

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134322.21 - phase: 0
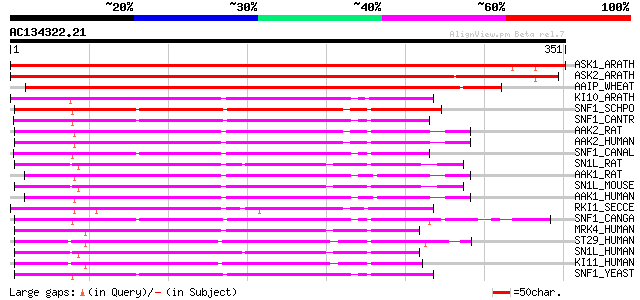
(351 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ASK1_ARATH (P43291) Serine/threonine-protein kinase ASK1 (EC 2.7... 602 e-172
ASK2_ARATH (P43292) Serine/threonine-protein kinase ASK2 (EC 2.7... 572 e-163
AAIP_WHEAT (Q02066) Abscisic acid-inducible protein kinase (EC 2... 463 e-130
KI10_ARATH (Q38997) SNF1-related protein kinase KIN10 (EC 2.7.1.... 219 1e-56
SNF1_SCHPO (O74536) SNF1-like protein kinase ssp2 (EC 2.7.1.-) 212 1e-54
SNF1_CANTR (O94168) Carbon catabolite derepressing protein kinas... 203 5e-52
AAK2_RAT (Q09137) 5'-AMP-activated protein kinase, catalytic alp... 201 2e-51
AAK2_HUMAN (P54646) 5'-AMP-activated protein kinase, catalytic a... 201 3e-51
SNF1_CANAL (P52497) Carbon catabolite derepressing protein kinas... 199 9e-51
SN1L_RAT (Q9R1U5) Probable serine/threonine-protein kinase SNF1L... 198 1e-50
AAK1_RAT (P54645) 5'-AMP-activated protein kinase, catalytic alp... 198 2e-50
SN1L_MOUSE (Q60670) Probable serine/threonine-protein kinase SNF... 197 3e-50
AAK1_HUMAN (Q13131) 5'-AMP-activated protein kinase, catalytic a... 196 6e-50
RKI1_SECCE (Q02723) Carbon catabolite derepressing protein kinas... 194 2e-49
SNF1_CANGA (Q00372) Carbon catabolite derepressing protein kinas... 193 5e-49
MRK4_HUMAN (Q96L34) MAP/microtubule affinity-regulating kinase 4... 192 1e-48
ST29_HUMAN (Q8IWQ3) Serine/threonine-protein kinase 29 (EC 2.7.1... 191 2e-48
SN1L_HUMAN (P57059) Probable serine/threonine-protein kinase SNF... 191 2e-48
KI11_HUMAN (Q8TDC3) Probable serine/threonine-protein kinase KIA... 190 4e-48
SNF1_YEAST (P06782) Carbon catabolite derepressing protein kinas... 190 5e-48
>ASK1_ARATH (P43291) Serine/threonine-protein kinase ASK1 (EC
2.7.1.37)
Length = 363
Score = 602 bits (1551), Expect = e-172
Identities = 293/363 (80%), Positives = 326/363 (89%), Gaps = 12/363 (3%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYE+VKDIG+GNFGVARLM+ K++K+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MDKYELVKDIGAGNFGVARLMKVKNSKELVAMKYIERGPKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTHL I MEYAAGGELF+RICSAGRFSEDEARYFFQQLISGVSYCH+MQIC
Sbjct: 61 IRFKEVVLTPTHLAIAMEYAAGGELFERICSAGRFSEDEARYFFQQLISGVSYCHAMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK+
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKM 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQ+DPKNFRKTI +IMAVQYKIPDYVHISQ+C++LLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQEDPKNFRKTIQKIMAVQYKIPDYVHISQDCKNLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FVA+ +RITI EIK H WFLKNLPRELTE AQA Y++KENPT+SLQ++E+IMKIV +AK
Sbjct: 241 FVANSLKRITIAEIKKHSWFLKNLPRELTETAQAAYFKKENPTFSLQTVEEIMKIVADAK 300
Query: 301 NPPQASRSVGGFGWGG-------EEDDDEINEAEAELE-----EDEYEKRVKEAQESGEI 348
PP SRS+GGFGWGG EED +++ E E E+E EDEY+K VKE SGE+
Sbjct: 301 TPPPVSRSIGGFGWGGNGDADGKEEDAEDVEEEEEEVEEEEDDEDEYDKTVKEVHASGEV 360
Query: 349 HVN 351
++
Sbjct: 361 RIS 363
>ASK2_ARATH (P43292) Serine/threonine-protein kinase ASK2 (EC
2.7.1.37)
Length = 353
Score = 572 bits (1474), Expect = e-163
Identities = 280/350 (80%), Positives = 314/350 (89%), Gaps = 4/350 (1%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KY+VVKD+G+GNFGVARL+RHKDTK+LVAMKYIERG KIDENVAREIINHRSL+HPNI
