

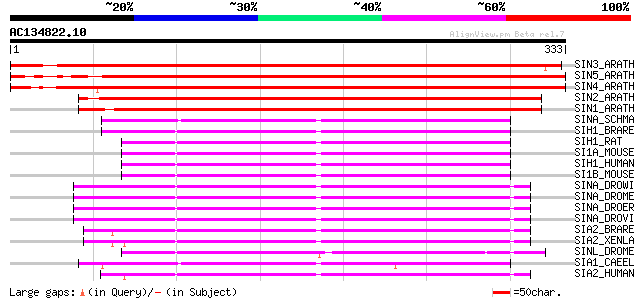
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.10 - phase: 0
(333 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SIN3_ARATH (Q84JL3) Ubiquitin ligase SINAT3 (EC 6.3.2.-) (Seven ... 569 e-162
SIN5_ARATH (Q8S3N1) Ubiquitin ligase SINAT5 (EC 6.3.2.-) (Seven ... 562 e-160
SIN4_ARATH (Q9STN8) Ubiquitin ligase SINAT4 (EC 6.3.2.-) (Seven ... 554 e-157
SIN2_ARATH (Q9M2P4) Ubiquitin ligase SINAT2 (EC 6.3.2.-) (Seven ... 469 e-132
SIN1_ARATH (P93748) Putative ubiquitin ligase SINAT1 (EC 6.3.2.-... 457 e-128
SINA_SCHMA (Q86MW9) Ubiquitin ligase sina (EC 6.3.2.-) (Seven in... 164 2e-40
SIH1_BRARE (Q7ZVG6) Ubiquitin ligase SIAH1 (EC 6.3.2.-) (Seven i... 160 5e-39
SIH1_RAT (Q920M9) Ubiquitin ligase SIAH1 (EC 6.3.2.-) (Seven in ... 159 9e-39
SI1A_MOUSE (P61092) Ubiquitin ligase SIAH1A (EC 6.3.2.-) (Seven ... 159 9e-39
SIH1_HUMAN (Q8IUQ4) Ubiquitin ligase SIAH1 (EC 6.3.2.-) (Seven i... 158 2e-38
SI1B_MOUSE (Q06985) Ubiquitin ligase SIAH1B (EC 6.3.2.-) (Seven ... 157 3e-38
SINA_DROWI (Q8I147) Ubiquitin ligase sina (EC 6.3.2.-) (Seven in... 153 5e-37
SINA_DROME (P21461) Ubiquitin ligase sina (EC 6.3.2.-) (Seven in... 153 5e-37
SINA_DROER (P61093) Ubiquitin ligase sina (EC 6.3.2.-) (Seven in... 153 5e-37
SINA_DROVI (P29304) Ubiquitin ligase sina (EC 6.3.2.-) (Seven in... 152 1e-36
SIA2_BRARE (Q7SYL3) Ubiquitin ligase SIAH2 (EC 6.3.2.-) (Seven i... 150 4e-36
SIA2_XENLA (Q9I8X5) Ubiquitin ligase SIAH2 (EC 6.3.2.-) (Seven i... 148 2e-35
SINL_DROME (Q8T3Y0) Probable ubiquitin ligase sina-like protein ... 148 2e-35
SIA1_CAEEL (Q965X6) Ubiquitin ligase sia-1 (EC 6.3.2.-) (Seven i... 147 3e-35
SIA2_HUMAN (O43255) Ubiquitin ligase SIAH2 (EC 6.3.2.-) (Seven i... 146 8e-35
>SIN3_ARATH (Q84JL3) Ubiquitin ligase SINAT3 (EC 6.3.2.-) (Seven in
absentia homolog 3)
Length = 326
Score = 569 bits (1467), Expect = e-162
Identities = 268/333 (80%), Positives = 282/333 (84%), Gaps = 10/333 (3%)
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIA 60
MDLDS++C S+ D D++EI HQ H ++ S + N S +
Sbjct: 1 MDLDSMDCTSTMDVTDDEEI--------HQDRHSYASVSKHHHTNNNTTNVNAAASGLLP 52
Query: 61 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK 120
TSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCK RVHNRCPTCRQELGDIRCLALEK
Sbjct: 53 TTTSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKARVHNRCPTCRQELGDIRCLALEK 112
Query: 121 VAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLR 180
VAESLELPCK+ SLGCPEIFPYYSKLKHET+CNFRPY+CPYAGSECS GDI FLVAHLR
Sbjct: 113 VAESLELPCKHMSLGCPEIFPYYSKLKHETVCNFRPYSCPYAGSECSVTGDIPFLVAHLR 172
Query: 181 DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF 240
DDHKVDMH+GCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF
Sbjct: 173 DDHKVDMHSGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF 232
Query: 241 LRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS 300
LRFMGDE EARNY YSLEVG GRKLIWEGTPRS+RDSHRKVRDSHDGLIIQRNMALFFS
Sbjct: 233 LRFMGDETEARNYNYSLEVGGYGRKLIWEGTPRSVRDSHRKVRDSHDGLIIQRNMALFFS 292
Query: 301 GGDRKELKLRVTGRIWKEQQ--NPDGGVCIPNL 331
GGDRKELKLRVTGRIWKEQQ GG CIPNL
Sbjct: 293 GGDRKELKLRVTGRIWKEQQQSGEGGGACIPNL 325
>SIN5_ARATH (Q8S3N1) Ubiquitin ligase SINAT5 (EC 6.3.2.-) (Seven in
absentia homolog 5)
