

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134322.18 - phase: 0
(488 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
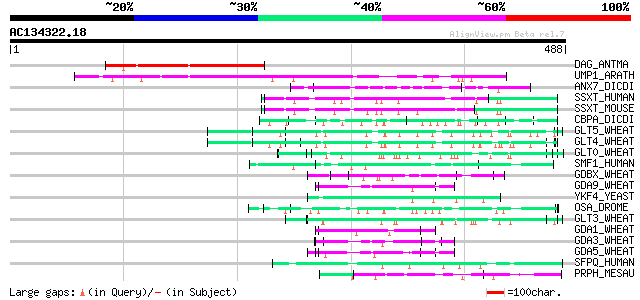
Score E
Sequences producing significant alignments: (bits) Value
DAG_ANTMA (Q38732) DAG protein, chloroplast precursor 145 2e-34
UMP1_ARATH (Q9LKA5) Unknown mitochondrial protein At3g15000 141 4e-33
ANX7_DICDI (P24639) Annexin A7 (Annexin VII) (Synexin) 88 4e-17
SSXT_HUMAN (Q15532) SSXT protein (Synovial sarcoma, translocated... 75 4e-13
SSXT_MOUSE (Q62280) SSXT protein (SYT protein) (Synovial sarcoma... 72 4e-12
CBPA_DICDI (P35085) Calcium-binding protein 68 6e-11
GLT5_WHEAT (P10388) Glutenin, high molecular weight subunit DX5 ... 64 1e-09
GLT4_WHEAT (P08489) Glutenin, high molecular weight subunit PW21... 63 1e-09
GLT0_WHEAT (P10387) Glutenin, high molecular weight subunit DY10... 62 4e-09
SMF1_HUMAN (O14497) SWI/SNF-related, matrix-associated, actin-de... 59 2e-08
GDBX_WHEAT (P21292) Gamma-gliadin precursor 59 4e-08
GDA9_WHEAT (P18573) Alpha/beta-gliadin MM1 precursor (Prolamin) 59 4e-08
YKF4_YEAST (P35732) Hypothetical 84.0 kDa protein in NUP120-CSE4... 58 6e-08
OSA_DROME (Q8IN94) Trithorax group protein OSA (Eyelid protein) 58 6e-08
GLT3_WHEAT (P08488) Glutenin, high molecular weight subunit 12 p... 57 1e-07
GDA1_WHEAT (P04721) Alpha/beta-gliadin A-I precursor (Prolamin) 57 1e-07
GDA3_WHEAT (P04723) Alpha/beta-gliadin A-III precursor (Prolamin) 56 2e-07
GDA5_WHEAT (P04725) Alpha/beta-gliadin A-V precursor (Prolamin) 55 3e-07
SFPQ_HUMAN (P23246) Splicing factor, proline-and glutamine-rich ... 55 4e-07
PRPH_MESAU (P06680) Acidic proline-rich protein HP43A precursor 55 4e-07
>DAG_ANTMA (Q38732) DAG protein, chloroplast precursor
Length = 230
Score = 145 bits (366), Expect = 2e-34
Identities = 78/145 (53%), Positives = 98/145 (66%), Gaps = 7/145 (4%)
Query: 85 IQSRSFRSTSISLL-----SSRYGETSELSPEIGPDTILFEGCDYNHWLFVCDFPRDNKP 139
I+SRS ++ L SSR + S E +TI+ GCDYNHWL V +FP+D P
Sbjct: 43 IKSRSAAYPTVRALTDGEYSSRRNNNNNNSGE-ERETIMLPGCDYNHWLIVMEFPKDPAP 101
Query: 140 PPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFV 199
E+MI Y T A L S+EEAKK +YA STTTYTGFQ +TEE S+KF+G+PGV++V
Sbjct: 102 TREQMIDTYLNTLATVLG-SMEEAKKNMYAFSTTTYTGFQCTVTEETSEKFKGLPGVLWV 160
Query: 200 LPDSYIDPVNKQYGGDQYIEGQIIP 224
LPDSYID NK YGGD+Y+ G+IIP
Sbjct: 161 LPDSYIDVKNKDYGGDKYVNGEIIP 185
>UMP1_ARATH (Q9LKA5) Unknown mitochondrial protein At3g15000
Length = 395
Score = 141 bits (355), Expect = 4e-33
Identities = 128/413 (30%), Positives = 176/413 (41%), Gaps = 69/413 (16%)
Query: 58 ATSLSVPITGSISSHFSLSKSPNVVDGIQSRS----------FRSTSISLLSSRYGETSE 107
A SLS T S +S L+KSP + SRS FR +S+ TS
Sbjct: 15 AKSLSFLFTRSFASSAPLAKSP--ASSLLSRSRPLVAAFSSVFRGGLVSVKGLSTQATSS 72
Query: 108 LSPEIGP--------DTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNIS 159
+ P +TIL +GCD+ HWL V + P +P +E+I Y +T A+ + S
Sbjct: 73 SLNDPNPNWSNRPPKETILLDGCDFEHWLVVVE-PPQGEPTRDEIIDSYIKTLAQIVG-S 130
Query: 160 VEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIE 219
+EA+ KIY+ ST Y F A+++E+ S K + + V +VLPDSY+D NK YGG+ +I+
Sbjct: 131 EDEARMKIYSVSTRCYYAFGALVSEDLSHKLKELSNVRWVLPDSYLDVRNKDYGGEPFID 190
Query: 220 GQIIPRPPPV--QFGRNLGGRRDYRQNNQLP--NNRGNPSYNNRDSMPRDGRNYGPPQNF 275
G+ +P P ++ RN + + N P N+R R++M PP
Sbjct: 191 GKAVPYDPKYHEEWIRNNARANERNRRNDRPRNNDRSRNFERRRENMAGGPPPQRPPMGG 250
Query: 276 PPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGY 335
PP H S PP G P + Q + P + R+PP
Sbjct: 251 PPPPPHIGGSAPPPPHMGGSAPPPPHM---GQNYGPPPPNNMGGPRHPPP---------- 297
Query: 336 LPKQNYGQAPQ-NYSAPQAPQNYSQQQTYGPASPQY---PPQQSFGPLGEGERRNYAPQQ 391
YG PQ N P+ PQNY G P Y PP + G AP
Sbjct: 298 -----YGAPPQNNMGGPRPPQNYG-----GTPPPNYGGAPPANNMGG---------APPP 338
Query: 392 NF--GPP---GEVDRRNY--APQQNFGPPRQGERRNYVPQQNFGPPGQGERGN 437
N+ GPP G V Y AP QN +QG QN PP + GN
Sbjct: 339 NYGGGPPPQYGAVPPPQYGGAPPQNNNYQQQGSGMQQPQYQNNYPPNRDGSGN 391
>ANX7_DICDI (P24639) Annexin A7 (Annexin VII) (Synexin)
Length = 462
Score = 88.2 bits (217), Expect = 4e-17
Identities = 61/150 (40%), Positives = 71/150 (46%), Gaps = 20/150 (13%)
Query: 248 PNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQ 307
P N+G P +N P+ G+ P Q +PPQQ + PPQQ QQ Q
Sbjct: 4 PPNQGYPPQSNS---PQPGQYGAPQQGYPPQQGYPPQQGYPPQQGYPPQQGY----PPQQ 56
Query: 308 RFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPAS 367
+PPQQ Y Q YPPQQ Y QGY P+Q Y PQ PQ Q Y QQ Y P
Sbjct: 57 GYPPQQGY-PPQQGYPPQQGY-PPQQGYPPQQGY--PPQQGYPPQ--QGYPPQQGY-PPQ 109
Query: 368 PQYPPQQSFGPLGEGERRNYAPQQNFGPPG 397
YPPQQ + P G Y PQQ + P G
Sbjct: 110 QGYPPQQGYPPQG------YPPQQGYPPVG 133
Score = 85.1 bits (209), Expect = 4e-16
Identities = 71/195 (36%), Positives = 86/195 (43%), Gaps = 47/195 (24%)
Query: 268 NYGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQS 327
+Y P Q +PPQ N Q P Q QQ +PPQQ Y Q YPPQQ
Sbjct: 2 SYPPNQGYPPQSNSPQ----PGQYGAPQQG-----------YPPQQGYPP-QQGYPPQQG 45
Query: 328 YDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNY 387
Y QGY P+Q Y PQ Q Y QQ Y P YPPQQ + P ++ Y
Sbjct: 46 YPP-QQGYPPQQGY--------PPQ--QGYPPQQGY-PPQQGYPPQQGYPP-----QQGY 88
Query: 388 APQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFG----PPGQGERGNSVPSEG 443
PQQ + P ++ Y PQQ + PP+QG Y PQQ + PP QG VP
Sbjct: 89 PPQQGYPP-----QQGYPPQQGY-PPQQG----YPPQQGYPPQGYPPQQGYPPVGVPVGV 138
Query: 444 GWDFKPSYMEEFEQG 458
F P + + QG
Sbjct: 139 PVGFAPGMVVGYHQG 153
>SSXT_HUMAN (Q15532) SSXT protein (Synovial sarcoma, translocated to
X chromosome) (SYT protein)
Length = 418
Score = 75.1 bits (183), Expect = 4e-13
Identities = 69/212 (32%), Positives = 89/212 (41%), Gaps = 29/212 (13%)
Query: 225 RPPPVQFGRNLGGRRDYRQNNQLPN------NRGNPSYNNRDSMPRDGRNYGPPQNFPPQ 278
+PP Q+ GG + Y Q Q P N+GN R P Y PPQ PPQ
Sbjct: 209 QPPSQQYNMPQGGGQHY-QGQQPPMGMMGQVNQGNHMMGQRQIPP-----YRPPQQGPPQ 262
Query: 279 QNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQR--YPPQ---QSYDQASQ 333
Q GQ + Q + G Q P + +Q++ P D Q+ YP Q + Y+ +SQ
Sbjct: 263 QYSGQEDYYGDQYSHGGQGPP---EGMNQQYYPDGHNDYGYQQPSYPEQGYDRPYEDSSQ 319
Query: 334 GYLPKQN--YGQAPQNYSAPQAPQNYSQQQTYGPASPQYP-PQQSFGPLGEGERRNYAPQ 390
Y N YGQ Y P Q Y QQ P YP QQ +GP G PQ
Sbjct: 320 HYYEGGNSQYGQQQDAYQGPPPQQGYPPQQQQYPGQQGYPGQQQGYGPSQGGP----GPQ 375
Query: 391 QNFGPPGEVDR-RNYAPQQNFGPPRQGERRNY 421
P G+ + Y P Q GPP+ ++R Y
Sbjct: 376 YPNYPQGQGQQYGGYRPTQP-GPPQPPQQRPY 406
Score = 68.6 bits (166), Expect = 4e-11
Identities = 71/292 (24%), Positives = 106/292 (35%), Gaps = 53/292 (18%)
Query: 222 IIPRPPPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGR-NYGPPQNFPPQQN 280
++P PP GG N G P +MP DG GPP Q
Sbjct: 74 LLPAPPTQNMPMGPGGM----------NQSGPPPPPRSHNMPSDGMVGGGPPAPHMQNQM 123
Query: 281 HGQA---SHIPPQ----QNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQ 333