Sbjct: 1 MDKYDVVKDLGAGNFGVARLLRHKDTKELVAMKYIERGRKIDENVAREIINHRSLKHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEV+LTPTHL IVMEYA+GGELFDRIC+AGRFSE EARYFFQQLI GV YCHS+QIC
Sbjct: 61 IRFKEVILTPTHLAIVMEYASGGELFDRICTAGRFSEAEARYFFQQLICGVDYCHSLQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAP LKICDFGYSKSS+LHSRPKSTVGTPAYIAPEVLSRREYDGK
Sbjct: 121 HRDLKLENTLLDGSPAPLLKICDFGYSKSSILHSRPKSTVGTPAYIAPEVLSRREYDGKH 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFED +DPKNFRKTI RIMAVQYKIPDYVHISQEC+HLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDPNDPKNFRKTIQRIMAVQYKIPDYVHISQECKHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FV + A+RIT+KEIK+HPW+LKNLP+EL E AQA YY+++ ++SLQS+EDIMKIV EA+
Sbjct: 241 FVTNSAKRITLKEIKNHPWYLKNLPKELLESAQAAYYKRDT-SFSLQSVEDIMKIVGEAR 299
Query: 301 NPPQASRSVGGFGWGGEEDDDEINEAEAELE---EDEYEKRVKEAQESGE 347
NP ++ +V G G +E+++E EAE E E EDEYEK VKEAQ E
Sbjct: 300 NPAPSTSAVKSSGSGADEEEEEDVEAEVEEEEDDEDEYEKHVKEAQSCQE 349
>AAIP_WHEAT (Q02066) Abscisic acid-inducible protein kinase (EC
2.7.1.-) (Fragment)
Length = 332
Score = 463 bits (1192), Expect = e-130
Identities = 225/301 (74%), Positives = 259/301 (85%), Gaps = 1/301 (0%)
Query: 11 GSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNIIRFKEVVLTP 70
GSGNFGVA+L+R TK+ A+K+IERGHKIDE+V REI+NHRSL+HPNIIRFKEVVLTP
Sbjct: 1 GSGNFGVAKLVRDVRTKEHFAVKFIERGHKIDEHVQREIMNHRSLKHPNIIRFKEVVLTP 60
Query: 71 THLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQICHRDLKLENTL 130
THL IVMEYA+GGELF RIC+AGRFSEDE R+FFQQLISGVSYCHSMQ+CHRDLKLENTL
Sbjct: 61 THLAIVMEYASGGELFQRICNAGRFSEDEGRFFFQQLISGVSYCHSMQVCHRDLKLENTL 120
Query: 131 LDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKLADVWSCGVTL 190
LDGS APRLKICDFGYSKSS+LHS+PKSTVGTPAYIAPEVLSRREYDGK+ADVWSCGVTL
Sbjct: 121 LDGSVAPRLKICDFGYSKSSVLHSQPKSTVGTPAYIAPEVLSRREYDGKVADVWSCGVTL 180
Query: 191 YVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRIFVASPARRIT 250
YVMLVGAYPFED D+P+NFRKTI RI++VQY +PDYV +S +C HLLSRIFV +P +RIT
Sbjct: 181 YVMLVGAYPFEDPDEPRNFRKTITRILSVQYSVPDYVRVSMDCIHLLSRIFVGNPQQRIT 240
Query: 251 IKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAKNPPQASRSVG 310
I EIK+HPWFLK LP E+T+ Q + T S QS+E+ M I++EA+ P + V
Sbjct: 241 IPEIKNHPWFLKRLPVEMTDEYQRSMQLADMNTPS-QSLEEAMAIIQEAQKPGDNALGVA 299
Query: 311 G 311
G
Sbjct: 300 G 300
>KI10_ARATH (Q38997) SNF1-related protein kinase KIN10 (EC 2.7.1.-)
(AKIN10)
Length = 535
Score = 219 bits (557), Expect = 1e-56
Identities = 122/272 (44%), Positives = 160/272 (57%), Gaps = 12/272 (4%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIER----GHKIDENVAREIINHRSLR 56
+ Y++ + +G G+FG ++ H T VA+K + R +++E V REI R
Sbjct: 39 LPNYKLGRTLGIGSFGRVKIAEHALTGHKVAIKILNRRKIKNMEMEEKVRREIKILRLFM 98
Query: 57 HPNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHS 116
HP+IIR EV+ TPT + +VMEY GELFD I GR EDEAR FFQQ+ISGV YCH
Sbjct: 99 HPHIIRLYEVIETPTDIYLVMEYVNSGELFDYIVEKGRLQEDEARNFFQQIISGVEYCHR 158
Query: 117 MQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREY 176
+ HRDLK EN LLD +KI DFG S K++ G+P Y APEV+S + Y
Sbjct: 159 NMVVHRDLKPENLLLDSK--CNVKIADFGLSNIMRDGHFLKTSCGSPNYAAPEVISGKLY 216
Query: 177 DGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHL 236
G DVWSCGV LY +L G PF+D++ P F+K I Y +P H+S R L
Sbjct: 217 AGPEVDVWSCGVILYALLCGTLPFDDENIPNLFKKIKGGI----YTLPS--HLSPGARDL 270