Length = 309
Score = 562 bits (1449), Expect = e-160
Identities = 268/333 (80%), Positives = 290/333 (86%), Gaps = 25/333 (7%)
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIA 60
M+ DSI+ V +D+DEI HQ+H +FSS K + G + I+
Sbjct: 1 METDSIDSV-----IDDDEI--------HQKH----QFSSTKSQGGA--------TVVIS 35
Query: 61 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK 120
PATSV+ELLECPVCTNSMYPPIHQCHNGHTLCSTCK+RVHNRCPTCRQELGDIRCLALEK
Sbjct: 36 PATSVYELLECPVCTNSMYPPIHQCHNGHTLCSTCKSRVHNRCPTCRQELGDIRCLALEK 95
Query: 121 VAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLR 180
VAESLELPCKYY+LGC IFPYYSKLKHE+ CNFRPY+CPYAGSEC+AVGDI FLVAHLR
Sbjct: 96 VAESLELPCKYYNLGCLGIFPYYSKLKHESQCNFRPYSCPYAGSECAAVGDITFLVAHLR 155
Query: 181 DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF 240
DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVF CFGQYFCLHFEAFQLGMAPVYMAF
Sbjct: 156 DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFQCFGQYFCLHFEAFQLGMAPVYMAF 215
Query: 241 LRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS 300
LRFMGDE++ARNYTYSLEVG +GRK WEGTPRS+RDSHRKVRDSHDGLIIQRNMALFFS
Sbjct: 216 LRFMGDEDDARNYTYSLEVGGSGRKQTWEGTPRSVRDSHRKVRDSHDGLIIQRNMALFFS 275
Query: 301 GGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
GGD+KELKLRVTGRIWKEQQNPD GVCI ++CS
Sbjct: 276 GGDKKELKLRVTGRIWKEQQNPDSGVCITSMCS 308
>SIN4_ARATH (Q9STN8) Ubiquitin ligase SINAT4 (EC 6.3.2.-) (Seven in
absentia homolog 4)
Length = 327
Score = 554 bits (1428), Expect = e-157
Identities = 266/337 (78%), Positives = 288/337 (84%), Gaps = 15/337 (4%)
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHS-EFSSLKPRSGGNNNH---GVIGS 56
M+ DS+ECVSS+ +EI H + H S +FSS K G ++G
Sbjct: 1 METDSMECVSSTG----NEI-------HQNGNGHQSYQFSSTKTHGGAAAAAVVTNIVGP 49
Query: 57 TAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCL 116
TA APATSV+ELLECPVCT SMYPPIHQCHNGHTLCSTCK RVHNRCPTCRQELGDIRCL
Sbjct: 50 TATAPATSVYELLECPVCTYSMYPPIHQCHNGHTLCSTCKVRVHNRCPTCRQELGDIRCL 109
Query: 117 ALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLV 176
ALEKVAESLELPCK+Y+LGCPEIFPYYSKLKHE++CNFRPY+CPYAGSEC VGDI FLV
Sbjct: 110 ALEKVAESLELPCKFYNLGCPEIFPYYSKLKHESLCNFRPYSCPYAGSECGIVGDIPFLV 169
Query: 177 AHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPV 236
AHLRDDHKVDMH G TFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGM PV
Sbjct: 170 AHLRDDHKVDMHAGSTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMGPV 229
Query: 237 YMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMA 296
YMAFLRFMGDE +AR+Y+YSLEVG +GRKL WEGTPRSIRDSHRKVRDS+DGLIIQRNMA
Sbjct: 230 YMAFLRFMGDEEDARSYSYSLEVGGSGRKLTWEGTPRSIRDSHRKVRDSNDGLIIQRNMA 289
Query: 297 LFFSGGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
LFFSGGDRKELKLRVTG+IWKEQ +PD G+ IPNL S
Sbjct: 290 LFFSGGDRKELKLRVTGKIWKEQHSPDSGLSIPNLSS 326
>SIN2_ARATH (Q9M2P4) Ubiquitin ligase SINAT2 (EC 6.3.2.-) (Seven in
absentia homolog 2)
Length = 308
Score = 469 bits (1207), Expect = e-132
Identities = 219/278 (78%), Positives = 237/278 (84%), Gaps = 6/278 (2%)
Query: 42 KPRSGGNNNHGVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHN 101
KP S G IG I V+ELLECPVCTN MYPPIHQC NGHTLCS CK RV N
Sbjct: 37 KPGSAG------IGKYGIHSNNGVYELLECPVCTNLMYPPIHQCPNGHTLCSNCKLRVQN 90
Query: 102 RCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPY 161
