW0) Translation initiation factor IF-2 36 0.096
WRK7_ARATH (Q9STX0) Probable WRKY transcription factor 7 (WRKY D... 36 0.13
DSRA_RAT (P55266) Double-stranded RNA-specific adenosine deamina... 36 0.13
MST2_DROHY (Q08696) Axoneme-associated protein mst101(2) 35 0.16
H12_VOLCA (Q08865) Histone H1-II 35 0.16
GIT7_SCHPO (O59709) Glucose insensitive transcription protein 7 35 0.16
DSRA_MOUSE (Q99MU3) Double-stranded RNA-specific adenosine deami... 35 0.16
TOLA_PSEAE (P50600) TolA protein 35 0.21
IF2_PASMU (P57873) Translation initiation factor IF-2 35 0.21
H12_ARATH (P26569) Histone H1.2 35 0.21
FAU_DROME (Q9VGX3) Anoxia up-regulated protein 35 0.21
SCE3_SCHPO (O14369) Probable RNA-binding protein sce3 35 0.28
FUTS_DROME (Q9W596) Microtubule-associated protein futsch 35 0.28
CN1C_HUMAN (Q14123) Calcium/calmodulin-dependent 3',5'-cyclic nu... 35 0.28
>NUDC_EMENI (P17624) Nuclear movement protein nudC
Length = 198
Score = 144 bits (364), Expect = 2e-34
Identities = 79/206 (38%), Positives = 118/206 (56%), Gaps = 17/206 (8%)
Query: 90 IAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPNQGNGMDLEKYSWTQTLQELNVNVPVP 149
++ +E + A + +EK E+ ++ P Y WTQT+++++V +PV
Sbjct: 1 MSEQEPSSADLAAREAEEKQRKAAEEAEQATLP----------YKWTQTIRDVDVTIPVS 50
Query: 150 NGTKSGFVICEIKKNHLKVGLKGQ--PPIIDRELYKSIKPDECYWSIEDQNT-----VSI 202
K + +KK+ +KV +KG+ ID + IKP E W++E + VSI
Sbjct: 51 ANLKGRDLDVVLKKDSIKVKVKGENGEVFIDGQFPHPIKPSESSWTLETTSKPPGKEVSI 110
Query: 203 LLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLGDLDSETRMTVEKMMFDQRQKSMGLPT 262
L K +QM+WW +V P+I+ K+ P +S L DLD ETR VEKMM+DQRQK MG PT
Sbjct: 111 HLDKVNQMEWWAHVVTTAPKIDVSKITPENSSLSDLDGETRAMVEKMMYDQRQKEMGAPT 170
Query: 263 SEELQKEEMMKKFMSQHPNMDFSGAK 288
S+E +K +++KKF +HP MDFS AK
Sbjct: 171 SDEQRKMDILKKFQKEHPEMDFSNAK 196
>TOLA_ECOLI (P19934) TolA protein
Length = 421
Score = 40.4 bits (93), Expect = 0.005
Identities = 27/63 (42%), Positives = 34/63 (53%), Gaps = 3/63 (4%)
Query: 62 DSAEKVVLSAVRAARVKKA---KSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDE 118
++AEK A + A +KA K AAEKA K KAAAEK D+K+ A D+
Sbjct: 214 EAAEKAKAEAEKKAAAEKAAADKKAAAEKAAADKKAAEKAAAEKAAADKKAAAEKAAADK 273
Query: 119 KAA 121
KAA
Sbjct: 274 KAA 276
Score = 38.1 bits (87), Expect = 0.025
Identities = 28/61 (45%), Positives = 35/61 (56%), Gaps = 4/61 (6%)
Query: 61 KDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKA 120
K AE +A A KKA + AAEKAK A++K AAAEK D+K+ A D+KA
Sbjct: 194 KKKAEAAEAAAAEAR--KKAATEAAEKAKAEAEKK--AAAEKAAADKKAAAEKAAADKKA 249
Query: 121 A 121
A
Sbjct: 250 A 250
Score = 37.7 bits (86), Expect = 0.033
Identities = 24/68 (35%), Positives = 30/68 (43%)
Query: 60 DKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEK 119
DK +AEK A + A+ AA+K AAK A+ AA K E + E K
Sbjct: 246 DKKAAEKAAAEKAAADKKAAAEKAAADKKAAAAKAAAEKAAAAKAAAEADDIFGELSSGK 305
Query: 120 AAPNQGNG 127
AP G G
Sbjct: 306 NAPKTGGG 313
Score = 37.0 bits (84), Expect = 0.056
Identities = 30/71 (42%), Positives = 39/71 (54%), Gaps = 7/71 (9%)
Query: 61 KDSAEKVVLSAVRAARVKK----AKSVAAE---KAKIAAKEKAKAAAEKKLNDEKSEAVT 113
K +AE + AA +KK A++ AAE KA A EKAKA AEKK EK+ A
Sbjct: 177 KAAAEAQKKAEAAAAALKKKAEAAEAAAAEARKKAATEAAEKAKAEAEKKAAAEKAAADK 236
Query: 114 EKIDEKAAPNQ 124
+ EKAA ++
Sbjct: 237 KAAAEKAAADK 247
Score = 33.5 bits (75), Expect = 0.62
Identities = 25/62 (40%), Positives = 35/62 (56%), Gaps = 4/62 (6%)
Query: 60 DKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEK 119
+K +A+K + AA K A+ AAEK AA +K KAAAEK D+K+ A ++
Sbjct: 230 EKAAADKKAAAEKAAADKKAAEKAAAEK---AAADK-KAAAEKAAADKKAAAAKAAAEKA 285
Query: 120 AA 121
AA
Sbjct: 286 AA 287
Score = 31.6 bits (70), Expect = 2.4
Identities = 23/76 (30%), Positives = 36/76 (47%), Gaps = 3/76 (3%)
Query: 66 KVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPNQG 125