Query: 237 LSRIFVASPARRITIKEIKSHPWFLKNLPREL 268
+ R+ V P +R+TI EI+ HPWF +LPR L
Sbjct: 271 IPRMLVVDPMKRVTIPEIRQHPWFQAHLPRYL 302
>SNF1_SCHPO (O74536) SNF1-like protein kinase ssp2 (EC 2.7.1.-)
Length = 576
Score = 212 bits (539), Expect = 1e-54
Identities = 121/274 (44%), Positives = 167/274 (60%), Gaps = 13/274 (4%)
Query: 4 YEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERG----HKIDENVAREIINHRSLRHPN 59
Y + + +G G+FG +L H T+Q VA+K+I R + V REI + LRHP+
Sbjct: 34 YIIRETLGEGSFGKVKLATHYKTQQKVALKFISRQLLKKSDMHMRVEREISYLKLLRHPH 93
Query: 60 IIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQI 119
II+ +V+ TPT + +V+EYA GGELFD I R +EDE R FFQQ+I + YCH +I
Sbjct: 94 IIKLYDVITTPTDIVMVIEYA-GGELFDYIVEKKRMTEDEGRRFFQQIICAIEYCHRHKI 152
Query: 120 CHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK 179
HRDLK EN LLD + +KI DFG S + K++ G+P Y APEV++ + Y G
Sbjct: 153 VHRDLKPENLLLDDNL--NVKIADFGLSNIMTDGNFLKTSCGSPNYAAPEVINGKLYAGP 210
Query: 180 LADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSR 239
DVWSCG+ LYVMLVG PF+D+ P F+K + + Y +PD+ +S + L+ R
Sbjct: 211 EVDVWSCGIVLYVMLVGRLPFDDEFIPNLFKK----VNSCVYVMPDF--LSPGAQSLIRR 264
Query: 240 IFVASPARRITIKEIKSHPWFLKNLPRELTEMAQ 273
+ VA P +RITI+EI+ PWF NLP L M +
Sbjct: 265 MIVADPMQRITIQEIRRDPWFNVNLPDYLRPMEE 298
>SNF1_CANTR (O94168) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 619
Score = 203 bits (517), Expect = 5e-52
Identities = 117/267 (43%), Positives = 161/267 (59%), Gaps = 13/267 (4%)
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERG----HKIDENVAREIINHRSLRHP 58
+Y+++K +G G+FG +L +H T Q VA+K I R + V REI R LRHP
Sbjct: 51 RYQIIKTLGEGSFGKVKLAQHVGTGQKVALKIINRKTLAKSDMQGRVEREISYLRLLRHP 110
Query: 59 NIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQ 118
+II+ +V+ + + +V+E+A G ELFD I G+ EDEAR FFQQ+I+ V YCH +
Sbjct: 111 HIIKLYDVIKSKDEIIMVIEFA-GKELFDYIVQRGKMPEDEARRFFQQIIAAVEYCHRHK 169
Query: 119 ICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDG 178
I HRDLK EN LLD +KI DFG S + K++ G+P Y APEV+S + Y G
Sbjct: 170 IVHRDLKPENLLLDDQL--NVKIADFGLSNIMTDGNFLKTSCGSPNYAAPEVISGKLYAG 227
Query: 179 KLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLS 238
DVWS GV LYVML G PF+D+ P F+K N + Y +P+Y +S +HLL+
Sbjct: 228 PEVDVWSSGVILYVMLCGRLPFDDEFIPALFKKISNGV----YTLPNY--LSPGAKHLLT 281
Query: 239 RIFVASPARRITIKEIKSHPWFLKNLP 265
R+ V +P RITI EI WF +++P
Sbjct: 282 RMLVVNPLNRITIHEIMEDEWFKQDMP 308
>AAK2_RAT (Q09137) 5'-AMP-activated protein kinase, catalytic
alpha-2 chain (EC 2.7.1.-) (AMPK alpha-2 chain)
Length = 552
Score = 201 bits (511), Expect = 2e-51
Identities = 116/292 (39%), Positives = 159/292 (53%), Gaps = 21/292 (7%)
Query: 4 YEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGH----KIDENVAREIINHRSLRHPN 59
Y + +G G FG ++ H+ T VA+K + R + + REI N + RHP+
Sbjct: 16 YVLGDTLGVGTFGKVKIGEHQLTGHKVAVKILNRQKIRSLDVVGKIKREIQNLKLFRHPH 75
Query: 60 IIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQI 119
II+ +V+ TPT +VMEY +GGELFD IC GR E EAR FQQ++S V YCH +
Sbjct: 76 IIKLYQVISTPTDFFMVMEYVSGGELFDYICKHGRVEEVEARRLFQQILSAVDYCHRHMV 135
Query: 120 CHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK 179
HRDLK EN LLD KI DFG S +++ G+P Y APEV+S R Y G
Sbjct: 136 VHRDLKPENVLLDAQ--MNAKIADFGLSNMMSDGEFLRTSCGSPNYAAPEVISGRLYAGP 193
Query: 180 LADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSR 239