CPTCR ELG+IRCLALEKVAESLE+PC+Y +LGC +IFPYYSKLKHE C FRPYTCPY
Sbjct: 91 TCPTCRYELGNIRCLALEKVAESLEVPCRYQNLGCHDIFPYYSKLKHEQHCRFRPYTCPY 150
Query: 162 AGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQY 221
AGSECS GDI LV HL+DDHKVDMH GCTFNHRYVKSNP EVENATWMLTVF+CFG+
Sbjct: 151 AGSECSVTGDIPTLVVHLKDDHKVDMHDGCTFNHRYVKSNPHEVENATWMLTVFNCFGRQ 210
Query: 222 FCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRK 281
FCLHFEAFQLGMAPVYMAFLRFMGDENEA+ ++YSLEVGA+GRKL W+G PRSIRDSHRK
Sbjct: 211 FCLHFEAFQLGMAPVYMAFLRFMGDENEAKKFSYSLEVGAHGRKLTWQGIPRSIRDSHRK 270
Query: 282 VRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 319
VRDS DGLII RN+AL+FSGGDR+ELKLRVTGRIWKE+
Sbjct: 271 VRDSQDGLIIPRNLALYFSGGDRQELKLRVTGRIWKEE 308
>SIN1_ARATH (P93748) Putative ubiquitin ligase SINAT1 (EC 6.3.2.-)
(Seven in absentia homolog 1)
Length = 305
Score = 457 bits (1177), Expect = e-128
Identities = 212/278 (76%), Positives = 236/278 (84%), Gaps = 5/278 (1%)
Query: 42 KPRSGGNNNHGVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHN 101
KP G+ + G S+ V+ELLECPVCTN MYPPIHQC NGHTLCS+CK RV N
Sbjct: 33 KPTKSGSGSIGKFHSS-----NGVYELLECPVCTNLMYPPIHQCPNGHTLCSSCKLRVQN 87
Query: 102 RCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPY 161
CPTCR ELG+IRCLALEKVAESLE+PC+Y +LGC +IFPYYSKLKHE C FR Y+CPY
Sbjct: 88 TCPTCRYELGNIRCLALEKVAESLEVPCRYQNLGCQDIFPYYSKLKHEQHCRFRSYSCPY 147
Query: 162 AGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQY 221
AGSECS GDI LV HL+DDHKVDMH GCTFNHRYVKSNP EVENATWMLTVF+CFG+
Sbjct: 148 AGSECSVTGDIPTLVDHLKDDHKVDMHDGCTFNHRYVKSNPHEVENATWMLTVFNCFGRQ 207
Query: 222 FCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRK 281
FCLHFEAFQLGMAPVYMAFLRFMGDENEA+ ++YSLEVGA+ RKL W+G PRSIRDSHRK
Sbjct: 208 FCLHFEAFQLGMAPVYMAFLRFMGDENEAKKFSYSLEVGAHSRKLTWQGIPRSIRDSHRK 267
Query: 282 VRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 319
VRDS DGLII RN+AL+FSG D++ELKLRVTGRIWKE+
Sbjct: 268 VRDSQDGLIIPRNLALYFSGSDKEELKLRVTGRIWKEE 305
>SINA_SCHMA (Q86MW9) Ubiquitin ligase sina (EC 6.3.2.-) (Seven in
absentia homolog) (SmSINA)
Length = 371
Score = 164 bits (416), Expect = 2e-40
Identities = 85/245 (34%), Positives = 131/245 (52%), Gaps = 3/245 (1%)
Query: 56 STAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRC 115
+T+ + + + L ECPVC + PPI QC +GH +C++C++++ + CPTCR L +IR
Sbjct: 111 NTSDSSSIDLASLFECPVCMDYALPPIMQCQSGHIVCASCRSKLSS-CPTCRGNLDNIRN 169
Query: 116 LALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFL 175
LA+EK+A S+ PCKY + GCPE F Y SK +HE C +RPY CP G+ C +G++ +
Sbjct: 170 LAMEKLASSVLFPCKYSTSGCPETFHYTSKSEHEAACEYRPYDCPCPGASCKWLGELEQV 229
Query: 176 VAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAP 235
+ HL HK T + + A + + CFG F L E +
Sbjct: 230 MPHLVHHHK--SITTLQGEDIVFLATDISLPGAVDWVMMQSCFGHSFMLVLEKQERVPDQ 287
Query: 236 VYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNM 295
++ A ++ +G +A + Y LE+ + R+L WE PRSI D + D L+ N
Sbjct: 288 IFFALVQLIGTRKQADQFVYRLELNGHRRRLTWEACPRSIHDGVQSAIAVSDCLVFDSNT 347
Query: 296 ALFFS 300
A F+
Sbjct: 348 AHSFA 352
>SIH1_BRARE (Q7ZVG6) Ubiquitin ligase SIAH1 (EC 6.3.2.-) (Seven in
absentia homolog 1) (Siah-1)
Length = 282
Score = 160 bits (404), Expect = 5e-39
Identities = 87/246 (35%), Positives = 128/246 (51%), Gaps = 4/246 (1%)