K A A + AK AA+ K A E AKAAAE + +K+EA + +KA +
Sbjct: 146 KAKAEADAKAAEEAAKKAAADAKKKAEAEAAKAAAEAQ---KKAEAAAAALKKKAEAAEA 202
Query: 126 NGMDLEKYSWTQTLQE 141
+ K + T+ ++
Sbjct: 203 AAAEARKKAATEAAEK 218
Score = 31.2 bits (69), Expect = 3.1
Identities = 19/45 (42%), Positives = 26/45 (57%)
Query: 70 SAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTE 114
+A +AA KAK+ A KA A +KA A A+KK E ++A E
Sbjct: 137 AAAKAAADAKAKAEADAKAAEEAAKKAAADAKKKAEAEAAKAAAE 181
>YDF3_SCHPO (Q10475) Probable eukaryotic initiation factor C17C9.03
Length = 1403
Score = 39.7 bits (91), Expect = 0.009
Identities = 24/69 (34%), Positives = 36/69 (51%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E +++A++ + +KAK A EKAK A+EKAK AE+K E E
Sbjct: 589 AEEKAKREAEENAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEK 648
Query: 112 VTEKIDEKA 120
+ +EKA
Sbjct: 649 AKREAEEKA 657
Score = 38.9 bits (89), Expect = 0.015
Identities = 24/69 (34%), Positives = 35/69 (49%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E ++ A++ + +KAK A EKAK A+EKAK AE+K E E
Sbjct: 581 AEEKAKREAEEKAKREAEENAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEK 640
Query: 112 VTEKIDEKA 120
+ +EKA
Sbjct: 641 AKREAEEKA 649
Score = 38.9 bits (89), Expect = 0.015
Identities = 23/69 (33%), Positives = 35/69 (50%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E +++A++ + +KAK A EKAK A+EKAK AE+ E E
Sbjct: 549 AEEKARLEAEENAKREAEEQAKREAEEKAKREAEEKAKREAEEKAKREAEENAKREAEEK 608
Query: 112 VTEKIDEKA 120
+ +EKA
Sbjct: 609 AKREAEEKA 617
Score = 38.9 bits (89), Expect = 0.015
Identities = 24/69 (34%), Positives = 35/69 (49%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E ++ A++ + +KAK A EKAK A+EKAK AE+K E E
Sbjct: 605 AEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEN 664
Query: 112 VTEKIDEKA 120
+ +EKA
Sbjct: 665 AKREAEEKA 673
Score = 38.5 bits (88), Expect = 0.019
Identities = 26/99 (26%), Positives = 44/99 (44%), Gaps = 5/99 (5%)
Query: 22 QPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLSAVRAARVKKAK 81
Q S P P + PT ++ + A + KD+ + + ++ +AK
Sbjct: 492 QKNASSPNPSETNSRAETPTAAPPQISEEEASQR-----KDAIKLAIQQRIQEKAEAEAK 546
Query: 82 SVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKA 120
A EKA++ A+E AK AE++ E E + +EKA
Sbjct: 547 RKAEEKARLEAEENAKREAEEQAKREAEEKAKREAEEKA 585
Score = 37.7 bits (86), Expect = 0.033
Identities = 23/69 (33%), Positives = 35/69 (50%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E+ ++ A++ + +KAK A EKAK A+EKAK AE+K E E
Sbjct: 597 AEENAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEK 656
Query: 112 VTEKIDEKA 120
+ +E A
Sbjct: 657 AKREAEENA 665
Score = 37.7 bits (86), Expect = 0.033
Identities = 23/69 (33%), Positives = 35/69 (50%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E+ ++ A++ + +KAK A EKAK A+E AK AE+K E E
Sbjct: 557 AEENAKREAEEQAKREAEEKAKREAEEKAKREAEEKAKREAEENAKREAEEKAKREAEEK 616
Query: 112 VTEKIDEKA 120
+ +EKA
Sbjct: 617 AKREAEEKA 625
Score = 37.4 bits (85), Expect = 0.043
Identities = 23/69 (33%), Positives = 34/69 (48%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E ++ A++ + + AK A EKAK A+EKAK AE+K E E
Sbjct: 573 AEEKAKREAEEKAKREAEEKAKREAEENAKREAEEKAKREAEEKAKREAEEKAKREAEEK 632
Query: 112 VTEKIDEKA 120
+ +EKA
Sbjct: 633 AKREAEEKA 641
Score = 37.4 bits (85), Expect = 0.043
Identities = 23/69 (33%), Positives = 34/69 (48%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E ++ A++ + +KAK A E AK A+EKAK AE+K E E
Sbjct: 565 AEEQAKREAEEKAKREAEEKAKREAEEKAKREAEENAKREAEEKAKREAEEKAKREAEEK 624
Query: 112 VTEKIDEKA 120
+ +EKA
Sbjct: 625 AKREAEEKA 633
Score = 35.8 bits (81), Expect = 0.13
Identities = 30/116 (25%), Positives = 47/116 (39%), Gaps = 1/116 (0%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E ++ A++ + + AK A EKAK A+E AK AE+K+ E E