D+WSCGV LY +L G PF+D+ P F+K I + IP+Y +++ LL
Sbjct: 194 EVDIWSCGVILYALLCGTLPFDDEHVPTLFKK----IRGGVFYIPEY--LNRSIATLLMH 247
Query: 240 IFVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIED 291
+ P +R TIK+I+ H WF ++LP Y E+P+Y I+D
Sbjct: 248 MLQVDPLKRATIKDIREHEWFKQDLPS---------YLFPEDPSYDANVIDD 290
>AAK2_HUMAN (P54646) 5'-AMP-activated protein kinase, catalytic
alpha-2 chain (EC 2.7.1.-) (AMPK alpha-2 chain)
Length = 552
Score = 201 bits (510), Expect = 3e-51
Identities = 116/292 (39%), Positives = 159/292 (53%), Gaps = 21/292 (7%)
Query: 4 YEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGH----KIDENVAREIINHRSLRHPN 59
Y + +G G FG ++ H+ T VA+K + R + + REI N + RHP+
Sbjct: 16 YVLGDTLGVGTFGKVKIGEHQLTGHKVAVKILNRQKIRSLDVVGKIKREIQNLKLFRHPH 75
Query: 60 IIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQI 119
II+ +V+ TPT +VMEY +GGELFD IC GR E EAR FQQ++S V YCH +
Sbjct: 76 IIKLYQVISTPTDFFMVMEYVSGGELFDYICKHGRVEEMEARRLFQQILSAVDYCHRHMV 135
Query: 120 CHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK 179
HRDLK EN LLD KI DFG S +++ G+P Y APEV+S R Y G
Sbjct: 136 VHRDLKPENVLLDAH--MNAKIADFGLSNMMSDGEFLRTSCGSPNYAAPEVISGRLYAGP 193
Query: 180 LADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSR 239
D+WSCGV LY +L G PF+D+ P F+K I + IP+Y +++ LL
Sbjct: 194 EVDIWSCGVILYALLCGTLPFDDEHVPTLFKK----IRGGVFYIPEY--LNRSVATLLMH 247
Query: 240 IFVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIED 291
+ P +R TIK+I+ H WF ++LP Y E+P+Y I+D
Sbjct: 248 MLQVDPLKRATIKDIREHEWFKQDLPS---------YLFPEDPSYDANVIDD 290
>SNF1_CANAL (P52497) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 620
Score = 199 bits (506), Expect = 9e-51
Identities = 117/268 (43%), Positives = 162/268 (59%), Gaps = 14/268 (5%)
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERG----HKIDENVAREIINHRSLRHP 58
+Y+++K +G G+FG +L +H T Q VA+K I R + V REI R LRHP
Sbjct: 52 RYQILKTLGEGSFGKVKLAQHLGTGQKVALKIINRKTLAKSDMQGRVEREISYLRLLRHP 111
Query: 59 NIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQ 118
+II+ +V+ + + +V+E+A G ELFD I G+ EDEAR FFQQ+I+ V YCH +
Sbjct: 112 HIIKLYDVIKSKDEIIMVIEFA-GKELFDYIVQRGKMPEDEARRFFQQIIAAVEYCHRHK 170
Query: 119 ICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYI-APEVLSRREYD 177
I HRDLK EN LLD +KI DFG S + K++ G+P Y+ APEV+S + Y
Sbjct: 171 IVHRDLKPENLLLDDQ--LNVKIADFGLSNIMTDGNFLKTSCGSPNYMPAPEVISGKLYA 228
Query: 178 GKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLL 237
G DVWS GV LYVML G PF+D+ P F+K N + Y +P+Y +S +HLL
Sbjct: 229 GPEVDVWSAGVILYVMLCGRLPFDDEFIPALFKKISNGV----YTLPNY--LSAGAKHLL 282
Query: 238 SRIFVASPARRITIKEIKSHPWFLKNLP 265
+R+ V +P RITI EI WF +++P
Sbjct: 283 TRMLVVNPLNRITIHEIMEDDWFKQDMP 310
>SN1L_RAT (Q9R1U5) Probable serine/threonine-protein kinase SNF1LK
(EC 2.7.1.37) (Salt-inducible protein kinase) (Protein
kinase KID2)
Length = 776
Score = 198 bits (504), Expect = 1e-50
Identities = 115/289 (39%), Positives = 169/289 (57%), Gaps = 25/289 (8%)
Query: 4 YEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKID----ENVAREIINHRSLRHPN 59
Y+V + +G GNF V +L RH+ TK VA+K I++ ++D E + RE+ + L HPN
Sbjct: 27 YDVERTLGKGNFAVVKLARHRVTKTQVAIKIIDKT-RLDSSNLEKIYREVQLMKLLNHPN 85
Query: 60 IIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQI 119
II+ +V+ T L IV E+A GE+FD + S G SE+EAR F Q++S V YCH+ I