Query: 56 STAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRC 115
S A + + L ECPVC + + PPI QC +GH +CS C+ ++ CPTCR LG IR
Sbjct: 26 SGTTASNSDLASLFECPVCFDYVLPPILQCQSGHLVCSNCRPKL-TCCPTCRGPLGSIRN 84
Query: 116 LALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFL 175
LA+EKVA S+ PCKY S GC P+ K +HE +C FRPY+CP G+ C G ++ +
Sbjct: 85 LAMEKVANSVLFPCKYASSGCEVTLPHTDKAEHEELCEFRPYSCPCPGASCKWQGSLDAV 144
Query: 176 VAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQ-LGMA 234
+ HL HK T + + A + + CFG +F L E +
Sbjct: 145 MPHLLHQHK--SITTLQGEDIVFLATDINLPGAVDWVMMQSCFGFHFMLVLEKQEKYDGH 202
Query: 235 PVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRN 294
+ A ++ +G +A N+ Y LE+ + R+L WE TPRSI + + D L+ +
Sbjct: 203 QQFFAIVQLIGTRKQAENFAYRLELNGHRRRLTWEATPRSIHEGIATAIMNSDCLVFDTS 262
Query: 295 MALFFS 300
+A F+
Sbjct: 263 IAQLFA 268
>SIH1_RAT (Q920M9) Ubiquitin ligase SIAH1 (EC 6.3.2.-) (Seven in
absentia homolog 1) (Siah-1) (Siah-1a)
Length = 282
Score = 159 bits (402), Expect = 9e-39
Identities = 85/234 (36%), Positives = 124/234 (52%), Gaps = 4/234 (1%)
Query: 68 LLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLEL 127
L ECPVC + + PPI QC +GH +CS C+ ++ CPTCR LG IR LA+EKVA S+
Sbjct: 38 LFECPVCFDYVLPPILQCQSGHLVCSNCRPKL-TCCPTCRGPLGSIRNLAMEKVANSVLF 96
Query: 128 PCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVDM 187
PCKY S GC P+ K +HE +C FRPY+CP G+ C G ++ ++ HL HK
Sbjct: 97 PCKYASSGCEITLPHTEKAEHEELCEFRPYSCPCPGASCKWQGSLDAVMPHLMHQHK--S 154
Query: 188 HTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQ-LGMAPVYMAFLRFMGD 246
T + + A + + CFG +F L E + + A ++ +G
Sbjct: 155 ITTLQGEDIVFLATDINLPGAVDWVMMQSCFGFHFMLVLEKQEKYDGHQQFFAIVQLIGT 214
Query: 247 ENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS 300
+A N+ Y LE+ + R+L WE TPRSI + + D L+ ++A F+
Sbjct: 215 RKQAENFAYRLELNGHRRRLTWEATPRSIHEGIATAIMNSDCLVFDTSIAQLFA 268
>SI1A_MOUSE (P61092) Ubiquitin ligase SIAH1A (EC 6.3.2.-) (Seven in
absentia homolog 1a) (Siah1a) (Siah-1a) (mSiah-1a)
Length = 282
Score = 159 bits (402), Expect = 9e-39
Identities = 85/234 (36%), Positives = 124/234 (52%), Gaps = 4/234 (1%)
Query: 68 LLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLEL 127
L ECPVC + + PPI QC +GH +CS C+ ++ CPTCR LG IR LA+EKVA S+
Sbjct: 38 LFECPVCFDYVLPPILQCQSGHLVCSNCRPKL-TCCPTCRGPLGSIRNLAMEKVANSVLF 96
Query: 128 PCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVDM 187
PCKY S GC P+ K +HE +C FRPY+CP G+ C G ++ ++ HL HK
Sbjct: 97 PCKYASSGCEITLPHTEKAEHEELCEFRPYSCPCPGASCKWQGSLDAVMPHLMHQHK--S 154
Query: 188 HTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQ-LGMAPVYMAFLRFMGD 246
T + + A + + CFG +F L E + + A ++ +G
Sbjct: 155 ITTLQGEDIVFLATDINLPGAVDWVMMQSCFGFHFMLVLEKQEKYDGHQQFFAIVQLIGT 214
Query: 247 ENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS 300
+A N+ Y LE+ + R+L WE TPRSI + + D L+ ++A F+
Sbjct: 215 RKQAENFAYRLELNGHRRRLTWEATPRSIHEGIATAIMNSDCLVFDTSIAQLFA 268
>SIH1_HUMAN (Q8IUQ4) Ubiquitin ligase SIAH1 (EC 6.3.2.-) (Seven in
absentia homolog 1) (Siah-1) (Siah-1a)
Length = 282
Score = 158 bits (400), Expect = 2e-38
Identities = 85/234 (36%), Positives = 123/234 (52%), Gaps = 4/234 (1%)
Query: 68 LLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLEL 127