Sbjct: 637 AEEKAKREAEEKAKREAEEKAKREAEENAKREAEEKAKREAEENAKREAEEKVKRETEEN 696
Query: 112 VTEKIDEKAAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLK 167
K +E+ ++ K S E NV+ V+ + K LK
Sbjct: 697 AKRKAEEEGKREADKNPEI-KSSAPLASSEANVDTSKQTNATEPEVVDKTKVEKLK 751
Score = 35.4 bits (80), Expect = 0.16
Identities = 22/68 (32%), Positives = 33/68 (48%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E ++ A++ + +KAK A EKAK A+E AK AE+K E E
Sbjct: 621 AEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEENAKREAEEKAKREAEEN 680
Query: 112 VTEKIDEK 119
+ +EK
Sbjct: 681 AKREAEEK 688
Score = 35.4 bits (80), Expect = 0.16
Identities = 22/69 (31%), Positives = 33/69 (46%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E ++ A++ + +KAK A EKAK A+EKAK AE+ E E
Sbjct: 613 AEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEENAKREAEEK 672
Query: 112 VTEKIDEKA 120
+ +E A
Sbjct: 673 AKREAEENA 681
Score = 35.0 bits (79), Expect = 0.21
Identities = 22/69 (31%), Positives = 33/69 (46%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E ++ A++ + +KAK A E AK A+EKAK AE+ E E
Sbjct: 629 AEEKAKREAEEKAKREAEEKAKREAEEKAKREAEENAKREAEEKAKREAEENAKREAEEK 688
Query: 112 VTEKIDEKA 120
V + +E A
Sbjct: 689 VKRETEENA 697
Score = 33.1 bits (74), Expect = 0.82
Identities = 24/63 (38%), Positives = 29/63 (45%), Gaps = 7/63 (11%)
Query: 65 EKVVLSAVRAARVK-------KAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKID 117
EK A R A K AK A E+AK A+EKAK AE+K E E + +
Sbjct: 539 EKAEAEAKRKAEEKARLEAEENAKREAEEQAKREAEEKAKREAEEKAKREAEEKAKREAE 598
Query: 118 EKA 120
E A
Sbjct: 599 ENA 601
>TOLA_HAEIN (P44678) TolA protein
Length = 372
Score = 39.3 bits (90), Expect = 0.011
Identities = 30/75 (40%), Positives = 44/75 (58%), Gaps = 7/75 (9%)
Query: 63 SAEKVVLSAVRAARVK---KAKSVAAEKAKIAAKEKAKAAAEKKLN-DEKSEAVTE---K 115
+A+K A A+++ KAK+VA KAK A+ KAKAAAE K D +++A TE K
Sbjct: 180 AAQKAKQEAEAKAKLEAEAKAKAVAEAKAKAEAEAKAKAAAEAKAKADAEAKAATEAKRK 239
Query: 116 IDEKAAPNQGNGMDL 130
D+ + + NG D+
Sbjct: 240 ADQASLDDFLNGGDI 254
Score = 32.7 bits (73), Expect = 1.1
Identities = 22/60 (36%), Positives = 28/60 (46%), Gaps = 1/60 (1%)
Query: 61 KDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKA 120
K + E+ A A +KAK A KAK+ A+ KAKA AE K E + KA
Sbjct: 166 KQAEEEAKAKAAEIA-AQKAKQEAEAKAKLEAEAKAKAVAEAKAKAEAEAKAKAAAEAKA 224
Score = 30.8 bits (68), Expect = 4.0
Identities = 25/75 (33%), Positives = 40/75 (53%), Gaps = 5/75 (6%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKS--VAAEKAKIAAKEKAKAAAE---KKLND 106
A E+ A+++ +A +A KAK+ +AA+KAK A+ KAK AE K + +
Sbjct: 146 AAEAAKLKADAEAKRLAAAAKQAEEEAKAKAAEIAAQKAKQEAEAKAKLEAEAKAKAVAE 205
Query: 107 EKSEAVTEKIDEKAA 121
K++A E + AA
Sbjct: 206 AKAKAEAEAKAKAAA 220
>SMI1_YARLI (Q6CDX0) KNR4/SMI1 homolog
Length = 713
Score = 36.2 bits (82), Expect = 0.096
Identities = 25/68 (36%), Positives = 36/68 (52%), Gaps = 1/68 (1%)
Query: 65 EKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPNQ 124
E+V +A +A +KA + AAE AK AK A+A A KK+ EK+ A K + +
Sbjct: 591 EEVAKAAAKAEEEQKA-TAAAEAAKAEAKRAAEADASKKVEAEKAAAEESKESKAESEES 649
Query: 125 GNGMDLEK 132
DLE+
Sbjct: 650 KVERDLEE 657
Score = 33.9 bits (76), Expect = 0.48
Identities = 21/44 (47%), Positives = 25/44 (56%)
Query: 78 KKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAA 121
+KAK A EK K AAKE A AE K EK+ K ++KAA
Sbjct: 524 EKAKVEATEKTKKAAKEAADKEAELKKAAEKAAEEKAKAEKKAA 567
Score = 31.6 bits (70), Expect = 2.4
Identities = 22/60 (36%), Positives = 33/60 (54%), Gaps = 3/60 (5%)
Query: 62 DSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAA 121
++AEK + A + KKA AA+K + K+ A+ AAE+K EK A + +EK A
Sbjct: 521 EAAEKAKVEATE--KTKKAAKEAADK-EAELKKAAEKAAEEKAKAEKKAAEAREKEEKEA 577
Score = 30.8 bits (68), Expect = 4.0
Identities = 21/57 (36%), Positives = 31/57 (53%), Gaps = 2/57 (3%)