Sbjct: 86 IIKLYQVMETKDMLYIVTEFAKNGEMFDYLTSNGHLSENEARKKFWQILSAVEYCHNHHI 145
Query: 120 CHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKST-VGTPAYIAPEVLSRREYDG 178
HRDLK EN LLDG+ +K+ DFG+ + P ST G+P Y APEV +EY+G
Sbjct: 146 VHRDLKTENLLLDGN--MDIKLADFGFG-NFYKPGEPLSTWCGSPPYAAPEVFEGKEYEG 202
Query: 179 KLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLS 238
D+WS GV LYV++ G+ PF D N R++ +++IP + +SQ+C L+
Sbjct: 203 PQLDIWSLGVVLYVLVCGSLPF----DGPNLPTLRQRVLEGRFRIPFF--MSQDCETLIR 256
Query: 239 RIFVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQ 287
R+ V PA+RITI +I+ H W A ++++P +S+Q
Sbjct: 257 RMLVVDPAKRITIAQIRQHRWM----------QADPTLLQQDDPAFSMQ 295
>AAK1_RAT (P54645) 5'-AMP-activated protein kinase, catalytic
alpha-1 chain (EC 2.7.1.-) (AMPK alpha-1 chain)
Length = 548
Score = 198 bits (503), Expect = 2e-50
Identities = 114/286 (39%), Positives = 159/286 (54%), Gaps = 21/286 (7%)
Query: 10 IGSGNFGVARLMRHKDTKQLVAMKYIERGH----KIDENVAREIINHRSLRHPNIIRFKE 65
+G G FG ++ +H+ T VA+K + R + + REI N + RHP+II+ +
Sbjct: 22 LGVGTFGKVKVGKHELTGHKVAVKILNRQKIRSLDVVGKIRREIQNLKLFRHPHIIKLYQ 81
Query: 66 VVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQICHRDLK 125
V+ TP+ + +VMEY +GGELFD IC GR E E+R FQQ++SGV YCH + HRDLK
Sbjct: 82 VISTPSDIFMVMEYVSGGELFDYICKNGRLDEKESRRLFQQILSGVDYCHRHMVVHRDLK 141
Query: 126 LENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKLADVWS 185
EN LLD KI DFG S +++ G+P Y APEV+S R Y G D+WS
Sbjct: 142 PENVLLDAH--MNAKIADFGLSNMMSDGEFLRTSCGSPNYAAPEVISGRLYAGPEVDIWS 199
Query: 186 CGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRIFVASP 245
GV LY +L G PF+D P F+K + I + P Y++ S LL + P
Sbjct: 200 SGVILYALLCGTLPFDDDHVPTLFKKICDGI----FYTPQYLNPS--VISLLKHMLQVDP 253
Query: 246 ARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIED 291
+R TIK+I+ H WF ++LP+ Y E+P+YS I+D
Sbjct: 254 MKRATIKDIREHEWFKQDLPK---------YLFPEDPSYSSTMIDD 290
>SN1L_MOUSE (Q60670) Probable serine/threonine-protein kinase SNF1LK
(EC 2.7.1.37) (HRT-20) (Myocardial SNF1-like kinase)
Length = 779
Score = 197 bits (501), Expect = 3e-50
Identities = 112/288 (38%), Positives = 165/288 (56%), Gaps = 23/288 (7%)
Query: 4 YEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKID----ENVAREIINHRSLRHPN 59
Y+V + +G GNF V +L RH+ TK VA+K I++ ++D E + RE+ + L HPN
Sbjct: 27 YDVERTLGKGNFAVVKLARHRVTKTQVAIKIIDKT-RLDSSNLEKIYREVQLMKLLNHPN 85
Query: 60 IIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQI 119
II+ +V+ T L IV E+A GE+FD + S G SE+EAR F Q++S V YCH+ I
Sbjct: 86 IIKLYQVMETKDMLYIVTEFAKNGEMFDYLTSNGHLSENEARQKFWQILSAVEYCHNHHI 145
Query: 120 CHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK 179
HRDLK EN LLD + +K+ DFG+ + VG+P Y APEV +EY+G
Sbjct: 146 VHRDLKTENLLLDSN--MDIKLADFGFGNFYKPGEPLSTCVGSPPYAAPEVFEGKEYEGP 203
Query: 180 LADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSR 239
DVWS GV LYV++ G+ PF D N R++ +++IP + +SQ+C L+ R
Sbjct: 204 QLDVWSLGVVLYVLVCGSLPF----DGPNLPTLRQRVLEGRFRIPFF--MSQDCETLIRR 257
Query: 240 IFVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQ 287
+ V PA+RITI +I+ H W A ++++P + +Q
Sbjct: 258 MLVVDPAKRITIAQIRQHRWM----------QADPTLLQQDDPAFDMQ 295
>AAK1_HUMAN (Q13131) 5'-AMP-activated protein kinase, catalytic
alpha-1 chain (EC 2.7.1.-) (AMPK alpha-1 chain)
Length = 550
Score = 196 bits (499), Expect = 6e-50
Identities = 113/286 (39%), Positives = 159/286 (55%), Gaps = 21/286 (7%)