L ECPVC + + PPI QC +GH +CS C+ ++ CPTCR LG IR LA+EKVA S+
Sbjct: 38 LFECPVCFDYVLPPILQCQSGHLVCSNCRPKL-TCCPTCRGPLGSIRNLAMEKVANSVLF 96
Query: 128 PCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVDM 187
PCKY S GC P+ K HE +C FRPY+CP G+ C G ++ ++ HL HK
Sbjct: 97 PCKYASSGCEITLPHTEKADHEELCEFRPYSCPCPGASCKWQGSLDAVMPHLMHQHK--S 154
Query: 188 HTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQ-LGMAPVYMAFLRFMGD 246
T + + A + + CFG +F L E + + A ++ +G
Sbjct: 155 ITTLQGEDIVFLATDINLPGAVDWVMMQSCFGFHFMLVLEKQEKYDGHQQFFAIVQLIGT 214
Query: 247 ENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS 300
+A N+ Y LE+ + R+L WE TPRSI + + D L+ ++A F+
Sbjct: 215 RKQAENFAYRLELNGHRRRLTWEATPRSIHEGIATAIMNSDCLVFDTSIAQLFA 268
>SI1B_MOUSE (Q06985) Ubiquitin ligase SIAH1B (EC 6.3.2.-) (Seven in
absentia homolog 1b) (Siah1b) (Siah-1b)
Length = 282
Score = 157 bits (398), Expect = 3e-38
Identities = 84/234 (35%), Positives = 124/234 (52%), Gaps = 4/234 (1%)
Query: 68 LLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLEL 127
L ECPVC + + PPI QC +GH +CS C+ ++ CPTCR LG IR LA+EKVA S+
Sbjct: 38 LFECPVCFDYVLPPILQCQSGHLVCSNCRPKL-TCCPTCRGPLGSIRNLAVEKVANSVLF 96
Query: 128 PCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVDM 187
PCKY + GC P+ K +HE +C FRPY+CP G+ C G ++ ++ HL HK
Sbjct: 97 PCKYSASGCEITLPHTKKAEHEELCEFRPYSCPCPGASCKWQGSLDAVMPHLMHQHK--S 154
Query: 188 HTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQ-LGMAPVYMAFLRFMGD 246
T + + A + + CFG +F L E + + A ++ +G
Sbjct: 155 ITTLQGEDIVFLATDINLPGAVDWVMMQSCFGFHFMLVLEKQEKYDGHQQFFAIVQLIGT 214
Query: 247 ENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS 300
+A N+ Y LE+ + R+L WE TPRSI + + D L+ ++A F+
Sbjct: 215 RKQAENFAYRLELNGHRRRLTWEATPRSIHEGIATAIMNSDCLVFDTSIAQLFA 268
>SINA_DROWI (Q8I147) Ubiquitin ligase sina (EC 6.3.2.-) (Seven in
absentia protein)
Length = 331
Score = 153 bits (387), Expect = 5e-37
Identities = 88/275 (32%), Positives = 134/275 (48%), Gaps = 6/275 (2%)
Query: 39 SSLKPRSGGNNNHGVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTR 98
SS S +++ G + + L ECPVC + + PPI QC +GH +C +C+++
Sbjct: 58 SSANTSSSSSSSLSSAGGGDAGMSADLTSLFECPVCFDYVLPPILQCSSGHLVCVSCRSK 117
Query: 99 VHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYT 158
+ CPTCR L +IR LA+EKVA +++ PCK+ GC Y K +HE C RPY
Sbjct: 118 L-TCCPTCRGPLANIRNLAMEKVASNVKFPCKHSGYGCTASLVYTEKTEHEETCECRPYL 176
Query: 159 CPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCF 218
CP G+ C G ++ ++ HL HK T + + A + + CF
Sbjct: 177 CPCPGASCKWQGPLDLVMQHLMMSHK--SITTLQGEDIVFLATDINLPGAVDWVMMQSCF 234
Query: 219 GQYFCLHFEAFQ-LGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRD 277
G +F L E + + A ++ +G EA N+ Y LE+ N R+L WE PRSI +
Sbjct: 235 GHHFMLVLEKQEKYDGHQQFFAIVQLIGSRKEAENFVYRLELNGNRRRLTWEAMPRSIHE 294
Query: 278 SHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVT 312
+ D L+ ++A F+ D L + VT
Sbjct: 295 GVASAIHNSDCLVFDTSIAQLFA--DNGNLGINVT 327
>SINA_DROME (P21461) Ubiquitin ligase sina (EC 6.3.2.-) (Seven in
absentia protein)
Length = 314
Score = 153 bits (387), Expect = 5e-37
Identities = 88/275 (32%), Positives = 134/275 (48%), Gaps = 6/275 (2%)
Query: 39 SSLKPRSGGNNNHGVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTR 98
SS S +++ G + + L ECPVC + + PPI QC +GH +C +C+++