+GQ +H+P Q + ++ + S+S ++ S + P Q+ SQ
Sbjct: 124 NGQMPGPNHMPMQGPGPNQLNMTNSSMNMPSSSHGSMGGYNHSVPSSQSMPVQNQMTMSQ 183
Query: 334 GYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNF 393
G P NYG P N S Q GP Q PP Q + G + Q
Sbjct: 184 GQ-PMGNYGPRP----------NMSMQPNQGPMMHQQPPSQQYNMPQGGGQHYQGQQPPM 232
Query: 394 GPPGEVDRRNYAPQQN----FGPPRQGERRNYVPQQNF----------GPP--------- 430
G G+V++ N+ Q + PP+QG + Y Q+++ GPP
Sbjct: 233 GMMGQVNQGNHMMGQRQIPPYRPPQQGPPQQYSGQEDYYGDQYSHGGQGPPEGMNQQYYP 292
Query: 431 -GQGERGNSVPSEGGWDFKPSYMEEFEQGQKGNNHAEEQKESQQRFPPPGPG 481
G + G PS + Y + + +G N Q++ + PPP G
Sbjct: 293 DGHNDYGYQQPSYPEQGYDRPYEDSSQHYYEGGNSQYGQQQDAYQGPPPQQG 344
>SSXT_MOUSE (Q62280) SSXT protein (SYT protein) (Synovial sarcoma
associated Ss18-alpha)
Length = 418
Score = 71.6 bits (174), Expect = 4e-12
Identities = 66/212 (31%), Positives = 87/212 (40%), Gaps = 38/212 (17%)
Query: 225 RPPPVQFGRNLGGRRDYRQNNQLPN------NRGNPSYNNRDSMPRDGRNYGPPQNFPPQ 278
+PP Q+ GG + Y Q Q P N+G+ R P Y PPQ PPQ
Sbjct: 209 QPPSQQYNMPPGGAQHY-QGQQAPMGLMGQVNQGSHMMGQRQMPP-----YRPPQQGPPQ 262
Query: 279 QNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQR--YPPQ---QSYDQASQ 333
Q GQ + Q + G Q P + +Q++ P D Q+ YP Q + Y+ +SQ
Sbjct: 263 QYSGQEDYYGDQYSHGGQGPP---EGMNQQYYPDGHNDYGYQQPSYPEQGYDRPYEDSSQ 319
Query: 334 GYLPKQN--YGQAPQNYSAPQAPQNY--------------SQQQTYGPASPQYPPQQSFG 377
Y N YGQ Y P Q Y QQQ+YGP+ PQ
Sbjct: 320 HYYEGGNSQYGQQQDAYQGPPPQQGYPPQQQQYPGQQGYPGQQQSYGPSQGGPGPQYPNY 379
Query: 378 PLGEGER-RNYAPQQNFGPPGEVDRRNYAPQQ 408
P G+G++ Y P Q GPP +R Y Q
Sbjct: 380 PQGQGQQYGGYRPTQP-GPPQPPQQRPYGYDQ 410
Score = 62.0 bits (149), Expect = 3e-09
Identities = 69/295 (23%), Positives = 106/295 (35%), Gaps = 59/295 (20%)
Query: 222 IIPRPPPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGR-NYGPPQNFPPQQN 280
++P PP GG + G P +MP DG GPP Q
Sbjct: 74 LLPAPPTQTMPMGPGGM----------SQSGPPPPPRSHNMPSDGMVGGGPPAPHMQNQM 123
Query: 281 HGQA---SHIPPQQNIGQQQPNIRIDSTSQRFPPQQ-------SYDQASQRYPPQQSYDQ 330
+GQ +H+P Q G + + ++S P ++ S + P Q+
Sbjct: 124 NGQMPGPNHMPMQ---GPGPSQLSMTNSSMNMPSSNHGSMGGYNHSVPSSQSMPVQNQMT 180
Query: 331 ASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQ 390
SQG P NYG P N + Q GP Q PP Q + G + Q
Sbjct: 181 MSQGQ-PMGNYGPRP----------NMNMQPNQGPMMHQQPPSQQYNMPPGGAQHYQGQQ 229
Query: 391 QNFGPPGEVDRRNYAPQQN----FGPPRQGERRNYVPQQNF----------GPP------ 430
G G+V++ ++ Q + PP+QG + Y Q+++ GPP
Sbjct: 230 APMGLMGQVNQGSHMMGQRQMPPYRPPQQGPPQQYSGQEDYYGDQYSHGGQGPPEGMNQQ 289
Query: 431 ----GQGERGNSVPSEGGWDFKPSYMEEFEQGQKGNNHAEEQKESQQRFPPPGPG 481
G + G PS + Y + + +G N Q++ + PPP G
Sbjct: 290 YYPDGHNDYGYQQPSYPEQGYDRPYEDSSQHYYEGGNSQYGQQQDAYQGPPPQQG 344
>CBPA_DICDI (P35085) Calcium-binding protein
Length = 467
Score = 67.8 bits (164), Expect = 6e-11
Identities = 68/235 (28%), Positives = 83/235 (34%), Gaps = 49/235 (20%)
Query: 270 GPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQR------YP 323
G P +PPQQ S++PP G QQP ++PPQQ Q+ YP
Sbjct: 41 GAPGQYPPQQPGAPGSNLPPYP--GTQQPGA--PGAPGQYPPQQPGQYPPQQPGAPGQYP 96
Query: 324 PQQS-------YDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQ----TYGPASP---- 368
PQQ G P Q GQ P AP Y QQ Y P P
Sbjct: 97 PQQPGQPGYPPQQPGQSGQYPPQQPGQPGYPPQQPGAPGQYPPQQGQPGQYPPQQPGQPG 156
Query: 369 --------QYPPQQ-----SFGPLGEGERRNYAPQQNF----------GPPGEVDRRNYA 405
QYPPQQ ++ P G+ Y PQQ G PG +
Sbjct: 157 QYPPQQQGQYPPQQPGQPGAYPPQQPGQPGAYPPQQGVQNTLAKTGAPGQPGVPPPQGAY 216
Query: 406 PQQNFGPPRQGERRNYVPQQNFGPPGQGERGNSVPSEGGWDFKPSYMEEFEQGQK 460
P Q PP+QG P PP QG+ G P + P + GQ+
Sbjct: 217 PGQPGVPPQQGAYPGQQPPMGAYPP-QGQPGAYPPQGQPGAYPPQQQQVAYPGQQ 270
Score = 58.9 bits (141), Expect = 3e-08
Identities = 73/248 (29%), Positives = 93/248 (37%), Gaps = 49/248 (19%)
Query: 220 GQIIPRPP--PVQFGRNLGGRRDYR-----QNNQLPNNR-GNPSYNNRDSMPRDGRNYGP 271
GQ P+ P P Q+ G+ Y Q+ Q P + G P Y + G
Sbjct: 82 GQYPPQQPGAPGQYPPQQPGQPGYPPQQPGQSGQYPPQQPGQPGYPPQQP--------GA 133
Query: 272 PQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQA 331
P +PPQQ GQ PPQQ QP ++PPQQ + YPPQQ
Sbjct: 134 PGQYPPQQ--GQPGQYPPQQ---PGQPGQYPPQQQGQYPPQQPGQPGA--YPPQQPGQPG 186
Query: 332 SQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQ-----------SFGPLG 380
+ Y P+Q AP P Q Y P P PPQQ ++ P G
Sbjct: 187 A--YPPQQGVQNTLAKTGAPGQPGVPPPQGAY-PGQPGVPPQQGAYPGQQPPMGAYPPQG 243
Query: 381 E-------GERRNYAPQQN-FGPPGEVDRRN-YAPQQNFGPPRQGE---RRNYVPQQNFG 428
+ G+ Y PQQ PG+ Y PQQ P +QG ++ P Q
Sbjct: 244 QPGAYPPQGQPGAYPPQQQQVAYPGQQPPMGAYPPQQGAYPGQQGAYPGQQGAYPGQQGA 303
Query: 429 PPGQGERG 436
PGQ G
Sbjct: 304 YPGQPPMG 311
Score = 51.6 bits (122), Expect = 5e-06
Identities = 52/159 (32%), Positives = 63/159 (38%), Gaps = 41/159 (25%)
Query: 350 APQAPQNYSQQQ-------------TYGPASP----QYPPQQ--SFGPLGEGERRNYAPQ 390
AP AP Y QQ T P +P QYPPQQ + P G Y PQ
Sbjct: 39 APGAPGQYPPQQPGAPGSNLPPYPGTQQPGAPGAPGQYPPQQPGQYPPQQPGAPGQYPPQ 98
Query: 391 QNFGPPGEVDRRNYAPQQ----NFGPPRQGERRNYVPQQNFG----PPGQGERGNSVPSE 442
Q G PG Y PQQ PP+Q + Y PQQ PP QG+ G P +
Sbjct: 99 QP-GQPG------YPPQQPGQSGQYPPQQPGQPGYPPQQPGAPGQYPPQQGQPGQYPPQQ 151
Query: 443 GGWDFKPSYMEEFEQGQKGNNHAEEQKESQQRFPPPGPG 481
G +P +QGQ + +Q +PP PG
Sbjct: 152 PG---QPGQYPPQQQGQ----YPPQQPGQPGAYPPQQPG 183
Score = 47.8 bits (112), Expect = 7e-05
Identities = 56/190 (29%), Positives = 67/190 (34%), Gaps = 29/190 (15%)
Query: 246 QLPNNRGNPSYNNRDSMPRDGRNYGPPQN--FPPQQNHGQASHIPPQQ--NIGQQQPNIR 301
Q P +G P + G+ Y P Q +PPQQ GQ PPQQ G P
Sbjct: 136 QYPPQQGQPGQYPPQQPGQPGQ-YPPQQQGQYPPQQP-GQPGAYPPQQPGQPGAYPPQQG 193
Query: 302 IDSTSQR--------FPPQQSYDQASQRYPPQQSY---DQASQGYLPKQNYGQAPQNYSA 350
+ +T + PP Q PPQQ Q G P Q A
Sbjct: 194 VQNTLAKTGAPGQPGVPPPQGAYPGQPGVPPQQGAYPGQQPPMGAYPPQGQPGAYPPQGQ 253
Query: 351 PQAPQNYSQQQTYGPASP---QYPPQQSFGPLGEGERRNYAPQQNFGP------PGEVDR 401
P A QQ Y P YPPQQ P G++ Y QQ P PG+
Sbjct: 254 PGAYPPQQQQVAYPGQQPPMGAYPPQQGAYP---GQQGAYPGQQGAYPGQQGAYPGQPPM 310
Query: 402 RNYAPQQNFG 411
Y QQ +G
Sbjct: 311 GAYPGQQQYG 320
Score = 37.7 bits (86), Expect = 0.067
Identities = 41/137 (29%), Positives = 50/137 (35%), Gaps = 12/137 (8%)
Query: 263 PRDGRNYG--PPQN-FPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQAS 319
P+ G G PP +PPQ GQ PPQ G P + + + PP +Y
Sbjct: 224 PQQGAYPGQQPPMGAYPPQ---GQPGAYPPQGQPGAYPPQQQQVAYPGQQPPMGAYPPQQ 280
Query: 320 QRYPPQQSYDQASQGYLPKQN---YGQAPQNYSAPQAPQNYS--QQQTYGPASPQYPPQQ 374
YP QQ QG P Q GQ P Q S Q T + YP Q
Sbjct: 281 GAYPGQQGAYPGQQGAYPGQQGAYPGQPPMGAYPGQQQYGVSAYSQTTAAYQTGAYPGQT 340
Query: 375 SFGPLGEGERRNYAPQQ 391
G + G+ YA Q
Sbjct: 341 PMG-VYPGQTTAYAAYQ 356
>GLT5_WHEAT (P10388) Glutenin, high molecular weight subunit DX5
precursor
Length = 839
Score = 63.5 bits (153), Expect = 1e-09
Identities = 75/257 (29%), Positives = 96/257 (37%), Gaps = 23/257 (8%)
Query: 243 QNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQ-QNIGQQQPNIR 301