Query: 63 SAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEK 119
+AE A RAA +K V AEKA AA+E ++ AE + + + + KIDE+
Sbjct: 609 AAEAAKAEAKRAAEADASKKVEAEKA--AAEESKESKAESEESKVERDLEELKIDEE 663
Score = 30.0 bits (66), Expect = 6.9
Identities = 34/114 (29%), Positives = 56/114 (48%), Gaps = 12/114 (10%)
Query: 44 LEKVFDFIAKESTNFFDK--DSAEKVVLSAVRAARVKKAKSVAAEKAKIAAK----EKAK 97
L+K + A+E K ++ EK A AA+ K+ + E AK AAK +KA
Sbjct: 548 LKKAAEKAAEEKAKAEKKAAEAREKEEKEAKAAAKAKEEELKKEEVAKAAAKAEEEQKAT 607
Query: 98 AAAEKKLNDEKSEA---VTEKID-EKAAPNQG--NGMDLEKYSWTQTLQELNVN 145
AAAE + K A ++K++ EKAA + + + E+ + L+EL ++
Sbjct: 608 AAAEAAKAEAKRAAEADASKKVEAEKAAAEESKESKAESEESKVERDLEELKID 661
>IF2_VIBVY (Q7MI09) Translation initiation factor IF-2
Length = 907
Score = 36.2 bits (82), Expect = 0.096
Identities = 21/60 (35%), Positives = 33/60 (55%)
Query: 61 KDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKA 120
+++A++ A + A + AK A E AK A+EKAK AE+ E ++V +EKA
Sbjct: 112 EEAAQREAEEAAKRAAEEAAKREAEEAAKREAEEKAKREAEEAAKREAEKSVDRDAEEKA 171
Score = 34.3 bits (77), Expect = 0.37
Identities = 21/69 (30%), Positives = 35/69 (50%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E+ +++A++ A + + AK A EKAK A+E AK AEK ++ + E
Sbjct: 111 AEEAAQREAEEAAKRAAEEAAKREAEEAAKREAEEKAKREAEEAAKREAEKSVDRDAEEK 170
Query: 112 VTEKIDEKA 120
+ KA
Sbjct: 171 AKRDAEGKA 179
Score = 34.3 bits (77), Expect = 0.37
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 3/70 (4%)
Query: 53 KESTNFFDKDSAEKVVLSAVRAARVKKAKSV---AAEKAKIAAKEKAKAAAEKKLNDEKS 109
+E+ +++ EK A AA+ + KSV A EKAK A+ KAK AE+K+ E +
Sbjct: 133 REAEEAAKREAEEKAKREAEEAAKREAEKSVDRDAEEKAKRDAEGKAKRDAEEKVKQEAA 192
Query: 110 EAVTEKIDEK 119
E++ +
Sbjct: 193 RKEAEELKRR 202
Score = 33.1 bits (74), Expect = 0.82
Identities = 20/48 (41%), Positives = 27/48 (55%)
Query: 73 RAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKA 120
R+A +AK A E A+ A+E AK AAE+ E EA + +EKA
Sbjct: 100 RSAIEDEAKREAEEAAQREAEEAAKRAAEEAAKREAEEAAKREAEEKA 147
>IF2_VIBVU (Q8DBW0) Translation initiation factor IF-2
Length = 907
Score = 36.2 bits (82), Expect = 0.096
Identities = 21/60 (35%), Positives = 33/60 (55%)
Query: 61 KDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKA 120
+++A++ A + A + AK A E AK A+EKAK AE+ E ++V +EKA
Sbjct: 112 EEAAQREAEEAAKRAAEEAAKREAEEAAKREAEEKAKREAEEAAKREAEKSVDRDAEEKA 171
Score = 34.3 bits (77), Expect = 0.37
Identities = 21/69 (30%), Positives = 35/69 (50%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A+E+ +++A++ A + + AK A EKAK A+E AK AEK ++ + E
Sbjct: 111 AEEAAQREAEEAAKRAAEEAAKREAEEAAKREAEEKAKREAEEAAKREAEKSVDRDAEEK 170
Query: 112 VTEKIDEKA 120
+ KA
Sbjct: 171 AKRDAEGKA 179
Score = 34.3 bits (77), Expect = 0.37
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 3/70 (4%)
Query: 53 KESTNFFDKDSAEKVVLSAVRAARVKKAKSV---AAEKAKIAAKEKAKAAAEKKLNDEKS 109
+E+ +++ EK A AA+ + KSV A EKAK A+ KAK AE+K+ E +
Sbjct: 133 REAEEAAKREAEEKAKREAEEAAKREAEKSVDRDAEEKAKRDAEGKAKRDAEEKVKQEAA 192
Query: 110 EAVTEKIDEK 119
E++ +
Sbjct: 193 RKEAEELKRR 202
Score = 33.1 bits (74), Expect = 0.82
Identities = 20/48 (41%), Positives = 27/48 (55%)
Query: 73 RAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKA 120
R+A +AK A E A+ A+E AK AAE+ E EA + +EKA
Sbjct: 100 RSAIEDEAKREAEEAAQREAEEAAKRAAEEAAKREAEEAAKREAEEKA 147
>WRK7_ARATH (Q9STX0) Probable WRKY transcription factor 7 (WRKY
DNA-binding protein 7)
Length = 353
Score = 35.8 bits (81), Expect = 0.13
Identities = 33/143 (23%), Positives = 56/143 (39%), Gaps = 4/143 (2%)
Query: 9 DDNQTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVV 68
+ NQTK SSSSS P + P + F PS P + ++ + E
Sbjct: 164 NQNQTKNGSSSSSPPMLANGAPSTINFAPSPPVSATNSFMSSHRCDTDSTHMSSGFEFTN 223
Query: 69 LSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPNQGNGM 128