Query: 10 IGSGNFGVARLMRHKDTKQLVAMKYIERGH----KIDENVAREIINHRSLRHPNIIRFKE 65
+G G FG ++ +H+ T VA+K + R + + REI N + RHP+II+ +
Sbjct: 24 LGVGTFGKVKVGKHELTGHKVAVKILNRQKIRSLDVVGKIRREIQNLKLFRHPHIIKLYQ 83
Query: 66 VVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQICHRDLK 125
V+ TP+ + +VMEY +GGELFD IC GR E E+R FQQ++SGV YCH + HRDLK
Sbjct: 84 VISTPSDIFMVMEYVSGGELFDYICKNGRLDEKESRRLFQQILSGVDYCHRHMVVHRDLK 143
Query: 126 LENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKLADVWS 185
EN LLD KI DFG S +++ G+P Y APEV+S R Y G D+WS
Sbjct: 144 PENVLLDAH--MNAKIADFGLSNMMSDGEFLRTSCGSPNYAAPEVISGRLYAGPEVDIWS 201
Query: 186 CGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRIFVASP 245
GV LY +L G PF+D P F+K + I + P Y++ S LL + P
Sbjct: 202 SGVILYALLCGTLPFDDDHVPTLFKKICDGI----FYTPQYLNPS--VISLLKHMLQVDP 255
Query: 246 ARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIED 291
+R +IK+I+ H WF ++LP+ Y E+P+YS I+D
Sbjct: 256 MKRASIKDIREHEWFKQDLPK---------YLFPEDPSYSSTMIDD 292
>RKI1_SECCE (Q02723) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 502
Score = 194 bits (494), Expect = 2e-49
Identities = 116/277 (41%), Positives = 159/277 (56%), Gaps = 19/277 (6%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGH----KIDENVAREIINHR--- 53
++ Y + K +G G F + HK T+ VA+K + R +++E REI R
Sbjct: 11 LKNYYLGKILGVGTFAKVIIAEHKHTRHKVAIKVLNRRQMRAPEMEEKAKREIKILRLFI 70
Query: 54 SLRHPNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSY 113
L HP+IIR EV++TP + +VMEY G+L D I R EDEAR FQQ+IS V Y
Sbjct: 71 DLIHPHIIRVYEVIVTPKDIFVVMEYCQNGDLLDYILEKRRLQEDEARRTFQQIISAVEY 130
Query: 114 CHSMQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRP--KSTVGTPAYIAPEVL 171
CH ++ HRDLK EN LLD +K+ DFG S+++H K++ G+ Y APEV+
Sbjct: 131 CHRNKVVHRDLKPENLLLDSK--YNVKLADFGL--SNVMHDGHFLKTSCGSLNYAAPEVI 186
Query: 172 SRREYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQ 231
S + Y G DVWSCGV LY +L GA PF+D + P F+K I Y +P +++S
Sbjct: 187 SGKLYAGPEIDVWSCGVILYALLCGAVPFDDDNIPNLFKK----IKGGTYILP--IYLSD 240
Query: 232 ECRHLLSRIFVASPARRITIKEIKSHPWFLKNLPREL 268
R L+SR+ + P +RITI EI+ H WF LPR L
Sbjct: 241 LVRDLISRMLIVDPMKRITIGEIRKHSWFQNRLPRYL 277
>SNF1_CANGA (Q00372) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 612
Score = 193 bits (491), Expect = 5e-49
Identities = 128/348 (36%), Positives = 184/348 (52%), Gaps = 35/348 (10%)
Query: 4 YEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERG----HKIDENVAREIINHRSLRHPN 59
Y++VK +G G+FG +L H T Q VA+K I + + + REI R LRHP+
Sbjct: 39 YQIVKTLGEGSFGKVKLAYHVTTGQKVALKIINKKVLAKSDMQGRIEREISYLRLLRHPH 98
Query: 60 IIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQI 119
II+ +V+ + + +V+EYA G ELFD I + SE EAR FFQQ+IS V YCH +I
Sbjct: 99 IIKLYDVIKSKDEIIMVIEYA-GNELFDYIVQRNKMSEQEARRFFQQIISAVEYCHRHKI 157
Query: 120 CHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK 179
HRDLK EN LLD +KI DFG S + K++ G+P Y APEV+S + Y G
Sbjct: 158 VHRDLKPENLLLDEHL--NVKIADFGLSNIMTDGNFLKTSCGSPNYAAPEVISGKLYAGP 215
Query: 180 LADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSR 239
DVWSCGV LYVML PF+D+ P F+ N + Y +P + +S L+ R
Sbjct: 216 EVDVWSCGVILYVMLCRRLPFDDESIPVLFKNISNGV----YTLPKF--LSPGASDLIKR 269
Query: 240 IFVASPARRITIKEIKSHPWFLKNL-----PRELTEMAQAVYYRKENPTYSLQSIEDIMK 294
+ + +P RI+I EI WF +L P++L + Q + +K +++ I+D M