Q Q P P + P G+ G Q Q GQ + P Q GQ QP
Sbjct: 232 QQGQQPGQGQQPGQGQQGQQPGQGQQPGQGQQ-GQQLGQGQQGYYPTSLQQSGQGQPGYY 290
Query: 302 IDSTSQRFPPQQSYDQASQRYPPQQSY----DQASQGYLPKQNY-----GQAPQNYSAPQ 352
S Q Q Y S + P Q Q +QG P Q GQ Q Q
Sbjct: 291 PTSLQQLGQGQSGYYPTSPQQPGQGQQPGQLQQPAQGQQPGQGQQGQQPGQGQQGQQPGQ 350
Query: 353 APQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQ-NFGPPGE-VDRRNYAPQQNF 410
Q Q Y P SPQ Q G ++ QQ G G+ V + A Q
Sbjct: 351 GQQPGQGQPGYYPTSPQQSGQGQPGYYPTSSQQPTQSQQPGQGQQGQQVGQGQQAQQPGQ 410
Query: 411 GP-PRQGERRNYV--PQQNFGPPGQGERGNSV--PSEGGWDFKPSYMEEFEQGQKGNNHA 465
G P QG+ Y PQQ+ GQG+ G + P + G +P +++ QGQKG
Sbjct: 411 GQQPGQGQPGYYPTSPQQS----GQGQPGYYLTSPQQSGQGQQPGQLQQSAQGQKGQQPG 466
Query: 466 EEQKESQ-QRFPPPGPG 481
+ Q+ Q Q+ PG G
Sbjct: 467 QGQQPGQGQQGQQPGQG 483
Score = 58.9 bits (141), Expect = 3e-08
Identities = 87/325 (26%), Positives = 117/325 (35%), Gaps = 38/325 (11%)
Query: 175 YTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQFGRN 234
Y G + + + F GIP ++ S P Y GQ P+ P
Sbjct: 87 YPGETTPPQQLQQRIFWGIPALLKRYYPSVTCPQQVSY-----YPGQASPQRPGQGQQPG 141
Query: 235 LGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQNIG 294
G + Y + Q P P + P + G Q Q GQ Q G
Sbjct: 142 QGQQGYYPTSPQQPGQWQQPEQGQQGYYPTSPQQPGQLQQPAQGQQPGQGQQ---GQQPG 198
Query: 295 QQQPNIRIDSTSQRFPPQ----------QSYDQASQRYPPQQSYD--QASQGYLPKQNY- 341
Q QP ++SQ P Q Q QA Q P Q Q QG P Q
Sbjct: 199 QGQPGY-YPTSSQLQPGQLQQPAQGQQGQQPGQAQQGQQPGQGQQPGQGQQGQQPGQGQQ 257
Query: 342 -GQAPQNYSAPQAPQNY--SQQQTYGPASPQYPPQQSFGPLGEGERRNY--APQQNFGPP 396
GQ Q Q Q Y + Q G P Y P S LG+G+ Y +PQQ P
Sbjct: 258 PGQGQQGQQLGQGQQGYYPTSLQQSGQGQPGYYP-TSLQQLGQGQSGYYPTSPQQ----P 312
Query: 397 GEVDRRNYAPQQNFG-PPRQGERRNYVPQQNFG-PPGQGERGNSVPSEGGWDFKPSYMEE 454
G+ + Q G P QG++ Q G PGQG++ P +G + P+ ++
Sbjct: 313 GQGQQPGQLQQPAQGQQPGQGQQGQQPGQGQQGQQPGQGQQ----PGQGQPGYYPTSPQQ 368
Query: 455 FEQGQKGNNHAEEQKESQQRFPPPG 479
QGQ G Q+ +Q + P G
Sbjct: 369 SGQGQPGYYPTSSQQPTQSQQPGQG 393
Score = 54.3 bits (129), Expect = 7e-07
Identities = 82/300 (27%), Positives = 115/300 (38%), Gaps = 52/300 (17%)
Query: 214 GDQYIEGQIIPRPPPVQFGRNLG-------GRRDYRQNNQLPNNRGNPSYNNRDSM-PRD 265
G Q +GQ +P Q G+ G G+ Y + + +G P Y S P
Sbjct: 327 GQQPGQGQQGQQPGQGQQGQQPGQGQQPGQGQPGYYPTSPQQSGQGQPGYYPTSSQQPTQ 386
Query: 266 GRNYGPPQNFPPQQNHGQASHIPPQ-QNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPP 324
+ G Q Q GQ + P Q Q GQ QP S Q Q Y S P
Sbjct: 387 SQQPGQGQQ-GQQVGQGQQAQQPGQGQQPGQGQPGYYPTSPQQSGQGQPGYYLTS----P 441
Query: 325 QQSYD--------QASQGYLPKQ-NYGQAPQNYSAPQAPQNYSQQQTYGPASPQY---PP 372
QQS Q++QG +Q GQ P Q P Q Q G P Y P
Sbjct: 442 QQSGQGQQPGQLQQSAQGQKGQQPGQGQQPGQGQQGQQPGQGQQGQQPGQGQPGYYPTSP 501
Query: 373 QQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAP---QQNFGP-----PRQGERRNYVPQ 424
QQS G+G++ Q G PG P Q + P P QG++ + Q
Sbjct: 502 QQS----GQGQQPGQWQQPGQGQPGYYPTSPLQPGQGQPGYDPTSPQQPGQGQQPGQLQQ 557
Query: 425 QNFGPPGQGERGNSVPSEGGWDFKPSYMEEFE---QGQKGNNHAEEQKESQQRFPPPGPG 481
P QG++G + ++G +P+ +++ + QGQ+G + Q+ Q PG G
Sbjct: 558 -----PAQGQQGQQL-AQGQQGQQPAQVQQGQQPAQGQQGQQLGQGQQGQQ-----PGQG 606
Score = 52.4 bits (124), Expect = 3e-06
Identities = 73/268 (27%), Positives = 97/268 (35%), Gaps = 41/268 (15%)
Query: 243 QNNQLPNNRGNPSYNNRDSMPRDGRN-----YGPPQNFP--PQQNHGQASHIPPQQNIGQ 295
Q Q P P + P G+ G P +P PQQ+ GQ Q GQ
Sbjct: 460 QKGQQPGQGQQPGQGQQGQQPGQGQQGQQPGQGQPGYYPTSPQQS-GQGQQPGQWQQPGQ 518
Query: 296 QQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQ 355
QP S Q Q YD S + P Q G L + GQ Q + Q Q
Sbjct: 519 GQPGYYPTSPLQPGQGQPGYDPTSPQQPGQGQ----QPGQLQQPAQGQQGQQLAQGQQGQ 574
Query: 356 NYSQ-QQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPR 414
+Q QQ PA Q Q LG+G++ Q G G+ + QQ P
Sbjct: 575 QPAQVQQGQQPAQGQQGQQ-----LGQGQQ---GQQPGQGQQGQQPAQGQQGQQ----PG 622
Query: 415 QGERRNYVPQQNFG-PPGQGERGN--------SVPSEGGWDFKPSYMEEFEQGQKGN--N 463
QG+ Q G PGQG++ + P E G +P ++ QGQ G
Sbjct: 623 QGQHGQQPGQGQQGQQPGQGQQPGQGQPWYYPTSPQESGQGQQPGQWQQPGQGQPGYYLT 682
Query: 464 HAEEQKESQQRFPP-----PGPGNFTGE 486
+ + QQ + P PG G G+
Sbjct: 683 FSVAARTGQQGYYPTSLQQPGQGQQPGQ 710
Score = 43.1 bits (100), Expect = 0.002
Identities = 52/189 (27%), Positives = 73/189 (38%), Gaps = 18/189 (9%)
Query: 214 GDQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRN--YGP 271
G Q +GQ +P Q G+ + +Q Q + + P + P G+ G
Sbjct: 591 GQQLGQGQQGQQPGQGQQGQQPAQGQQGQQPGQGQHGQ-QPGQGQQGQQPGQGQQPGQGQ 649
Query: 272 PQNFPPQ-QNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQS--- 327
P +P Q GQ Q GQ QP + + QQ Y S + P Q
Sbjct: 650 PWYYPTSPQESGQGQQPGQWQQPGQGQPGYYLTFSVAARTGQQGYYPTSLQQPGQGQQPG 709
Query: 328 -YDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQ----QQTYGPASPQYPPQ-QSFGPL-- 379
+ Q+ QG Y +P+ Q P + Q QQ Y P SPQ PPQ Q G
Sbjct: 710 QWQQSGQGQ--HWYYPTSPKLSGQGQRPGQWLQPGQGQQGYYPTSPQQPPQGQQLGQWLQ 767
Query: 380 -GEGERRNY 387
G+G++ Y
Sbjct: 768 PGQGQQGYY 776
>GLT4_WHEAT (P08489) Glutenin, high molecular weight subunit PW212
precursor
Length = 838
Score = 63.2 bits (152), Expect = 1e-09
Identities = 72/260 (27%), Positives = 94/260 (35%), Gaps = 29/260 (11%)
Query: 243 QNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQ-QNIGQQQPNIR 301
Q Q P P + P G+ G Q Q GQ + P Q GQ QP
Sbjct: 237 QQGQQPGQGQQPGQGQQGQQPGQGQQPGQGQQ-GQQLGQGQQGYYPTSLQQSGQGQPGYY 295
Query: 302 IDSTSQRFPPQQSYDQASQRYPPQQSY----DQASQGYLPKQNYGQAPQNYSAPQAPQNY 357
S Q Q Y S + P Q Q +QG P+Q GQ Q Q Q
Sbjct: 296 PTSLQQLGQGQSGYYPTSPQQPGQGQQPGQLQQPAQGQQPEQ--GQQGQQPGQGQQGQQP 353
Query: 358 SQQQTYGPASPQY---PPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPR 414
Q Q G P Y PQQS G+G+ Y P + P Q G +
Sbjct: 354 GQGQQPGQGQPGYYPTSPQQS----GQGQP-GYYPTSSQQPTQSQQPGQGQQGQQVGQGQ 408
Query: 415 QGERRNYVPQQNFGPPG----------QGERGNSV--PSEGGWDFKPSYMEEFEQGQKGN 462
Q ++ Q G PG QG+ G + P + G +P +++ QGQKG
Sbjct: 409 QAQQPGQGQQPGQGQPGYYPTSPLQSGQGQPGYYLTSPQQSGQGQQPGQLQQSAQGQKGQ 468
Query: 463 NHAEEQKESQ-QRFPPPGPG 481
+ Q+ Q Q+ PG G
Sbjct: 469 QPGQGQQPGQGQQGQQPGQG 488
Score = 55.1 bits (131), Expect = 4e-07
Identities = 87/331 (26%), Positives = 120/331 (35%), Gaps = 44/331 (13%)
Query: 175 YTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPP-----PV 229
Y G + + + F GIP ++ S P Y GQ P+ P P
Sbjct: 86 YPGETTPPQQLQQRIFWGIPALLKRYYPSVTSPQQVSY-----YPGQASPQRPGQGQQPG 140
Query: 230 QFGRNLGGRRDYRQNN-QLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIP 288
Q ++ G++ Y + Q P P P + G Q Q GQ
Sbjct: 141 QGQQSGQGQQGYYPTSPQQPGQWQQPEQGQPGYYPTSPQQPGQLQQPAQGQQPGQGQQ-- 198
Query: 289 PQQNIGQQQPNIRIDSTSQRFPPQ----------QSYDQASQRYPPQQSYD--QASQGYL 336
+ GQ QP ++SQ P Q Q Q Q P Q Q QG
Sbjct: 199 -GRQPGQGQPGY-YPTSSQLQPGQLQQPAQGQQGQQPGQGQQGQQPGQGQQPGQGQQGQQ 256
Query: 337 PKQNY--GQAPQNYSAPQAPQNY--SQQQTYGPASPQYPPQQSFGPLGEGERRNY--APQ 390
P Q GQ Q Q Q Y + Q G P Y P S LG+G+ Y +PQ