S + +R K S A+ K + + ++ KK + V I A ++ +
Sbjct: 224 PSQLSGSRGKPPLSSASLKRRCNSSPSSRCHCSKK----RKSRVKRVIRVPAVSSKMADI 279
Query: 129 DLEKYSWTQTLQELNVNVPVPNG 151
+++SW + Q+ P P G
Sbjct: 280 PSDEFSWRKYGQKPIKGSPHPRG 302
>DSRA_RAT (P55266) Double-stranded RNA-specific adenosine deaminase
(EC 3.5.4.-) (DRADA)
Length = 1175
Score = 35.8 bits (81), Expect = 0.13
Identities = 41/137 (29%), Positives = 60/137 (42%), Gaps = 24/137 (17%)
Query: 8 QDDNQTKTSSSSSSQPKPSKPI----PFSST---FDPSNP-TTFLEKV--------FDFI 51
+D Q + S+ + P KP P SS F +P TT LE + F +
Sbjct: 526 KDSGQPEELSNCPMEEDPEKPAESQPPSSSATSLFSGKSPVTTLLECMHKLGNSCEFRLL 585
Query: 52 AKESTNFFDKDSAEKVV----LSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDE 107
+KE K V +V A K AK +AAE+A A +E+A +A+ D+
Sbjct: 586 SKEGPAHDPKFQYCVAVGAQTFPSVSAPSKKVAKQMAAEEAMKALQEEAANSAD----DQ 641
Query: 108 KSEAVTEKIDEKAAPNQ 124
A T+ +DE APN+
Sbjct: 642 SGGANTDSLDESVAPNK 658
>MST2_DROHY (Q08696) Axoneme-associated protein mst101(2)
Length = 1391
Score = 35.4 bits (80), Expect = 0.16
Identities = 23/82 (28%), Positives = 39/82 (47%), Gaps = 5/82 (6%)
Query: 51 IAKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSE 110
+AK+ +K EK AA KK + A ++ + A K+K + +A+K+ E
Sbjct: 514 LAKKEKETAEKKKCEKAAKKRKEAAEKKKCEKAAKKRKEAAEKKKCEKSAKKR-----KE 568
Query: 111 AVTEKIDEKAAPNQGNGMDLEK 132
A +K EKAA + + +K
Sbjct: 569 AAEKKKCEKAAKERKEAAEKKK 590
Score = 32.3 bits (72), Expect = 1.4
Identities = 19/59 (32%), Positives = 29/59 (48%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSE 110
AK+ +K E+ AA KK + +A + K A K+K K A+KK EK++
Sbjct: 691 AKKEKEAAEKKKCEEAAKKEKEAAERKKCEELAKKIKKAAEKKKCKKLAKKKKAGEKNK 749
Score = 32.0 bits (71), Expect = 1.8
Identities = 23/84 (27%), Positives = 37/84 (43%)
Query: 44 LEKVFDFIAKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKK 103
L K IAK++ +K EK+ A A KK +A +K + K+ AK A ++K
Sbjct: 1297 LRKKCAIIAKKAKMAAEKKECEKLAKKAKEAIEWKKCAKLAKKKREAEKKKCAKLAKKEK 1356
Query: 104 LNDEKSEAVTEKIDEKAAPNQGNG 127
EK + + K ++ G
Sbjct: 1357 EAAEKKKRCKDLAKNKKKGHKKKG 1380
Score = 31.2 bits (69), Expect = 3.1
Identities = 20/51 (39%), Positives = 27/51 (52%), Gaps = 5/51 (9%)
Query: 60 DKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSE 110
+K++AEK + A K + AAEK K A K + AEKK EK+E
Sbjct: 1231 EKEAAEKK-----KCAEAAKKEQEAAEKKKCAEAAKKEKEAEKKRKCEKAE 1276
Score = 30.8 bits (68), Expect = 4.0
Identities = 24/82 (29%), Positives = 35/82 (42%), Gaps = 1/82 (1%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKA-KAAAEKKLNDEKSE 110
AK+ +K EK AA KK + A ++ + A K+K KAA E+K EK +
Sbjct: 531 AKKRKEAAEKKKCEKAAKKRKEAAEKKKCEKSAKKRKEAAEKKKCEKAAKERKEAAEKKK 590
Query: 111 AVTEKIDEKAAPNQGNGMDLEK 132
EK + +L K
Sbjct: 591 CEEAAKKEKEVAERKKCEELAK 612
Score = 30.0 bits (66), Expect = 6.9
Identities = 19/66 (28%), Positives = 34/66 (50%), Gaps = 7/66 (10%)
Query: 61 KDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKA-------AAEKKLNDEKSEAVT 113
K +K+ + RAA KK A ++ + A K+K + AAEKK +E+++ +
Sbjct: 965 KKKCKKLGKKSKRAAEKKKCAEAAKKEKEAATKKKCEERAKKQKEAAEKKQCEERAKKLK 1024
Query: 114 EKIDEK 119
E ++K
Sbjct: 1025 EAAEQK 1030
Score = 29.6 bits (65), Expect = 9.0
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 1/83 (1%)
Query: 51 IAKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKA-KAAAEKKLNDEKS 109
+AK+ +K + +++ A KK + A ++ K A K+K KAA E+K EK
Sbjct: 354 LAKKQKEEDEKKACKELAKKKKEADEKKKCEEAANKEKKAAEKKKCEKAAKERKEAAEKK 413
Query: 110 EAVTEKIDEKAAPNQGNGMDLEK 132
+ EK A + +L K
Sbjct: 414 KCEEAAKKEKEAAERKKCEELAK 436
Score = 29.6 bits (65), Expect = 9.0
Identities = 26/92 (28%), Positives = 44/92 (47%), Gaps = 11/92 (11%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVA------AEKAKI--AAKEKAKAAAEKK 103
AK+ ++ E++ +A KK K +A AEK K AAK++ +AA +KK
Sbjct: 483 AKKGKEVAERKKCEELAKKIKKAEIKKKCKKLAKKEKETAEKKKCEKAAKKRKEAAEKKK 542