Sbjct: 41 SSANTSSSSSSSLSSAGGGDAGMSADLTSLFECPVCFDYVLPPILQCSSGHLVCVSCRSK 100
Query: 99 VHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYT 158
+ CPTCR L +IR LA+EKVA +++ PCK+ GC Y K +HE C RPY
Sbjct: 101 L-TCCPTCRGPLANIRNLAMEKVASNVKFPCKHSGYGCTASLVYTEKTEHEETCECRPYL 159
Query: 159 CPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCF 218
CP G+ C G ++ ++ HL HK T + + A + + CF
Sbjct: 160 CPCPGASCKWQGPLDLVMQHLMMSHK--SITTLQGEDIVFLATDINLPGAVDWVMMQSCF 217
Query: 219 GQYFCLHFEAFQ-LGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRD 277
G +F L E + + A ++ +G EA N+ Y LE+ N R+L WE PRSI +
Sbjct: 218 GHHFMLVLEKQEKYDGHQQFFAIVQLIGSRKEAENFVYRLELNGNRRRLTWEAMPRSIHE 277
Query: 278 SHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVT 312
+ D L+ ++A F+ D L + VT
Sbjct: 278 GVASAIHNSDCLVFDTSIAQLFA--DNGNLGINVT 310
>SINA_DROER (P61093) Ubiquitin ligase sina (EC 6.3.2.-) (Seven in
absentia protein)
Length = 314
Score = 153 bits (387), Expect = 5e-37
Identities = 88/275 (32%), Positives = 134/275 (48%), Gaps = 6/275 (2%)
Query: 39 SSLKPRSGGNNNHGVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTR 98
SS S +++ G + + L ECPVC + + PPI QC +GH +C +C+++
Sbjct: 41 SSANTSSSSSSSLSSAGGGDAGMSADLTSLFECPVCFDYVLPPILQCSSGHLVCVSCRSK 100
Query: 99 VHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYT 158
+ CPTCR L +IR LA+EKVA +++ PCK+ GC Y K +HE C RPY
Sbjct: 101 L-TCCPTCRGPLANIRNLAMEKVASNVKFPCKHSGYGCTASLVYTEKTEHEETCECRPYL 159
Query: 159 CPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCF 218
CP G+ C G ++ ++ HL HK T + + A + + CF
Sbjct: 160 CPCPGASCKWQGPLDLVMQHLMMSHK--SITTLQGEDIVFLATDINLPGAVDWVMMQSCF 217
Query: 219 GQYFCLHFEAFQ-LGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRD 277
G +F L E + + A ++ +G EA N+ Y LE+ N R+L WE PRSI +
Sbjct: 218 GHHFMLVLEKQEKYDGHQQFFAIVQLIGSRKEAENFVYRLELNGNRRRLTWEAMPRSIHE 277
Query: 278 SHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVT 312
+ D L+ ++A F+ D L + VT
Sbjct: 278 GVASAIHNSDCLVFDTSIAQLFA--DNGNLGINVT 310
>SINA_DROVI (P29304) Ubiquitin ligase sina (EC 6.3.2.-) (Seven in
absentia protein)
Length = 314
Score = 152 bits (383), Expect = 1e-36
Identities = 87/275 (31%), Positives = 134/275 (48%), Gaps = 6/275 (2%)
Query: 39 SSLKPRSGGNNNHGVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTR 98
SS S +++ G + + L ECPVC + + PPI QC +GH +C +C+++
Sbjct: 41 SSANTSSSSSSSLSSAGGGDAGMSADLTSLFECPVCFDYVLPPILQCSSGHLVCVSCRSK 100
Query: 99 VHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYT 158
+ CPTCR L +IR LA+E+VA +++ PCK+ GC Y K +HE C RPY
Sbjct: 101 L-TCCPTCRGPLANIRNLAMEEVASNVKFPCKHSGYGCTASLVYTEKTEHEETCECRPYL 159
Query: 159 CPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCF 218
CP G+ C G ++ ++ HL HK T + + A + + CF
Sbjct: 160 CPCPGASCKWQGPLDLVMQHLMMSHK--SITTLQGEDIVFLATDINLPGAVDWVMMQSCF 217
Query: 219 GQYFCLHFEAFQ-LGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRD 277
G +F L E + + A ++ +G EA N+ Y LE+ N R+L WE PRSI +
Sbjct: 218 GHHFMLVLEKQEKYDGHQQFFAIVQLIGSRKEAENFVYRLELNGNRRRLTWEAMPRSIHE 277
Query: 278 SHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVT 312
+ D L+ ++A F+ D L + VT
Sbjct: 278 GVASAIHNSDCLVFDTSIAQLFA--DNGNLGINVT 310
>SIA2_BRARE (Q7SYL3) Ubiquitin ligase SIAH2 (EC 6.3.2.-) (Seven in
absentia homolog 2) (Siah-2)