Sbjct: 257 PGQGQQPGQGQQGQQLGQGQQGYYPTSLQQSGQGQPGYYP-TSLQQLGQGQSGYYPTSPQ 315
Query: 391 QNFGPPGEVDRRNYAPQQNFG-PPRQGERRNYVPQQNFG-PPGQGERGNSVPSEGGWDFK 448
Q PG+ + Q G P QG++ Q G PGQG++ P +G +
Sbjct: 316 Q----PGQGQQPGQLQQPAQGQQPEQGQQGQQPGQGQQGQQPGQGQQ----PGQGQPGYY 367
Query: 449 PSYMEEFEQGQKGNNHAEEQKESQQRFPPPG 479
P+ ++ QGQ G Q+ +Q + P G
Sbjct: 368 PTSPQQSGQGQPGYYPTSSQQPTQSQQPGQG 398
Score = 53.9 bits (128), Expect = 9e-07
Identities = 82/300 (27%), Positives = 115/300 (38%), Gaps = 52/300 (17%)
Query: 214 GDQYIEGQIIPRPPPVQFGRNLG-------GRRDYRQNNQLPNNRGNPSYNNRDSM-PRD 265
G Q +GQ +P Q G+ G G+ Y + + +G P Y S P
Sbjct: 332 GQQPEQGQQGQQPGQGQQGQQPGQGQQPGQGQPGYYPTSPQQSGQGQPGYYPTSSQQPTQ 391
Query: 266 GRNYGPPQNFPPQQNHGQASHIPPQ-QNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPP 324
+ G Q Q GQ + P Q Q GQ QP S Q Q Y S P
Sbjct: 392 SQQPGQGQQ-GQQVGQGQQAQQPGQGQQPGQGQPGYYPTSPLQSGQGQPGYYLTS----P 446
Query: 325 QQSYD--------QASQGYLPKQ-NYGQAPQNYSAPQAPQNYSQQQTYGPASPQY---PP 372
QQS Q++QG +Q GQ P Q P Q Q G P Y P
Sbjct: 447 QQSGQGQQPGQLQQSAQGQKGQQPGQGQQPGQGQQGQQPGQGQQGQQPGQGQPGYYPTSP 506
Query: 373 QQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAP---QQNFGP-----PRQGERRNYVPQ 424
QQS G+G++ Q G PG P Q + P P QG++ + Q
Sbjct: 507 QQS----GQGQQPGQWQQPGQGQPGYYPTSPLQPGQGQPGYDPTSPQQPGQGQQPGQLQQ 562
Query: 425 QNFGPPGQGERGNSVPSEGGWDFKPSYMEEFE---QGQKGNNHAEEQKESQQRFPPPGPG 481
P QG++G + ++G +P+ +++ + QGQ+G + Q+ Q PG G
Sbjct: 563 -----PAQGQQGQQL-AQGQQGQQPAQVQQGQQPAQGQQGQQLGQGQQGQQ-----PGQG 611
Score = 53.5 bits (127), Expect = 1e-06
Identities = 61/226 (26%), Positives = 87/226 (37%), Gaps = 42/226 (18%)
Query: 256 YNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSY 315
Y + +P+ G Y P + PPQQ QQ+ I + +R+ P +
Sbjct: 73 YEQQIVVPKGGSFY-PGETTPPQQL--------------QQRIFWGIPALLKRYYPSVTS 117
Query: 316 DQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQS 375
Q YP Q S + QG P Q GQ Q+ QQ Y P SPQ P Q
Sbjct: 118 PQQVSYYPGQASPQRPGQGQQPGQ--GQ-----------QSGQGQQGYYPTSPQQPGQWQ 164
Query: 376 FGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFG----PPRQGERRNYVPQQNFGP-- 429
G+ +PQQ PG++ + Q G P QG+ Y P
Sbjct: 165 QPEQGQPGYYPTSPQQ----PGQLQQPAQGQQPGQGQQGRQPGQGQPGYYPTSSQLQPGQ 220
Query: 430 ---PGQGERGNSVPSEGGWDFKPSYMEEFEQGQKGNNHAEEQKESQ 472
P QG++G P +G +P ++ QGQ+G + Q+ Q
Sbjct: 221 LQQPAQGQQGQQ-PGQGQQGQQPGQGQQPGQGQQGQQPGQGQQPGQ 265
Score = 49.3 bits (116), Expect = 2e-05
Identities = 63/263 (23%), Positives = 89/263 (32%), Gaps = 27/263 (10%)
Query: 243 QNNQLPNNRGNPSYNNRDSMPRDGRN-----YGPPQNFP--PQQNHGQASHIPPQQNIGQ 295
Q Q P P + P G+ G P +P PQQ+ GQ Q GQ
Sbjct: 465 QKGQQPGQGQQPGQGQQGQQPGQGQQGQQPGQGQPGYYPTSPQQS-GQGQQPGQWQQPGQ 523
Query: 296 QQPNIRIDSTSQRFPPQQSYDQASQRYPPQ-------QSYDQASQG-YLPKQNYGQAPQN 347
QP S Q Q YD S + P Q Q Q QG L + GQ P
Sbjct: 524 GQPGYYPTSPLQPGQGQPGYDPTSPQQPGQGQQPGQLQQPAQGQQGQQLAQGQQGQQPAQ 583
Query: 348 YSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQ 407
Q P Q Q G P Q P + + Q PG+ +
Sbjct: 584 VQQGQQPAQGQQGQQLGQGQQGQQPGQGQQPAQGQQGQQPGQGQQGQQPGQGQQPGQGQP 643
Query: 408 QNFGPPRQGERRNYVPQQ-----NFGPPGQGERGNSVPS-----EGGWDFKPSYMEEFEQ 457
+ Q + P Q + PGQG+ G + S +G + P+ +++ Q
Sbjct: 644 WYYPTSPQESGQGQQPGQWQQPGQWQQPGQGQPGYYLTSPLQLGQGQQGYYPTSLQQPGQ 703
Query: 458 GQKGNNHAEEQKESQQRFPPPGP 480
GQ+ ++ + Q + P P
Sbjct: 704 GQQPGQW-QQSGQGQHGYYPTSP 725
>GLT0_WHEAT (P10387) Glutenin, high molecular weight subunit DY10
precursor
Length = 648
Score = 61.6 bits (148), Expect = 4e-09
Identities = 73/276 (26%), Positives = 102/276 (36%), Gaps = 32/276 (11%)
Query: 237 GRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPP--QQNHGQASHIPPQ-QNI 293
G+R Q P P + P + G Q Q GQ H P Q
Sbjct: 213 GQRQQPVQGQQPEQGQQPGQWQQGYYPTSPQQLGQGQQPRQWQQSGQGQQGHYPTSLQQP 272
Query: 294 GQQQPNIRIDSTSQRFPPQQSYDQASQRYP----------PQQSYDQASQGYLP--KQNY 341
GQ Q + S Q QQ + ASQ+ P QQ Q QG+ P +Q
Sbjct: 273 GQGQQGHYLASQQQPGQGQQGHYPASQQQPGQGQQGHYPASQQQPGQGQQGHYPASQQEP 332
Query: 342 GQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFG-------PLGEGERRNYAPQQNFG 394
GQ Q Q QQ + PAS Q P Q G LG+G++ Q+
Sbjct: 333 GQGQQGQIPASQQQPGQGQQGHYPASLQQPGQGQQGHYPTSLQQLGQGQQTGQPGQKQQP 392
Query: 395 PPGEVDRRNYAPQQNFGPPRQGERRNY---VPQQNFGPP-GQGERGN-----SVPSEGGW 445
G+ + P+Q P QG++ Y + Q G GQG++G P +G
Sbjct: 393 GQGQQTGQGQQPEQE-QQPGQGQQGYYPTSLQQPGQGQQQGQGQQGYYPTSLQQPGQGQQ 451
Query: 446 DFKPSYMEEFEQGQKGNNHAEEQKESQQRFPPPGPG 481
P+ +++ QGQ G Q + ++ PG G
Sbjct: 452 GHYPASLQQPGQGQPGQRQQPGQGQHPEQGKQPGQG 487
Score = 54.3 bits (129), Expect = 7e-07
Identities = 72/262 (27%), Positives = 97/262 (36%), Gaps = 20/262 (7%)
Query: 236 GGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQ-QNIG 294
G + Y + Q P Y P G+ P + Q GQ H P Q G
Sbjct: 305 GQQGHYPASQQQPGQGQQGHYPASQQEPGQGQQGQIPAS-QQQPGQGQQGHYPASLQQPG 363
Query: 295 QQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNY--GQAPQNYSAPQ 352
Q Q S Q QQ+ Q Q+ P Q Q QG P+Q GQ Q Y P
Sbjct: 364 QGQQGHYPTSLQQLGQGQQT-GQPGQKQQPGQG-QQTGQGQQPEQEQQPGQGQQGY-YPT 420
Query: 353 APQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNY---APQQNFGPPGEVDRRNYAPQQN 409
+ Q Q Q G Y P P G+G++ +Y Q G PG+ +
Sbjct: 421 SLQQPGQGQQQGQGQQGYYPTSLQQP-GQGQQGHYPASLQQPGQGQPGQRQQPGQGQHPE 479
Query: 410 FG-PPRQGERRNY--VPQQNFGPPGQGERGNSVPSEGGWDFKPSYMEEFEQ-GQKGNNHA 465
G P QG++ Y PQQ PGQG++ +G + P + +Q GQ H
Sbjct: 480 QGKQPGQGQQGYYPTSPQQ----PGQGQQLGQ-GQQGYYPTSPQQPGQGQQPGQGQQGHC 534
Query: 466 EEQKESQQRFPPPGPGNFTGEV 487
+ + PG G G+V
Sbjct: 535 PTSPQQSGQAQQPGQGQQIGQV 556
Score = 52.8 bits (125), Expect = 2e-06
Identities = 54/229 (23%), Positives = 85/229 (36%), Gaps = 20/229 (8%)
Query: 262 MPRDGRNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQR 321
+P G ++ P + P QQ Q + P + +P Q S Q Q
Sbjct: 93 VPPKGGSFYPGETTPLQQLQQGIFWGTSSQTVQGYYPGVTSPRQGSYYPGQASPQQPGQG 152
Query: 322 YPPQ--QSYDQASQGYLPK--------QNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYP 371
P Q Q Q Y P Q G+ Q Y Q QQ Y P S Q+
Sbjct: 153 QQPGKWQEPGQGQQWYYPTSLQQPGQGQQIGKGQQGYYPTSLQQPGQGQQGYYPTSLQHT 212
Query: 372 PQQSFGPLGEGERRNYAP---QQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFG 428
Q+ G+ + P QQ + P + + QG++ +Y +
Sbjct: 213 GQRQQPVQGQQPEQGQQPGQWQQGYYPTSPQQLGQGQQPRQWQQSGQGQQGHY--PTSLQ 270
Query: 429 PPGQGERGNSV-----PSEGGWDFKPSYMEEFEQGQKGNNHAEEQKESQ 472
PGQG++G+ + P +G P+ ++ QGQ+G+ A +Q+ Q
Sbjct: 271 QPGQGQQGHYLASQQQPGQGQQGHYPASQQQPGQGQQGHYPASQQQPGQ 319
Score = 49.7 bits (117), Expect = 2e-05
Identities = 65/232 (28%), Positives = 88/232 (37%), Gaps = 30/232 (12%)
Query: 266 GRNYGPPQNFPPQQNHGQASHIPPQ-------QNIGQQQPNIRIDSTSQRFPPQQSYDQA 318
G+ P Q P Q GQ + P Q GQ Q S Q QQ + A
Sbjct: 399 GQGQQPEQEQQPGQ--GQQGYYPTSLQQPGQGQQQGQGQQGYYPTSLQQPGQGQQGHYPA 456
Query: 319 SQRYPPQQS---YDQASQGYLPKQNY--GQAPQNY--SAPQAPQNYSQ----QQTYGPAS 367
S + P Q Q QG P+Q GQ Q Y ++PQ P Q QQ Y P S