Query: 104 LN---DEKSEAVTEKIDEKAAPNQGNGMDLEK 132
++ EA +K EK+A + + +K
Sbjct: 543 CEKAAKKRKEAAEKKKCEKSAKKRKEAAEKKK 574
>H12_VOLCA (Q08865) Histone H1-II
Length = 240
Score = 35.4 bits (80), Expect = 0.16
Identities = 21/55 (38%), Positives = 29/55 (52%)
Query: 61 KDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEK 115
K + K + +AA KKAK+ +KAK A +KAKAAA+ K + A K
Sbjct: 179 KKATPKKAAAPKKAAAPKKAKAATPKKAKAATPKKAKAAAKPKAAAKPKAAAKPK 233
>GIT7_SCHPO (O59709) Glucose insensitive transcription protein 7
Length = 379
Score = 35.4 bits (80), Expect = 0.16
Identities = 35/156 (22%), Positives = 64/156 (40%), Gaps = 9/156 (5%)
Query: 132 KYSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGLKGQP----PIIDRELYKSIKP 187
+Y W+QT LN+++ K V ++KN LK+ +K + ++ LY+ I P
Sbjct: 185 RYDWSQTSFSLNIDI-YAKKVKDEDVSLLMEKNTLKIEIKLEDGSIFSLVLDPLYEEIVP 243
Query: 188 DECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLGDLDSETRMTVE 247
++ + + L+ K ++ W+ LVK P N+ V S T+ +
Sbjct: 244 EKSSFKLFSSKVEITLIKKVSEIK-WEALVK-SPANNSVNVYAKDSNHSSASGNTKNKAK 301
Query: 248 KMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMD 283
+D K L E + + F + + N D
Sbjct: 302 D--WDSLAKLADLEEDEPTGEAALANLFQNLYKNAD 335
>DSRA_MOUSE (Q99MU3) Double-stranded RNA-specific adenosine
deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase
1)
Length = 1178
Score = 35.4 bits (80), Expect = 0.16
Identities = 21/53 (39%), Positives = 32/53 (59%), Gaps = 4/53 (7%)
Query: 72 VRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPNQ 124
V A K AK +AAE+A A +E+A ++A+ D+ A T+ +DE APN+
Sbjct: 613 VSAPSKKVAKQMAAEEAMKALQEEAASSAD----DQSGGANTDSLDESMAPNK 661
>TOLA_PSEAE (P50600) TolA protein
Length = 347
Score = 35.0 bits (79), Expect = 0.21
Identities = 29/102 (28%), Positives = 46/102 (44%), Gaps = 5/102 (4%)
Query: 49 DFIAKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEK----AKAAAEKKL 104
D K++ K +AE A A+ K A A +KA + A +K A AAA K
Sbjct: 161 DEAKKKAAEDAKKKAAEDAKKKAAEEAKKKAAAEAAKKKAAVEAAKKKAAAAAAAARKAA 220
Query: 105 NDEKSEAVTEKIDEKAAPNQGNGMDLEKYSWTQTLQELNVNV 146
D+K+ A+ E + + Q ++ T +L +L VN+
Sbjct: 221 EDKKARALAELLSDTTERQQALADEVGS-EVTGSLDDLIVNL 261
Score = 34.3 bits (77), Expect = 0.37
Identities = 20/60 (33%), Positives = 34/60 (56%), Gaps = 2/60 (3%)
Query: 62 DSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAA 121
D A+K + +AA KK +A ++A+ AK+KA A+KK ++ + E+ +KAA
Sbjct: 135 DEAKKA--AEAKAAEQKKQADIAKKRAEDEAKKKAAEDAKKKAAEDAKKKAAEEAKKKAA 192
>IF2_PASMU (P57873) Translation initiation factor IF-2
Length = 833
Score = 35.0 bits (79), Expect = 0.21
Identities = 25/71 (35%), Positives = 41/71 (57%), Gaps = 4/71 (5%)
Query: 62 DSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAA 121
D+A+K + ++A + +A +AAEKAK A+EKA+ AEK + EA +K +A
Sbjct: 50 DAAQKAEEARLKAKQ--EADRLAAEKAKKDAEEKARLEAEKAKQAKAEEA--KKAQAVSA 105
Query: 122 PNQGNGMDLEK 132
P + ++ EK
Sbjct: 106 PTKAVDVEKEK 116
>H12_ARATH (P26569) Histone H1.2
Length = 273
Score = 35.0 bits (79), Expect = 0.21
Identities = 36/135 (26%), Positives = 57/135 (41%), Gaps = 3/135 (2%)
Query: 16 SSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLSAVRAA 75
S+ S++ PKP+ P+ +T V AK + K +A KVV A A
Sbjct: 130 SARSAATPKPAAPVKKKATVVAKPKGKVAAAVAPAKAKAAAKGTKKPAA-KVVAKAKVTA 188
Query: 76 RVKKAKSVAAEKAK-IAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPNQGNGMDLEKYS 134
+ K + A K+K +AA K KA A K E+ A + + +P + +K +
Sbjct: 189 KPKAKVTAAKPKSKSVAAVSKTKAVAAKPKAKER-PAKASRTSTRTSPGKKVAAPAKKVA 247
Query: 135 WTQTLQELNVNVPVP 149
T+ +V V P
Sbjct: 248 VTKKAPAKSVKVKSP 262
>FAU_DROME (Q9VGX3) Anoxia up-regulated protein
Length = 619
Score = 35.0 bits (79), Expect = 0.21
Identities = 24/63 (38%), Positives = 35/63 (55%), Gaps = 3/63 (4%)
Query: 61 KDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEK--AKAAAEKKLNDEKSEAVTEKIDE 118