Sbjct: 270 MLIVNPLNRISIHEIMQDEWFKVDLAEYLVPQDLKQQEQ--FNKKSGNEENVEEIDDEMV 327
Query: 295 IVEEAKNPPQASRSVGGFGWGGEEDDDEINEAEAELEEDEYEKRVKEA 342
+ S+++G D DEI EA E+ ++ A
Sbjct: 328 VT--------LSKTMG-------YDKDEIYEALESSEDTPAYNEIRNA 360
>MRK4_HUMAN (Q96L34) MAP/microtubule affinity-regulating kinase 4
(EC 2.7.1.37) (MAP/microtubule affinity-regulating
kinase like 1)
Length = 752
Score = 192 bits (488), Expect = 1e-48
Identities = 103/259 (39%), Positives = 152/259 (57%), Gaps = 11/259 (4%)
Query: 4 YEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVA---REIINHRSLRHPNI 60
Y +++ IG GNF +L RH T + VA+K I++ ++ RE+ + L HPNI
Sbjct: 59 YRLLRTIGKGNFAKVKLARHILTGREVAIKIIDKTQLNPSSLQKLFREVRIMKGLNHPNI 118
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
++ EV+ T L +VMEYA+ GE+FD + S GR E EAR F+Q++S V YCH I
Sbjct: 119 VKLFEVIETEKTLYLVMEYASAGEVFDYLVSHGRMKEKEARAKFRQIVSAVHYCHQKNIV 178
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLK EN LLD +KI DFG+S L S+ + G+P Y APE+ ++YDG
Sbjct: 179 HRDLKAENLLLDAE--ANIKIADFGFSNEFTLGSKLDTFCGSPPYAAPELFQGKKYDGPE 236
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
D+WS GV LY ++ G+ PF D N ++ R++ +Y++P Y +S +C +L R
Sbjct: 237 VDIWSLGVILYTLVSGSLPF----DGHNLKELRERVLRGKYRVPFY--MSTDCESILRRF 290
Query: 241 FVASPARRITIKEIKSHPW 259
V +PA+R T+++I W
Sbjct: 291 LVLNPAKRCTLEQIMKDKW 309
>ST29_HUMAN (Q8IWQ3) Serine/threonine-protein kinase 29 (EC
2.7.1.37) (BRSK2) (HUSSY-12)
Length = 736
Score = 191 bits (486), Expect = 2e-48
Identities = 111/295 (37%), Positives = 170/295 (57%), Gaps = 21/295 (7%)
Query: 4 YEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVA----REIINHRSLRHPN 59
Y + K +G G G+ +L H T Q VA+K + R K+ E+V REI + + HP+
Sbjct: 19 YRLEKTLGKGQTGLVKLGVHCVTCQKVAIKIVNR-EKLSESVLMKVEREIAILKLIEHPH 77
Query: 60 IIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQI 119
+++ +V +L +V+E+ +GGELFD + GR + EAR FF+Q+IS + +CHS I
Sbjct: 78 VLKLHDVYENKKYLYLVLEHVSGGELFDYLVKKGRLTPKEARKFFRQIISALDFCHSHSI 137
Query: 120 CHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK 179
CHRDLK EN LLD ++I DFG + + S +++ G+P Y PEV+ +YDG+
Sbjct: 138 CHRDLKPENLLLD--EKNNIRIADFGMASLQVGDSLLETSCGSPHYACPEVIRGEKYDGR 195
Query: 180 LADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSR 239
ADVWSCGV L+ +LVGA PF+D N R+ + ++ + +P + I +C+ LL
Sbjct: 196 KADVWSCGVILFALLVGALPFDDD----NLRQLLEKVKRGVFHMPHF--IPPDCQSLLRG 249
Query: 240 IFVASPARRITIKEIKSHPWFL--KNLPRELTEMAQAVYYRKENPTYSLQSIEDI 292
+ ARR+T++ I+ H W++ KN P + + V R SL S+EDI
Sbjct: 250 MSEVDAARRLTLEHIQKHIWYIGGKNEPEPEQPIPRKVQIR------SLPSLEDI 298
>SN1L_HUMAN (P57059) Probable serine/threonine-protein kinase SNF1LK
(EC 2.7.1.37)
Length = 786
Score = 191 bits (485), Expect = 2e-48
Identities = 110/262 (41%), Positives = 157/262 (58%), Gaps = 14/262 (5%)
Query: 4 YEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKID----ENVAREIINHRSLRHPN 59
Y++ + +G GNF V +L RH+ TK VA+K I++ ++D E + RE+ + L HP+
Sbjct: 27 YDIERTLGKGNFAVVKLARHRVTKTQVAIKIIDKT-RLDSSNLEKIYREVQLMKLLNHPH 85
Query: 60 IIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQI 119
II+ +V+ T L IV E+A GE+FD + S G SE+EAR F Q++S V YCH I
Sbjct: 86 IIKLYQVMETKDMLYIVTEFAKNGEMFDYLTSNGHLSENEARKKFWQILSAVEYCHDHHI 145
Query: 120 CHRDLKLENTLLDGSPAPRLK-ICDFGYSKSSLLHSRPKST-VGTPAYIAPEVLSRREYD 177
HRDLK EN LLDG+ +L DFG+ + P ST G+P Y APEV +EY+