Sbjct: 457 SLQQPGQGQPGQRQQPGQGQHPEQGKQPGQGQQGYYPTSPQQPGQGQQLGQGQQGYYPTS 516
Query: 368 PQYPPQQSFGPLGEGERRNYAPQQN--FGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQ 425
PQ P Q G+ +PQQ+ PG+ + Q P QG++ Y
Sbjct: 517 PQQPGQGQQPGQGQQGHCPTSPQQSGQAQQPGQGQQIGQVQQ-----PGQGQQGYY--PT 569
Query: 426 NFGPPGQGERGNSVPSEGGWDFKPSYMEEFEQGQKGNNHAEEQKESQQRFPP 477
+ PGQG++ + G +P ++ Q Q+G + QQ P
Sbjct: 570 SVQQPGQGQQSGQ-GQQSGQGHQPGQGQQSGQEQQGYDSPYHVSAEQQAASP 620
Score = 31.6 bits (70), Expect = 4.8
Identities = 34/134 (25%), Positives = 41/134 (30%), Gaps = 13/134 (9%)
Query: 243 QNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRI 302
Q P + P + + G PQ Q GQ I Q GQ Q
Sbjct: 509 QQGYYPTSPQQPGQGQQPGQGQQGHCPTSPQQSGQAQQPGQGQQIGQVQQPGQGQQGYYP 568
Query: 303 DSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNY--GQAPQNYSAPQAPQNYSQQ 360
S Q QQS Q+ QG+ P Q GQ Q Y +P Q
Sbjct: 569 TSVQQPGQGQQS-----------GQGQQSGQGHQPGQGQQSGQEQQGYDSPYHVSAEQQA 617
Query: 361 QTYGPASPQYPPQQ 374
+ A Q P Q
Sbjct: 618 ASPMVAKAQQPATQ 631
>SMF1_HUMAN (O14497) SWI/SNF-related, matrix-associated,
actin-dependent regulator of chromatin subfamily F
member 1 (SWI-SNF complex protein p270) (B120)
Length = 1902
Score = 59.3 bits (142), Expect = 2e-08
Identities = 55/210 (26%), Positives = 75/210 (35%), Gaps = 19/210 (9%)
Query: 212 YGGDQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQLP--NNRGNPSYNNRDSMPRDGRNY 269
Y G Y Q R P GR Q+P +G Y + P +
Sbjct: 34 YPGQPY-GSQTPQRYPMTVQGRAQSAMGGLSYTQQIPPYGQQGPSGYGQQGQTPYYNQQS 92
Query: 270 GPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPP------QQSYDQASQRYP 323
PQ P + S P Q QQQP + PP Q + Q+ YP
Sbjct: 93 PHPQQQQPPYSQQPPSQTPHAQPSYQQQPQSQPPQLQSSQPPYSQQPSQPPHQQSPAPYP 152
Query: 324 PQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSF-GPLGEG 382
QQS Q + Q+ YS PQA Y QQQ PA Q ++ P +
Sbjct: 153 SQQS---------TTQQHPQSQPPYSQPQAQSPYQQQQPQQPAPSTLSQQAAYPQPQSQQ 203
Query: 383 ERRNYAPQQNFGPPGEVDRRNYAPQQNFGP 412
++ QQ F PP E+ + ++ Q + P
Sbjct: 204 SQQTAYSQQRFPPPQELSQDSFGSQASSAP 233
Score = 51.6 bits (122), Expect = 5e-06
Identities = 58/213 (27%), Positives = 78/213 (36%), Gaps = 22/213 (10%)
Query: 270 GPPQNFPPQQNHGQASHIPPQQNIGQQQPN---IRIDSTSQRFPPQQSYDQASQRYPPQQ 326
GPP + PQQ HG P Q G Q P + + +Q SY Q Y Q
Sbjct: 23 GPPSD--PQQGHGY-----PGQPYGSQTPQRYPMTVQGRAQSAMGGLSYTQQIPPYGQQG 75
Query: 327 SYDQASQGYLPKQN-YGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERR 385
QG P N PQ P + Q SQ P+ Q P QS P + +
Sbjct: 76 PSGYGQQGQTPYYNQQSPHPQQQQPPYSQQPPSQTPHAQPSYQQQP--QSQPPQLQSSQP 133
Query: 386 NYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFGPPGQGERGNSVPSEGGW 445
Y+ QQ PP + Y QQ+ + Y Q P Q + PS
Sbjct: 134 PYS-QQPSQPPHQQSPAPYPSQQSTTQQHPQSQPPYSQPQAQSPYQQQQPQQPAPS---- 188
Query: 446 DFKPSYMEEFEQGQKGNNHAEEQKESQQRFPPP 478
+ ++ Q + +++ SQQRFPPP
Sbjct: 189 ----TLSQQAAYPQPQSQQSQQTAYSQQRFPPP 217
Score = 42.4 bits (98), Expect = 0.003
Identities = 68/274 (24%), Positives = 85/274 (30%), Gaps = 64/274 (23%)
Query: 252 GNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQ------------------ASHIPPQQNI 293
G P N S P G P +PPQQ Q S P QQ
Sbjct: 920 GAPQPNLMPSNPDSGMY--SPSRYPPQQQQQQQQRHDSYGNQFSTQGTPSGSPFPSQQTT 977
Query: 294 --GQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAP 351
QQQ N + PP + ++ P Q Q LP A Q +A
Sbjct: 978 MYQQQQQNYKRPMDGTYGPPAKRHEGEMYSVPYSTGQGQPQQQQLPPAQPQPASQQQAAQ 1037
Query: 352 QAPQN--YSQQQTYGPAS--------PQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDR 401
+PQ Y+Q PA+ P PQ F P G R A PPG +
Sbjct: 1038 PSPQQDVYNQYGNAYPATATAATERRPAGGPQNQF-PFQFGRDRVSA------PPGTNAQ 1090
Query: 402 RNYAPQQNFGP---------------PRQGERRNYVPQQNFGPPGQGERGNSVPSEGGWD 446
+N PQ GP R NY +Q+ G QG + V
Sbjct: 1091 QNMPPQMMGGPIQASAEVAQQGTMWQGRNDMTYNYANRQSTGSAPQGPAYHGV------- 1143
Query: 447 FKPSYMEEFEQGQKGNNHAEEQKESQQRFPPPGP 480
+ +E + NH R PP GP
Sbjct: 1144 ---NRTDEMLHTDQRANHEGSWPSHGTRQPPYGP 1174
Score = 39.7 bits (91), Expect = 0.018
Identities = 64/249 (25%), Positives = 89/249 (35%), Gaps = 36/249 (14%)
Query: 242 RQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIR 301
++N+ PN PS N D M R +Y P N P + +A P + GQ
Sbjct: 818 KRNSMTPNPGYQPSMNTSDMMGR--MSYEP--NKDPYGSMRKAPGSDPFMSSGQGPNGGM 873
Query: 302 IDSTSQRFPPQQSYDQAS--QRYPPQQSYD--QASQGYLPKQNYGQ-APQNYSAPQAPQN 356
D S+ P Q YP YD + G P+ N APQ P P +
Sbjct: 874 GDPYSRAAGPGLGNVAMGPRQHYPYGGPYDRVRTEPGIGPEGNMSTGAPQPNLMPSNPDS 933
Query: 357 YSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQG 416
Y P+ +YPPQQ + ++R+ + F G + QQ +Q
Sbjct: 934 ----GMYSPS--RYPPQQQ----QQQQQRHDSYGNQFSTQGTPSGSPFPSQQTTMYQQQQ 983
Query: 417 ERRNYVPQQNFGPPGQGERGN--SVPSEGGWDFKPSYMEEFEQGQKGNNH---AEEQKES 471
+ +GPP + G SVP G QGQ A+ Q S
Sbjct: 984 QNYKRPMDGTYGPPAKRHEGEMYSVPYSTG------------QGQPQQQQLPPAQPQPAS 1031
Query: 472 QQRFPPPGP 480
QQ+ P P
Sbjct: 1032 QQQAAQPSP 1040
>GDBX_WHEAT (P21292) Gamma-gliadin precursor
Length = 302
Score = 58.5 bits (140), Expect = 4e-08
Identities = 48/147 (32%), Positives = 64/147 (42%), Gaps = 16/147 (10%)
Query: 283 QASHIPPQQNIGQQQPNIRIDSTSQRF--PPQQSYDQASQRYP--PQQSYDQASQGYLPK 338
Q P Q QQP I Q F PQQ++ Q Q YP PQQ + Q Q P+
Sbjct: 33 QQQPFPQPQQPFCQQPQRTIPQPHQTFHHQPQQTFPQPQQTYPHQPQQQFPQTQQ---PQ 89
Query: 339 QNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGE 398
Q + Q Q + PQ PQ QQ P PQQ F + ++ PQQ F P +
Sbjct: 90 QPFPQPQQTF--PQQPQLPFPQQPQQPFPQPQQPQQPFPQSQQPQQPFPQPQQQFPQPQQ 147
Query: 399 VDRRNYAPQQNFGPPRQGERRNYVPQQ 425
PQQ+F +Q ++++ QQ
Sbjct: 148 -------PQQSFPQQQQPAIQSFLQQQ 167
Score = 48.5 bits (114), Expect = 4e-05
Identities = 45/142 (31%), Positives = 65/142 (45%), Gaps = 16/142 (11%)
Query: 299 NIRIDSTSQ-RFPPQQSYDQASQRY--PPQQSYDQASQGY--LPKQNYGQAPQNYSAPQA 353
N+++D + Q ++P QQ + Q Q + PQ++ Q Q + P+Q + Q Q Y P
Sbjct: 20 NMQVDPSGQVQWPQQQPFPQPQQPFCQQPQRTIPQPHQTFHHQPQQTFPQPQQTY--PHQ 77
Query: 354 PQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPP 413
PQ Q T P P PQQ+F P PQQ F P + PQQ F
Sbjct: 78 PQQQFPQ-TQQPQQPFPQPQQTF-PQQPQLPFPQQPQQPFPQPQQ-------PQQPFPQS 128
Query: 414 RQGERRNYVPQQNFGPPGQGER 435
+Q ++ PQQ F P Q ++
Sbjct: 129 QQPQQPFPQPQQQFPQPQQPQQ 150
Score = 47.0 bits (110), Expect = 1e-04
Identities = 44/133 (33%), Positives = 55/133 (41%), Gaps = 15/133 (11%)
Query: 263 PRDGRNYGPPQNFP-PQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQR 321
P ++ P Q FP PQQ + H P QQ QQP Q FP Q Q
Sbjct: 55 PHQTFHHQPQQTFPQPQQTY---PHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQ- 110
Query: 322 YPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYP-PQQSFGPLG 380
PQQ + Q Q P+Q + Q+ Q PQ P QQQ P PQ PQQ +
Sbjct: 111 --PQQPFPQPQQ---PQQPFPQSQQ----PQQPFPQPQQQFPQPQQPQQSFPQQQQPAIQ 161
Query: 381 EGERRNYAPQQNF 393
++ P +NF
Sbjct: 162 SFLQQQMNPCKNF 174
>GDA9_WHEAT (P18573) Alpha/beta-gliadin MM1 precursor (Prolamin)