K+ +++ A + A K AK A E AKIAA+E A+AAA+K + K+ E +
Sbjct: 350 KEERDRLTAEAEKQAAAK-AKKAAEEAAKIAAEEALLAEAAAQKAAEEAKALKAAEDAAQ 408
Query: 119 KAA 121
KAA
Sbjct: 409 KAA 411
Score = 32.7 bits (73), Expect = 1.1
Identities = 28/82 (34%), Positives = 42/82 (51%), Gaps = 6/82 (7%)
Query: 42 TFLEKVFDFIAKESTNFFDKDSAEKVVLSAVRAARVK---KAKSVAAEKAKIAAKEKAKA 98
T E+ D + E D A++ A++ R + +A+ AA KAK AA+E AK
Sbjct: 319 TVNEEALDEVDLEKKRAQKADEAKRREERALKEERDRLTAEAEKQAAAKAKKAAEEAAKI 378
Query: 99 AAEKKLNDEKSEAVTEKIDEKA 120
AAE+ L +EA +K E+A
Sbjct: 379 AAEEAL---LAEAAAQKAAEEA 397
Score = 30.8 bits (68), Expect = 4.0
Identities = 21/70 (30%), Positives = 35/70 (50%), Gaps = 1/70 (1%)
Query: 52 AKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEA 111
A E ++ +A+K A + A ++A AAE+A++A + KAA E L +
Sbjct: 433 AAEEARLAEEAAAQKAAEEAAQKA-AEEAALKAAEEARLAEEAAQKAAEEAALKAVEEAR 491
Query: 112 VTEKIDEKAA 121
E+ +KAA
Sbjct: 492 AAEEAAQKAA 501
Score = 30.8 bits (68), Expect = 4.0
Identities = 24/77 (31%), Positives = 38/77 (49%), Gaps = 8/77 (10%)
Query: 52 AKESTNFFDKDSAEKVVLSAVR--AARVKKAKSVAAEKAKIAAKEKA------KAAAEKK 103
A E ++ +A+KV A + A + A+ AA+KA A +KA KAA E +
Sbjct: 410 AAEEARLAEEAAAQKVAEEAAQKAAEEARLAEEAAAQKAAEEAAQKAAEEAALKAAEEAR 469
Query: 104 LNDEKSEAVTEKIDEKA 120
L +E ++ E+ KA
Sbjct: 470 LAEEAAQKAAEEAALKA 486
>SCE3_SCHPO (O14369) Probable RNA-binding protein sce3
Length = 388
Score = 34.7 bits (78), Expect = 0.28
Identities = 30/128 (23%), Positives = 58/128 (44%), Gaps = 2/128 (1%)
Query: 11 NQTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLS 70
N T S++++ K PF P + T+ L++V + +AK + +F +D+A + +
Sbjct: 255 NTEATPSATTTTSSKPKRDPFGGA-KPVDNTSVLQRVEEKLAKRTQSFRREDNANRERST 313
Query: 71 AVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSE-AVTEKIDEKAAPNQGNGMD 129
+ + + K K+ + + EKK ++ SE A T+ + AP G
Sbjct: 314 SRKPSADKAEKTDKTDAIAEKVSDIRLGDGEKKSSETDSEVAATKTPATEDAPATNAGEA 373
Query: 130 LEKYSWTQ 137
E+ WT+
Sbjct: 374 EEEEGWTK 381
>FUTS_DROME (Q9W596) Microtubule-associated protein futsch
Length = 5412
Score = 34.7 bits (78), Expect = 0.28
Identities = 35/129 (27%), Positives = 55/129 (42%), Gaps = 33/129 (25%)
Query: 12 QTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLSA 71
+++ S + P PSK + S PT+ E V D K + AEK L++
Sbjct: 3708 ESRRESVAEKSPLPSK--------EASRPTSVAESVKDEAEKSKEESRRESVAEKSSLAS 3759
Query: 72 VRAAR-VKKAKSV---------------AAEKAKIAAKEKAKAA---------AEKKLND 106
+A+R A+SV AEK+ +A+KE ++ A AEK +
Sbjct: 3760 KKASRPASVAESVKDEAEKSKEESRRESVAEKSPLASKEASRPASVAESVKDEAEKSKEE 3819
Query: 107 EKSEAVTEK 115
+ E+V EK
Sbjct: 3820 SRRESVAEK 3828
Score = 33.5 bits (75), Expect = 0.62
Identities = 30/104 (28%), Positives = 44/104 (41%), Gaps = 20/104 (19%)
Query: 12 QTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLSA 71
+++ S + P PSK + S PT+ E V D K + AEK L++
Sbjct: 3819 ESRRESVAEKSPLPSK--------EASRPTSVAESVKDEADKSKEESRRESGAEKSPLAS 3870
Query: 72 VRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEK 115
+ A+R SVA E K EK + + E+VTEK
Sbjct: 3871 MEASR---PTSVA---------ESVKDETEKSKEESRRESVTEK 3902
Score = 33.1 bits (74), Expect = 0.82
Identities = 35/129 (27%), Positives = 53/129 (40%), Gaps = 33/129 (25%)
Query: 12 QTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLSA 71
+++ S + P PSK + S PT+ E V D K AEK L++
Sbjct: 3449 ESRRESVAEKSPLPSK--------EASRPTSVAESVKDEAEKSKEESRRDSVAEKSPLAS 3500
Query: 72 VRAAR-VKKAKSV---------------AAEKAKIAAKEKAKAA---------AEKKLND 106
A+R A+SV AEK+ +A+KE ++ A AEK +
Sbjct: 3501 KEASRPASVAESVQDEAEKSKEESRRESVAEKSPLASKEASRPASVAESIKDEAEKSKEE 3560
Query: 107 EKSEAVTEK 115
+ E+V EK
Sbjct: 3561 SRRESVAEK 3569
Score = 32.3 bits (72), Expect = 1.4
Identities = 34/130 (26%), Positives = 55/130 (42%), Gaps = 26/130 (20%)