Length = 331
Score = 150 bits (379), Expect = 4e-36
Identities = 94/278 (33%), Positives = 135/278 (47%), Gaps = 15/278 (5%)
Query: 45 SGGNNNHGVIGSTAIA--------PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCK 96
SG G + + A+A + + L ECPVC + + PPI QC GH +C+ C+
Sbjct: 55 SGSVAGSGTVPAAAVALPVAALPGQSPELTALFECPVCFDYVLPPILQCQAGHLVCNQCR 114
Query: 97 TRVHNRCPTCRQELG-DIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFR 155
++ + CPTCR L IR LA+EKVA +L PCKY S GC + K +HE +C FR
Sbjct: 115 QKL-SCCPTCRGPLTPSIRNLAMEKVASTLPFPCKYSSAGCLLSLHHSEKPEHEEVCEFR 173
Query: 156 PYTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVF 215
PYTCP G+ C G + ++ HL HK T + + A + +
Sbjct: 174 PYTCPCPGASCKWQGSLEEVMPHLMHAHK--SITTLQGEDIVFLATDINLPGAVDWVMMQ 231
Query: 216 HCFGQYFCLHFEAFQLGMA-PVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRS 274
CFG +F L E + + A + +G +A N+ Y LE+ N R+L WE TPRS
Sbjct: 232 SCFGHHFMLVLEKQEKYEGHQQFFAIVLLIGTRKQAENFAYRLELNGNRRRLTWEATPRS 291
Query: 275 IRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVT 312
I D + D L+ ++A F+ D L + VT
Sbjct: 292 IHDGVAAAIMNSDCLVFDTSIAHLFA--DNGNLGINVT 327
>SIA2_XENLA (Q9I8X5) Ubiquitin ligase SIAH2 (EC 6.3.2.-) (Seven in
absentia homolog 2) (Xsiah-2)
Length = 313
Score = 148 bits (374), Expect = 2e-35
Identities = 95/277 (34%), Positives = 134/277 (48%), Gaps = 14/277 (5%)
Query: 45 SGGNNNHGVIGSTAIA---PATSVHE----LLECPVCTNSMYPPIHQCHNGHTLCSTCKT 97
SGG TA A P + H+ L ECPVC + + PPI QC GH +C+ C+
Sbjct: 36 SGGPGASAPPAPTAAAITGPLSQQHQELTSLFECPVCFDYVLPPILQCQAGHLVCNQCRQ 95
Query: 98 RVHNRCPTCRQELG-DIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRP 156
++ + CPTCR L IR LA+EKVA ++ PCKY S GC + K +HE IC +RP
Sbjct: 96 KL-SCCPTCRASLTPSIRNLAMEKVASAVLFPCKYASTGCSLSLHHTEKPEHEDICEYRP 154
Query: 157 YTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFH 216
Y+CP G+ C G + ++ HL HK T + + A + + +
Sbjct: 155 YSCPCPGASCKWQGSLENVMQHLTHSHK--SITTLQGEDIVFLATDINLPGAVDWVMMQY 212
Query: 217 CFGQYFCLHFEAFQLGMA-PVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSI 275
CF +F L E + + A + +G +A NY Y LE+ N R+L WE TPRSI
Sbjct: 213 CFNHHFMLVLEKQEKYEGHQQFFAIVLLIGTRKQAENYAYRLELNGNRRRLTWEATPRSI 272
Query: 276 RDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVT 312
D + D L+ +A F+ D L + VT
Sbjct: 273 HDGVAAAIMNSDCLVFDTAIAHLFA--DNGNLGINVT 307
>SINL_DROME (Q8T3Y0) Probable ubiquitin ligase sina-like protein
CG13030 (EC 6.3.2.-)
Length = 351
Score = 148 bits (373), Expect = 2e-35
Identities = 90/257 (35%), Positives = 127/257 (49%), Gaps = 11/257 (4%)
Query: 68 LLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLEL 127
LLECPVC + PPI QC GH +CSTC++++ CP CR + +IR LA+EKVA L
Sbjct: 103 LLECPVCFGYIMPPIMQCPRGHLICSTCRSKL-TICPVCRVFMTNIRSLAMEKVASKLIF 161
Query: 128 PCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHK--V 185
PCK+ GC Y K KHE C RPY CPY +CS G + + HL H+ +
Sbjct: 162 PCKHSHFGCRARLSYAEKTKHEEDCECRPYFCPYPDDKCSWQGPLRDVYQHLMSSHENVI 221
Query: 186 DMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGM-APVYMAFLRFM 244
M N + +E A V C G++F L E LG Y R +
Sbjct: 222 TMEG----NDIIFLATNVNLEGALDWTMVQSCHGRHFLLSLEKINLGEDCQQYFTACRMI 277
Query: 245 GDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDR 304
G +A + Y++ + A R L W+ PRSIR++ ++ D L++ ++ FS +