Length = 307
Score = 58.5 bits (140), Expect = 4e-08
Identities = 46/125 (36%), Positives = 56/125 (44%), Gaps = 19/125 (15%)
Query: 272 PQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQA 331
PQN QQ Q + QQ GQQQP FPPQQ Y Q Q +P QQ Y Q
Sbjct: 30 PQNPSQQQPQEQVPLVQQQQFPGQQQP----------FPPQQPYPQ-PQPFPSQQPYLQL 78
Query: 332 SQGYLPKQNYGQAPQNYSAPQ----APQNYSQQQTYGPASPQY-PPQQSFGPLGEGERRN 386
P+ Y Q Y PQ PQ + QQ Y + PQY PQQ P+ + +++
Sbjct: 79 QPFPQPQLPYPQPQLPYPQPQLPYPQPQPFRPQQPYPQSQPQYSQPQQ---PISQQQQQQ 135
Query: 387 YAPQQ 391
QQ
Sbjct: 136 QQQQQ 140
Score = 54.7 bits (130), Expect = 5e-07
Identities = 41/107 (38%), Positives = 47/107 (43%), Gaps = 13/107 (12%)
Query: 270 GPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFP-PQQSYDQASQRYP-PQQS 327
G Q FPPQQ + Q P QQ Q QP FP PQ Y Q YP PQ
Sbjct: 52 GQQQPFPPQQPYPQPQPFPSQQPYLQLQP----------FPQPQLPYPQPQLPYPQPQLP 101
Query: 328 YDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQ 374
Y Q Q + P+Q Y Q+ YS PQ P + QQQ + QQ
Sbjct: 102 YPQP-QPFRPQQPYPQSQPQYSQPQQPISQQQQQQQQQQQQKQQQQQ 147
Score = 32.7 bits (73), Expect = 2.2
Identities = 42/153 (27%), Positives = 55/153 (35%), Gaps = 19/153 (12%)
Query: 216 QYIEGQIIPRPP--PVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQ 273
Q GQ P PP P + ++ Y Q P + P + P+ Y PQ
Sbjct: 48 QQFPGQQQPFPPQQPYPQPQPFPSQQPYLQLQPFPQPQ-LPYPQPQLPYPQPQLPYPQPQ 106
Query: 274 NFPPQQNH--GQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQ- 330
F PQQ + Q + PQQ I QQQ QQ Q Q+ QQ Q
Sbjct: 107 PFRPQQPYPQSQPQYSQPQQPISQQQQQ-----------QQQQQQQKQQQQQQQQILQQI 155
Query: 331 ASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTY 363
Q +P ++ Q +S QQ TY
Sbjct: 156 LQQQLIPCRDV--VLQQHSIAYGSSQVLQQSTY 186
>YKF4_YEAST (P35732) Hypothetical 84.0 kDa protein in NUP120-CSE4
intergenic region
Length = 738
Score = 57.8 bits (138), Expect = 6e-08
Identities = 56/194 (28%), Positives = 70/194 (35%), Gaps = 28/194 (14%)
Query: 263 PRDGRNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRY 322
P D N P P QQ+ PQQ QQP + + P QQ Q Q+
Sbjct: 373 PEDQANTVPQ---PQQQSQQPQQPQQPQQPQQPQQPQQQQQPQQPQQPQQQLQQQQQQQQ 429
Query: 323 PPQQSYDQASQGYLPKQNYGQAPQNYSAPQ----APQNYSQQQTYGPASPQYP------P 372
P Q+ QA + L + Y Q Q A Q Q SQQQ Y Q+P P
Sbjct: 430 QPVQAQAQAQEEQLSQNYYTQQQQQQYAQQQHQLQQQYLSQQQQYAQQQQQHPQPQSQQP 489
Query: 373 QQSFGPLGEGERRNYAPQQ---------NFGPPGEVDRRNYAPQQNFGPPRQ------GE 417
Q P + + N A QQ + PG D + YA Q + Q G
Sbjct: 490 QSQQSPQSQKQGNNVAAQQYYMYQNQFPGYSYPGMFDSQGYAYGQQYQQLAQNNAQTSGN 549
Query: 418 RRNYVPQQNFGPPG 431
Y QQ +G G
Sbjct: 550 ANQYNFQQGYGQAG 563
Score = 40.4 bits (93), Expect = 0.010
Identities = 52/205 (25%), Positives = 77/205 (37%), Gaps = 35/205 (17%)
Query: 289 PQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQ----NYGQA 344
P++ + + + I + PP+ DQA+ PQQ Q Q P+Q Q
Sbjct: 352 PEEEVTVVEEKVEISAVISE-PPE---DQANTVPQPQQQSQQPQQPQQPQQPQQPQQPQQ 407
Query: 345 PQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNY 404
Q PQ PQ QQQ Q P Q E +NY QQ ++ Y
Sbjct: 408 QQQPQQPQQPQQQLQQQ---QQQQQQPVQAQAQAQEEQLSQNYYTQQ--------QQQQY 456
Query: 405 APQQN-FGPPRQGERRNYVPQQNFGPPGQGERGNSVPSEGGWDFKPSYMEEFEQGQKGNN 463
A QQ+ +++ Y QQ P Q + +P + + ++GNN
Sbjct: 457 AQQQHQLQQQYLSQQQQYAQQQQQHPQPQSQ-------------QPQSQQSPQSQKQGNN 503
Query: 464 HAEEQKES-QQRFPPPG-PGNFTGE 486
A +Q Q +FP PG F +
Sbjct: 504 VAAQQYYMYQNQFPGYSYPGMFDSQ 528
Score = 32.7 bits (73), Expect = 2.2
Identities = 49/227 (21%), Positives = 75/227 (32%), Gaps = 22/227 (9%)
Query: 210 KQYGGDQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNR---------D 260
+Q+ Q + Q P + G N+ ++ Y NQ P ++++
Sbjct: 479 QQHPQPQSQQPQSQQSPQSQKQGNNVAAQQYYMYQNQFPGYSYPGMFDSQGYAYGQQYQQ 538
Query: 261 SMPRDGRNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQ 320
+ + G + QQ +GQA N+ + PQQ
Sbjct: 539 LAQNNAQTSGNANQYNFQQGYGQAGANTAAANLTSAAAAAAASPATAHAQPQQQQPYGGS 598
Query: 321 RYPPQQSYDQASQGYLPKQNYGQAPQN-YSAPQAPQNYSQQQTYGPASPQYPPQQSFGPL 379
P + Q S Y Q YG A Q Y P+ NY Q Q Q P Q
Sbjct: 599 FMPYYAHFYQQSFPYGQPQ-YGVAGQYPYQLPKNNYNYYQTQN---GQEQQSPNQGVAQH 654
Query: 380 GEGERRNYAPQQ-----NFGPPGEVDRRNYAPQQNFGPPRQGERRNY 421
E ++ + QQ P EV +N P P +Q + + Y
Sbjct: 655 SEDSQQKQSQQQQQQQPQGQPQPEVQMQNGQP---VNPQQQMQFQQY 698
>OSA_DROME (Q8IN94) Trithorax group protein OSA (Eyelid protein)
Length = 2716
Score = 57.8 bits (138), Expect = 6e-08
Identities = 79/289 (27%), Positives = 102/289 (34%), Gaps = 76/289 (26%)
Query: 224 PRPPPVQFGRNLGGRRDY-----RQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQ 278
P PPP GG ++ + LP +P+Y GR + P P +
Sbjct: 118 PAPPPP------GGAPEHAPGVKEEYTHLPPPHPHPAY---------GRYHADPNMDPYR 162
Query: 279 QNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSY--DQASQGYL 336
PPQQ QQP+ Q+ PPQQ S PPQQ Y Q QG
Sbjct: 163 YGQPLPGGKPPQQ----QQPH------PQQQPPQQPGPGGSPNRPPQQRYIPGQPPQGPT 212
Query: 337 PKQNYGQAPQNYSAPQAPQN--------------------YSQQQTYGPASPQY---PPQ 373
P N Q+ + P PQ+ QQQ GP P + PPQ
Sbjct: 213 PTLN--SLLQSSNPPPPPQHRYANTYDPQQAAASAAAAAAAQQQQAGGPPPPGHGPPPPQ 270
Query: 374 QSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFGPPGQG 433
P G G++ +AP PP + QQ PP R P PP G
Sbjct: 271 HQPSPYG-GQQGGWAP-----PPRPYSPQLGPSQQYRTPPPTNTSRGQSPY----PPAHG 320
Query: 434 ERGNSVPSEGGWDFKPSYMEEFEQGQKGNNHAEEQKESQ-QRFPPPGPG 481
+ S PS S ++ +Q Q+ A +Q PPPG G
Sbjct: 321 QNSGSYPS--------SPQQQQQQQQQQQQQAGQQPGGPVPGGPPPGTG 361
Score = 56.6 bits (135), Expect = 1e-07
Identities = 78/311 (25%), Positives = 101/311 (32%), Gaps = 64/311 (20%)
Query: 211 QYGG-DQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQLPNNRGN-PSYNNRDSMPRDGRN 268
QYG DQY PP FG+ G P NR P Y P G N
Sbjct: 1370 QYGSSDQYNA----TGPPGQPFGQGPG--------QYPPQNRNMYPPYGPEGEAPPTGAN 1417
Query: 269 YGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDST--SQRFPPQQSYDQASQRYPPQQ 326
P P PP Q + P S +P + Q + PP Q
Sbjct: 1418 QYGPYGSRPYSQPPPGGPQPPTQTVAGGPPAGGAPGAPPSSAYPTGRPSQQDYYQPPPDQ 1477
Query: 327 S-----YDQASQGYLPKQNYGQAPQNYSAPQ--------------APQNYSQQQTYG--- 364
S + + P Y PQ Y A Q APQN+ G
Sbjct: 1478 SPQPRRHPDFIKDSQPYPGYNARPQIYGAWQSGTQQYRPQYPSSPAPQNWGGAPPRGAAP 1537
Query: 365 -PASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVP 423
P +P PP Q + + ++ Y PQQ PP PQQ P +Q ++ Y
Sbjct: 1538 PPGAPHGPPIQQPAGVAQWDQHRYPPQQGPPPP---------PQQQQQPQQQQQQPPY-- 1586
Query: 424 QQNFGPPGQ--------------GERGNSVPSEGGWDFKPSYMEEFEQGQKGNNHAEEQK 469
QQ GPPGQ G+ S + G +P + G ++Q
Sbjct: 1587 QQVAGPPGQQPPQAPPQWAQMNPGQTAQSGIAPPGSPLRPPSGPGQQNRMPGMPAQQQQS 1646
Query: 470 ESQQRFPPPGP 480
+ Q P P P
Sbjct: 1647 QQQGGVPQPPP 1657
Score = 46.6 bits (109), Expect = 1e-04
Identities = 62/256 (24%), Positives = 83/256 (32%), Gaps = 33/256 (12%)
Query: 248 PNNRGNPSYNNRDSMPRDGRNYG---PPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDS 304
P G P ++ S P G+ G PP+ + PQ Q PP N + Q