Sbjct: 146 VHRDLKTENLLLDGNMDIKLAGTEDFGFG-NFYKSGEPLSTWCGSPPYAAPEVFEGKEYE 204
Query: 178 GKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLL 237
G D+WS GV LYV++ G+ PF D N R++ +++IP + +SQ+C L+
Sbjct: 205 GPQLDIWSLGVVLYVLVCGSLPF----DGPNLPTLRQRVLEGRFRIPFF--MSQDCESLI 258
Query: 238 SRIFVASPARRITIKEIKSHPW 259
R+ V PARRITI +I+ H W
Sbjct: 259 RRMLVVDPARRITIAQIRQHRW 280
>KI11_HUMAN (Q8TDC3) Probable serine/threonine-protein kinase
KIAA1811 (EC 2.7.1.37)
Length = 794
Score = 190 bits (483), Expect = 4e-48
Identities = 98/262 (37%), Positives = 158/262 (59%), Gaps = 13/262 (4%)
Query: 4 YEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVA----REIINHRSLRHPN 59
Y + K +G G G+ +L H T Q VA+K + R K+ E+V REI + + HP+
Sbjct: 50 YRLEKTLGKGQTGLVKLGVHCITGQKVAIKIVNR-EKLSESVLMKVEREIAILKLIEHPH 108
Query: 60 IIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQI 119
+++ +V +L +V+E+ +GGELFD + GR + EAR FF+Q++S + +CHS I
Sbjct: 109 VLKLHDVYENKKYLYLVLEHVSGGELFDYLVKKGRLTPKEARKFFRQIVSALDFCHSYSI 168
Query: 120 CHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK 179
CHRDLK EN LLD ++I DFG + + S +++ G+P Y PEV+ +YDG+
Sbjct: 169 CHRDLKPENLLLD--EKNNIRIADFGMASLQVGDSLLETSCGSPHYACPEVIKGEKYDGR 226
Query: 180 LADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSR 239
AD+WSCGV L+ +LVGA PF+D N R+ + ++ + +P + I +C+ LL
Sbjct: 227 RADMWSCGVILFALLVGALPFDDD----NLRQLLEKVKRGVFHMPHF--IPPDCQSLLRG 280
Query: 240 IFVASPARRITIKEIKSHPWFL 261
+ P +R+++++I+ HPW+L
Sbjct: 281 MIEVEPEKRLSLEQIQKHPWYL 302
>SNF1_YEAST (P06782) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 633
Score = 190 bits (482), Expect = 5e-48
Identities = 113/269 (42%), Positives = 153/269 (56%), Gaps = 13/269 (4%)
Query: 4 YEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERG----HKIDENVAREIINHRSLRHPN 59
Y++VK +G G+FG +L H T Q VA+K I + + + REI R LRHP+
Sbjct: 55 YQIVKTLGEGSFGKVKLAYHTTTGQKVALKIINKKVLAKSDMQGRIEREISYLRLLRHPH 114
Query: 60 IIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQI 119
II+ +V+ + + +V+EYA G ELFD I + SE EAR FFQQ+IS V YCH +I
Sbjct: 115 IIKLYDVIKSKDEIIMVIEYA-GNELFDYIVQRDKMSEQEARRFFQQIISAVEYCHRHKI 173
Query: 120 CHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK 179
HRDLK EN LLD +KI DFG S + K++ G+P Y APEV+S + Y G
Sbjct: 174 VHRDLKPENLLLDEHL--NVKIADFGLSNIMTDGNFLKTSCGSPNYAAPEVISGKLYAGP 231
Query: 180 LADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSR 239
DVWSCGV LYVML PF+D+ P F+ N + Y +P + +S L+ R
Sbjct: 232 EVDVWSCGVILYVMLCRRLPFDDESIPVLFKNISNGV----YTLPKF--LSPGAAGLIKR 285
Query: 240 IFVASPARRITIKEIKSHPWFLKNLPREL 268
+ + +P RI+I EI WF +LP L
Sbjct: 286 MLIVNPLNRISIHEIMQDDWFKVDLPEYL 314
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,473,790
Number of Sequences: 164201
Number of extensions: 1829144
Number of successful extensions: 13472
Number of sequences better than 10.0: 1676
Number of HSP's better than 10.0 without gapping: 1044
Number of HSP's successfully gapped in prelim test: 632
Number of HSP's that attempted gapping in prelim test: 8530
Number of HSP's gapped (non-prelim): 2260
length of query: 351
length of database: 59,974,054
effective HSP length: 111
effective length of query: 240
effective length of database: 41,747,743
effective search space: 10019458320
effective search space used: 10019458320
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC134322.21