Sbjct: 278 GSMKDAAEFVYNISLEAYNRTLRWQSKPRSIRENFSSFTNA-DFLVLNKHTVELFS--ED 334
Query: 305 KELKLRVTGRIWKEQQN 321
L L V R +E+ N
Sbjct: 335 GNLALNVVIRKVEERTN 351
>SIA1_CAEEL (Q965X6) Ubiquitin ligase sia-1 (EC 6.3.2.-) (Seven in
absentia homolog 1)
Length = 339
Score = 147 bits (372), Expect = 3e-35
Identities = 83/267 (31%), Positives = 127/267 (47%), Gaps = 11/267 (4%)
Query: 42 KPRSGGNNNHGVI----GSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKT 97
+PR +H + G + + + ECPVC M PP QC +GH +CS C+
Sbjct: 57 RPRKESKASHHIFQSAGGGATDDSSAEILSVFECPVCLEYMLPPYMQCSSGHLVCSNCRP 116
Query: 98 RVHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPY 157
++ CPTCR +R L LEK+A ++ PCK+ + GCP F + K +HE +C FRPY
Sbjct: 117 KLQC-CPTCRGPTPSVRNLGLEKIANTVRFPCKFSTSGCPLNFHHADKTEHEELCEFRPY 175
Query: 158 TCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHC 217
CP G+ C G ++ ++ HL+ HK T + + A + + C
Sbjct: 176 CCPCPGASCKWQGGLSDVMEHLKKIHK--SITTLQGEDIVFLATDINLPGAVDWVMMQSC 233
Query: 218 FGQYFCLHFEAFQ----LGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPR 273
F F L E + ++ A ++ +G + EA N+ Y LE+ A+ R++ WE TPR
Sbjct: 234 FDYNFMLVLEKQEKYDPAQPTQMFYAVVQLIGSKKEADNFVYRLELSASRRRMSWEATPR 293
Query: 274 SIRDSHRKVRDSHDGLIIQRNMALFFS 300
SI + D L N A F+
Sbjct: 294 SIHEGVVVAIQQSDCLAFDSNAAQLFA 320
>SIA2_HUMAN (O43255) Ubiquitin ligase SIAH2 (EC 6.3.2.-) (Seven in
absentia homolog 2) (Siah-2) (hSiah2)
Length = 324
Score = 146 bits (368), Expect = 8e-35
Identities = 89/264 (33%), Positives = 129/264 (48%), Gaps = 11/264 (4%)
Query: 55 GSTAIAPATSVHE----LLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQEL 110
G P + H L ECPVC + + PPI QC GH +C+ C+ ++ + CPTCR L
Sbjct: 60 GGGGAGPVSPQHHELTSLFECPVCFDYVLPPILQCQAGHLVCNQCRQKL-SCCPTCRGAL 118
Query: 111 G-DIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAV 169
IR LA+EKVA ++ PCKY + GC + K +HE IC +RPY+CP G+ C
Sbjct: 119 TPSIRNLAMEKVASAVLFPCKYATTGCSLTLHHTEKPEHEDICEYRPYSCPCPGASCKWQ 178
Query: 170 GDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAF 229
G + +++HL HK T + + A + + CFG +F L E
Sbjct: 179 GSLEAVMSHLMHAHK--SITTLQGEDIVFLATDINLPGAVDWVMMQSCFGHHFMLVLEKQ 236
Query: 230 QLGMA-PVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDG 288
+ + A + +G +A N+ Y LE+ N R+L WE TPRSI D + D
Sbjct: 237 EKYEGHQQFFAIVLLIGTRKQAENFAYRLELNGNRRRLTWEATPRSIHDGVAAAIMNSDC 296
Query: 289 LIIQRNMALFFSGGDRKELKLRVT 312
L+ +A F+ D L + VT
Sbjct: 297 LVFDTAIAHLFA--DNGNLGINVT 318
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.138 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,763,081
Number of Sequences: 164201
Number of extensions: 1883411
Number of successful extensions: 6501
Number of sequences better than 10.0: 122
Number of HSP's better than 10.0 without gapping: 48
Number of HSP's successfully gapped in prelim test: 75
Number of HSP's that attempted gapping in prelim test: 6047
Number of HSP's gapped (non-prelim): 284
length of query: 333
length of database: 59,974,054
effective HSP length: 111
effective length of query: 222
effective length of database: 41,747,743
effective search space: 9267998946
effective search space used: 9267998946
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 66 (30.0 bits)
Medicago: description of AC134822.10