Sbjct: 261 PPGHGPPPPQHQPS-PYGGQQGGWAPPPRPYSPQLGPSQQYRTPPPTNTSRGQ------- 312
Query: 305 TSQRFPPQQ-----SYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQ-NYS 358
+PP SY + Q+ QQ Q G P P + Q PQ N
Sbjct: 313 --SPYPPAHGQNSGSYPSSPQQQQQQQQQQQQQAGQQPGGPVPGGPPPGTGQQPPQQNTP 370
Query: 359 QQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGER 418
Y P +YP G R Y+ Q + P +P G P
Sbjct: 371 PTSQYSPYPQRYPTPPGLPAGGSNHRTAYSTHQ-YPEPNRPWPGGSSPSPGSGHPLPPAS 429
Query: 419 RNYVPQQN-----------FGPPGQGERGNS-VPSEGGWDFKPSYMEEFEQGQKGNNHAE 466
++VP GPP G++ PS PS +E GQ N+ +
Sbjct: 430 PHHVPPLQQQPPPPPHVSAGGPPPSSSPGHAPSPSPQPSQASPSPHQEL-IGQNSNDSSS 488
Query: 467 EQKESQQRFPPPGPGN 482
S PPG N
Sbjct: 489 GGAHSGMGSGPPGTPN 504
Score = 44.7 bits (104), Expect = 6e-04
Identities = 65/281 (23%), Positives = 87/281 (30%), Gaps = 39/281 (13%)
Query: 224 PRPPPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMP------RDGRNYGPPQNFPP 277
P P P G G + + Q +N + + N+ P G N PP
Sbjct: 558 PPPMPPHPGMPGGPPQQQQSQQQQASNSASSASNSPQQTPPPAPPPNQGMNNMATPPPPP 617
Query: 278 QQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSY--------- 328
Q G +PP + G + +Q +PPQQ YPP+ Y
Sbjct: 618 QGAAGGGYPMPPHMHGGYKMGGPGQSPGAQGYPPQQPQQYPPGNYPPRPQYPPGAYATGP 677
Query: 329 -----DQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSF--GPLGE 381
QA G G Y P + Q Y P PQQ+ G G
Sbjct: 678 PPPPTSQAGAGGANSMPSGAQAGGYPGRGMPNHTGQYPPYQWVPPS--PQQTVPGGAPGG 735
Query: 382 GERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFGPPGQGERGNSVPS 441
N+ + PP V P Q G PR NY+ Q G G G+ P
Sbjct: 736 AMVGNHVQGKGTPPPPVVG--GPPPPQGSGSPRP---LNYLKQHLQHKGGYG--GSPTPP 788
Query: 442 EGGWDFKPSYMEEFEQGQKGNNHAEEQKESQQRFPPPGPGN 482
+G + + G G + PP GP N
Sbjct: 789 QG--------PQGYGNGPTGMHPGMPMGPPHHMGPPHGPTN 821
Score = 43.9 bits (102), Expect = 0.001
Identities = 72/325 (22%), Positives = 98/325 (30%), Gaps = 78/325 (24%)
Query: 226 PPPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQAS 285
PPP + LG + YR +RG Y P G+N G + P QQ Q
Sbjct: 285 PPPRPYSPQLGPSQQYRTPPPTNTSRGQSPYP-----PAHGQNSGSYPSSPQQQQQQQQQ 339
Query: 286 H-----------IP--PQQNIGQQQPNIRIDSTSQRFPPQQSYDQ--------------- 317
+P P GQQ P TSQ P Q Y
Sbjct: 340 QQQQAGQQPGGPVPGGPPPGTGQQPPQQNTPPTSQYSPYPQRYPTPPGLPAGGSNHRTAY 399
Query: 318 ASQRYP-PQQSYDQASQ---------------------------------GYLPKQNYGQ 343
++ +YP P + + S G P + G
Sbjct: 400 STHQYPEPNRPWPGGSSPSPGSGHPLPPASPHHVPPLQQQPPPPPHVSAGGPPPSSSPGH 459
Query: 344 APQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNF----GPPGEV 399
AP P Q+ G S + +G G PQQ P G
Sbjct: 460 APSPSPQPSQASPSPHQELIGQNSNDSSSGGAHSGMGSGPPGTPNPQQVMRPTPSPTGSS 519
Query: 400 DRRNYAP--QQN--FGPPRQGERRNYVPQQNFGPPGQGERGNSVPSEGGWDFKPSYMEEF 455
R+ +P QN P + + P Q PP +P G P ++
Sbjct: 520 GSRSMSPAVAQNHPISRPASNQSSSGGPMQQ--PPVGAGGPPPMPPHPGMPGGPPQQQQS 577
Query: 456 EQGQKGNNHAEEQKESQQRFPPPGP 480
+Q Q+ +N A S Q+ PPP P
Sbjct: 578 QQ-QQASNSASSASNSPQQTPPPAP 601
Score = 35.0 bits (79), Expect = 0.44
Identities = 56/241 (23%), Positives = 72/241 (29%), Gaps = 47/241 (19%)
Query: 224 PRPPPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNY-------GPPQNFP 276
P PP+Q + +R P +G P + P+ + GPP P
Sbjct: 1542 PHGPPIQQPAGVAQWDQHRY----PPQQGPPPPPQQQQQPQQQQQQPPYQQVAGPPGQQP 1597
Query: 277 PQ-------QNHGQASHI--------------PPQQNIGQQQPNIRIDSTSQRFPPQQSY 315
PQ N GQ + P QQN P + S Q PQ
Sbjct: 1598 PQAPPQWAQMNPGQTAQSGIAPPGSPLRPPSGPGQQNRMPGMPAQQQQSQQQGGVPQPPP 1657
Query: 316 DQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPAS---PQYPP 372
QAS P Q G + K Y P Q QQ GP Q PP
Sbjct: 1658 QQASHGGVPSPGLPQVGPGGMVKPPYAM------PPPPSQGVGQQVGQGPPGGMMSQKPP 1711
Query: 373 QQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFGPPGQ 432
+ + + P P + + + Q PP Q Y P G PG
Sbjct: 1712 PMPGQAMQQQPLQQQPPSHQHPHPHQHPQHQHPHQM---PPNQTAPGGYGPP---GMPGG 1765
Query: 433 G 433
G
Sbjct: 1766 G 1766
Score = 34.7 bits (78), Expect = 0.57
Identities = 64/274 (23%), Positives = 87/274 (31%), Gaps = 27/274 (9%)
Query: 224 PRPPPVQFGRNLGGRRDYRQNNQLPNNRG-NPSYNNRDSMPRDGRNYGPPQNFPPQQNHG 282
P PP + G Y + P G P Y M R P QN P + G
Sbjct: 1144 PAPPGSAPNAAIDGYPGYPGGSPYPVASGPQPDYATAGQMQRP-----PSQNNPQTPHPG 1198
Query: 283 QASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYG 342
A+ + NI P D + P + P + AS G G
Sbjct: 1199 AAAAVAAGDNISVSNPFE--DPIAAGGGPGSGTGPGPGQGPGPGA---ASGGAGAVGAVG 1253
Query: 343 QAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRR 402
PQ + P + + QQ G Q+P Q G G + QQ PP V
Sbjct: 1254 GGPQPHPPPPHSPHTAAQQAAGQHQQQHPQHQHPGLPGPPPPQQQQGQQGQQPPPSVGGG 1313
Query: 403 NYAPQQNFGP------PRQGER----RNYVP-QQNFGPPGQGERGNSVPSE----GGWDF 447
Q GP P+Q R Y P + P G+ PS+ G +
Sbjct: 1314 PPPAPQQHGPGQVPPSPQQHVRPAAGAPYPPGGSGYPTPVSRTPGSPYPSQPGAYGQYGS 1373
Query: 448 KPSYMEEFEQGQK-GNNHAEEQKESQQRFPPPGP 480
Y GQ G + +++ +PP GP
Sbjct: 1374 SDQYNATGPPGQPFGQGPGQYPPQNRNMYPPYGP 1407
Score = 32.7 bits (73), Expect = 2.2
Identities = 40/168 (23%), Positives = 63/168 (36%), Gaps = 31/168 (18%)
Query: 324 PQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGE 383
P + + S + +Q AP Y AP P + GP P P +
Sbjct: 45 PNNNSNNGSDPSIQQQQQNVAPHPYGAPPPPGS-------GPGGPPGPDPAAV----MHY 93
Query: 384 RRNYAPQQNFGPPGEVDRRNY----APQQNFGPPRQ--GERRNYVPQQNFGPPGQGERGN 437
+ QQ PP + ++ + AP G P G + Y + PP +
Sbjct: 94 HHLHQQQQQHPPPPHMQQQQHHGGPAPPPPGGAPEHAPGVKEEYT---HLPPP------H 144
Query: 438 SVPSEGGWDFKPSYMEEFEQGQ--KGNNHAEEQKESQQRFPP--PGPG 481
P+ G + P+ M+ + GQ G ++Q+ Q+ PP PGPG
Sbjct: 145 PHPAYGRYHADPN-MDPYRYGQPLPGGKPPQQQQPHPQQQPPQQPGPG 191
Score = 32.7 bits (73), Expect = 2.2
Identities = 33/133 (24%), Positives = 47/133 (34%), Gaps = 16/133 (12%)
Query: 352 QAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFG 411
++PQ QQQ PA PP P A GPP + N
Sbjct: 6 KSPQTQQQQQGGAPAPAATPPSAGAAP-------GAATPPTSGPPTPNNNSN----NGSD 54
Query: 412 PPRQGERRNYVPQQNFG---PPGQGERGNSVPSEGGWDFKPSYMEEFEQGQKGNNHAEEQ 468
P Q +++N P +G PPG G G P ++ + +Q H ++Q
Sbjct: 55 PSIQQQQQNVAPHP-YGAPPPPGSGPGGPPGPDPAA-VMHYHHLHQQQQQHPPPPHMQQQ 112
Query: 469 KESQQRFPPPGPG 481
+ PPP G
Sbjct: 113 QHHGGPAPPPPGG 125
>GLT3_WHEAT (P08488) Glutenin, high molecular weight subunit 12
precursor
Length = 660
Score = 57.0 bits (136), Expect = 1e-07
Identities = 71/270 (26%), Positives = 99/270 (36%), Gaps = 34/270 (12%)
Query: 243 QNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQ--NFPPQ-QNHGQASHIPPQ-QNIGQQQP 298
Q P ++ P + +P + G Q ++P Q GQ H P Q +GQ Q
Sbjct: 333 QQGHYPASQQEPGQGQQGQIPASQQQPGQGQQGHYPASLQQPGQQGHYPTSLQQLGQGQ- 391
Query: 299 NIRIDSTSQRFPPQQSYDQASQRYPPQ-QSYDQASQGYLPK--------QNYGQAPQNYS 349
+I Q+ P Q + P Q Q Q QGY P Q GQ Q Y
Sbjct: 392 --QIGQPGQKQQPGQGQQTGQGQQPEQEQQPGQGQQGYYPTSLQQPGQGQQQGQGQQGYY 449
Query: 350 APQAPQNYSQQQTYGPASPQYPPQQSFGP-----LGEGERRNYAPQQNFGPPGEVDRRNY 404
Q QQ + PAS Q P Q P G+G+ Q G G