Query: 11 NQTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLS 70
++ + S S + ++ P +S + S PT+ E V D K AEK L+
Sbjct: 3552 DEAEKSKEESRRESVAEKSPLASK-EASRPTSVAESVKDEAEKSKEESSRDSVAEKSPLA 3610
Query: 71 AVRAAR-VKKAKSV---------------AAEKAKIAAKEKAKAA---------AEKKLN 105
+ A+R A+SV AEK+ +A+KE ++ A AEK
Sbjct: 3611 SKEASRPASVAESVQDEAEKSKEESRRESVAEKSPLASKEASRPASVAESVKDDAEKSKE 3670
Query: 106 DEKSEAVTEK 115
+ + E+V EK
Sbjct: 3671 ESRRESVAEK 3680
Score = 31.6 bits (70), Expect = 2.4
Identities = 28/144 (19%), Positives = 61/144 (41%), Gaps = 22/144 (15%)
Query: 4 ISDYQDDNQTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDS 63
+ D D ++ ++ S ++ P ++ + S PT+ E V D K +
Sbjct: 3846 VKDEADKSKEESRRESGAEKSPL------ASMEASRPTSVAESVKDETEKSKEESRRESV 3899
Query: 64 AEKVVLSAVRAAR-----------VKKAKSVA-----AEKAKIAAKEKAKAAAEKKLNDE 107
EK L + A+R +K+K + AEK+ +A+KE ++ A+ + +
Sbjct: 3900 TEKSPLPSKEASRPTSVAESVKDEAEKSKEESRRESVAEKSPLASKESSRPASVAESIKD 3959
Query: 108 KSEAVTEKIDEKAAPNQGNGMDLE 131
++E ++ ++ P G ++
Sbjct: 3960 EAEGTKQESRRESMPESGKAESIK 3983
Score = 30.4 bits (67), Expect = 5.3
Identities = 29/130 (22%), Positives = 54/130 (41%), Gaps = 26/130 (20%)
Query: 11 NQTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLS 70
++ + S S + ++ P +S + S P + E + D K + AEK L+
Sbjct: 3515 DEAEKSKEESRRESVAEKSPLASK-EASRPASVAESIKDEAEKSKEESRRESVAEKSPLA 3573
Query: 71 AVRAAR----------------VKKAKSVAAEKAKIAAKEKAKAA---------AEKKLN 105
+ A+R + ++ AEK+ +A+KE ++ A AEK
Sbjct: 3574 SKEASRPTSVAESVKDEAEKSKEESSRDSVAEKSPLASKEASRPASVAESVQDEAEKSKE 3633
Query: 106 DEKSEAVTEK 115
+ + E+V EK
Sbjct: 3634 ESRRESVAEK 3643
Score = 30.0 bits (66), Expect = 6.9
Identities = 28/114 (24%), Positives = 48/114 (41%), Gaps = 13/114 (11%)
Query: 12 QTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLSA 71
+++ S + P PSK + S P + E + D K + AEK L +
Sbjct: 2191 ESRRESVAEKSPLPSK--------EASRPASVAESIKDEAEKSKEETRRESVAEKSPLPS 2242
Query: 72 VRAAR-VKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKS----EAVTEKIDEKA 120
A+R A+S+ E K + + ++AAEK K +V E + ++A
Sbjct: 2243 KEASRPASVAESIKDEAEKSKEESRRESAAEKSPLPSKEASRPASVAESVKDEA 2296
Score = 30.0 bits (66), Expect = 6.9
Identities = 28/114 (24%), Positives = 48/114 (41%), Gaps = 13/114 (11%)
Query: 12 QTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLSA 71
+++ S + P PSK + S P + E V D K + AEK L++
Sbjct: 3375 ESRRESVAEKSPLPSK--------EASRPASVAESVKDEADKSKEESRRESGAEKSPLAS 3426
Query: 72 VRAAR-VKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKS----EAVTEKIDEKA 120
A+R A+S+ E K + + ++ AEK K +V E + ++A
Sbjct: 3427 KEASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKEASRPTSVAESVKDEA 3480
>CN1C_HUMAN (Q14123) Calcium/calmodulin-dependent 3',5'-cyclic
nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE
1C) (hCam-3)
Length = 709
Score = 34.7 bits (78), Expect = 0.28
Identities = 20/60 (33%), Positives = 29/60 (48%)
Query: 73 RAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPNQGNGMDLEK 132
+ + +KAK A EKA++AA+E+ K K +E + EK NQ NG K
Sbjct: 524 KVPKEEKAKKEAEEKARLAAEEQQKEMEAKSQAEEGASGKAEKKTSGETKNQVNGTRANK 583
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.309 0.126 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,243,739
Number of Sequences: 164201
Number of extensions: 1390563
Number of successful extensions: 6266
Number of sequences better than 10.0: 166
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 115
Number of HSP's that attempted gapping in prelim test: 5745
Number of HSP's gapped (non-prelim): 466
length of query: 289
length of database: 59,974,054
effective HSP length: 109
effective length of query: 180
effective length of database: 42,076,145
effective search space: 7573706100
effective search space used: 7573706100
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC134822.6