Sbjct: 450 PTSLQQPGQGQQGHYPASLQQPGQGQGQPGQRQQPGQGQHPEQGQQPGQGQQGYYPTSPQ 509
Query: 405 AP--QQNFGPPRQGERRNYVPQQNFG-PPGQGERGNSVPSEGGWDFKPSYMEEFEQGQKG 461
P Q G +QG Q G PGQG++G+ S + ++ QGQ+
Sbjct: 510 QPGQGQQLGQGQQGYYPTSPQQPGQGQQPGQGQQGHCPMSPQ----QTGQAQQLGQGQQ- 564
Query: 462 NNHAEEQKESQQRFPP-----PGPGNFTGE 486
++ + QQ + P PG G +G+
Sbjct: 565 IGQVQQPGQGQQGYYPTSLQQPGQGQQSGQ 594
Score = 55.5 bits (132), Expect = 3e-07
Identities = 68/261 (26%), Positives = 99/261 (37%), Gaps = 51/261 (19%)
Query: 263 PRDGRNYGPPQN--FPPQ-QNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQAS 319
P G+ G Q +P Q GQ Q GQ Q S Q QQ + AS
Sbjct: 236 PEQGQQPGQWQQGYYPTSPQQLGQGQQPGQWQQSGQGQQGHYPTSLQQPGQGQQGHYLAS 295
Query: 320 QRYPPQQSYDQASQGYLP--KQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFG 377
Q+ P Q QG+ P +Q GQ Q + Q QQ + PAS Q P Q G
Sbjct: 296 QQQPAQ-----GQQGHYPASQQQPGQGQQGHYPASQQQPGQGQQGHYPASQQEPGQGQQG 350
Query: 378 PL-------GEGERRNYAPQ--------------------QNFGPPGEVDRRNYAPQQNF 410
+ G+G++ +Y Q G PG+ + Q
Sbjct: 351 QIPASQQQPGQGQQGHYPASLQQPGQQGHYPTSLQQLGQGQQIGQPGQKQQPGQGQQTGQ 410
Query: 411 GPPRQGERRNYVPQQNFGP-----PGQGERGNSVPSEGGWDFKPSYMEEFEQGQKGNNHA 465
G + E++ QQ + P PGQG++ +G + P+ +++ QGQ+G+ A
Sbjct: 411 GQQPEQEQQPGQGQQGYYPTSLQQPGQGQQ----QGQGQQGYYPTSLQQPGQGQQGHYPA 466
Query: 466 EEQKESQQRFPP-----PGPG 481
Q+ Q + P PG G
Sbjct: 467 SLQQPGQGQGQPGQRQQPGQG 487
Score = 53.9 bits (128), Expect = 9e-07
Identities = 60/242 (24%), Positives = 95/242 (38%), Gaps = 34/242 (14%)
Query: 262 MPRDGRNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQR 321
+P G ++ P + P QQ Q + P++ +P Q S Q Q
Sbjct: 93 VPPKGGSFYPGETTPLQQLQQGIFWGTSSQTVQGYYPSVTSPRQGSYYPGQASPQQPGQG 152
Query: 322 YPPQ--QSYDQASQGYLP-------------KQNYGQAPQNYSAPQAPQNYSQ-QQTYGP 365
P Q Q Q Y P K G P + P Q Q QQ Y P
Sbjct: 153 QQPGKWQEPGQGQQWYYPTSLQQPGQGQQIGKGKQGYYPTSLQQPGQGQQIGQGQQGYYP 212
Query: 366 ASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNY--APQQ--------NFGPPRQ 415
SPQ+ Q+ G+ + P+Q PG+ + Y +PQQ + Q
Sbjct: 213 TSPQHTGQRQQPVQGQQIGQGQQPEQG-QQPGQWQQGYYPTSPQQLGQGQQPGQWQQSGQ 271
Query: 416 GERRNYVPQQNFGPPGQGERGNSV-----PSEGGWDFKPSYMEEFEQGQKGNNHAEEQKE 470
G++ +Y + PGQG++G+ + P++G P+ ++ QGQ+G+ A +Q+
Sbjct: 272 GQQGHY--PTSLQQPGQGQQGHYLASQQQPAQGQQGHYPASQQQPGQGQQGHYPASQQQP 329
Query: 471 SQ 472
Q
Sbjct: 330 GQ 331
Score = 41.2 bits (95), Expect = 0.006
Identities = 50/182 (27%), Positives = 63/182 (34%), Gaps = 15/182 (8%)
Query: 236 GGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQ-QNHGQASHIPPQQNIG 294
G ++ Q P + P + P + G Q P Q Q GQ H Q G
Sbjct: 438 GQQQGQGQQGYYPTSLQQPGQGQQGHYPASLQQPGQGQGQPGQRQQPGQGQHPEQGQQPG 497
Query: 295 QQQPNIRIDSTSQRFPPQQ------SYDQASQRYPPQ-QSYDQASQGYLPK--QNYGQAP 345
Q Q S Q QQ Y S + P Q Q Q QG+ P Q GQA
Sbjct: 498 QGQQGYYPTSPQQPGQGQQLGQGQQGYYPTSPQQPGQGQQPGQGQQGHCPMSPQQTGQAQ 557
Query: 346 Q---NYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRR 402
Q Q Q QQ Y P S Q P Q G+G++ Q G +++
Sbjct: 558 QLGQGQQIGQVQQPGQGQQGYYPTSLQQPGQGQ--QSGQGQQSGQGHQPGQGQQSGQEKQ 615
Query: 403 NY 404
Y
Sbjct: 616 GY 617
>GDA1_WHEAT (P04721) Alpha/beta-gliadin A-I precursor (Prolamin)
Length = 262
Score = 56.6 bits (135), Expect = 1e-07
Identities = 42/115 (36%), Positives = 48/115 (41%), Gaps = 23/115 (20%)
Query: 272 PQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQA 331
PQN QQ Q + QQ +GQQQP FPPQQ Y Q Q +P QQ Y Q
Sbjct: 30 PQNPSQQQPQEQVPLVQQQQFLGQQQP----------FPPQQPYPQP-QPFPSQQPYLQL 78
Query: 332 S------------QGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQ 374
Q + P+Q Y Q YS PQ P + QQQ Q QQ
Sbjct: 79 QPFLQPQLPYSQPQPFRPQQPYPQPQPQYSQPQQPISQQQQQQQQQQQQQQQQQQ 133
Score = 46.2 bits (108), Expect = 2e-04
Identities = 36/96 (37%), Positives = 41/96 (42%), Gaps = 8/96 (8%)
Query: 270 GPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRID---STSQRFPPQQSYDQASQRY-PPQ 325
G Q FPPQQ + Q P QQ Q QP ++ S Q F PQQ Y Q +Y PQ
Sbjct: 52 GQQQPFPPQQPYPQPQPFPSQQPYLQLQPFLQPQLPYSQPQPFRPQQPYPQPQPQYSQPQ 111
Query: 326 QSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQ 361
Q Q Q +Q Q Q Q Q QQQ
Sbjct: 112 QPISQQQQ----QQQQQQQQQQQQQQQIIQQILQQQ 143
>GDA3_WHEAT (P04723) Alpha/beta-gliadin A-III precursor (Prolamin)
Length = 282
Score = 55.8 bits (133), Expect = 2e-07
Identities = 37/93 (39%), Positives = 42/93 (44%), Gaps = 17/93 (18%)
Query: 270 GPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYD 329
G + FPPQQ + P QQ Q QP FPPQ Y Q +Q +PPQQ Y
Sbjct: 53 GQQEQFPPQQPYPHQQPFPSQQPYPQPQP----------FPPQLPYPQ-TQPFPPQQPYP 101
Query: 330 QASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQT 362
Q P+ Y Q Q S QA Q QQQT
Sbjct: 102 Q------PQPQYPQPQQPISQQQAQQQQQQQQT 128
Score = 47.0 bits (110), Expect = 1e-04
Identities = 36/116 (31%), Positives = 49/116 (42%), Gaps = 16/116 (13%)
Query: 277 PQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYL 336
PQ S PQ+ + Q + ++FPPQQ Y Q +P QQ Y Q Q +
Sbjct: 26 PQLQPQNPSQQQPQEQVPLMQQQQQFPGQQEQFPPQQPYPH-QQPFPSQQPYPQ-PQPFP 83
Query: 337 PKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYP-PQQSFGPLGEGERRNYAPQQ 391
P+ Y Q Q + QQ Y PQYP PQQ P+ + + + QQ
Sbjct: 84 PQLPYPQT----------QPFPPQQPYPQPQPQYPQPQQ---PISQQQAQQQQQQQ 126
Score = 46.6 bits (109), Expect = 1e-04
Identities = 41/114 (35%), Positives = 45/114 (38%), Gaps = 14/114 (12%)
Query: 269 YGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYP-PQQ- 326
Y Q FP QQ + Q PPQ Q QP FPPQQ Y Q +YP PQQ
Sbjct: 64 YPHQQPFPSQQPYPQPQPFPPQLPYPQTQP----------FPPQQPYPQPQPQYPQPQQP 113
Query: 327 -SYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPL 379
S QA Q +Q Q Q P QQ AS Q Q S+ L
Sbjct: 114 ISQQQAQQQQQQQQTLQQILQQQLIP-CRDVVLQQHNIAHASSQVLQQSSYQQL 166
Score = 32.3 bits (72), Expect = 2.8
Identities = 31/106 (29%), Positives = 39/106 (36%), Gaps = 33/106 (31%)
Query: 291 QNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSA 350
QN QQQP ++ Q QQ + +++PPQQ Y Q P Q
Sbjct: 31 QNPSQQQPQEQVPLMQQ----QQQFPGQQEQFPPQQPYPH--QQPFPSQ----------- 73
Query: 351 PQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPP 396
Q Y Q Q + P P YP Q F PQQ + P
Sbjct: 74 ----QPYPQPQPFPPQLP-YPQTQPF-----------PPQQPYPQP 103
>GDA5_WHEAT (P04725) Alpha/beta-gliadin A-V precursor (Prolamin)
Length = 319
Score = 55.5 bits (132), Expect = 3e-07
Identities = 38/93 (40%), Positives = 41/93 (43%), Gaps = 6/93 (6%)
Query: 270 GPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYD 329
G Q FPPQQ + Q P QQ Q QP Q FPPQ Y Q Q +PPQQ Y
Sbjct: 52 GQQQQFPPQQPYPQPQPFPSQQPYLQLQP----FPQPQPFPPQLPYPQ-PQSFPPQQPYP 106
Query: 330 QASQGYL-PKQNYGQAPQNYSAPQAPQNYSQQQ 361
Q YL P+Q Q Q Q QQQ
Sbjct: 107 QQQPQYLQPQQPISQQQAQQQQQQQQQQQQQQQ 139
Score = 50.4 bits (119), Expect = 1e-05
Identities = 42/120 (35%), Positives = 48/120 (40%), Gaps = 21/120 (17%)
Query: 263 PRDGRNYGPPQNFP---PQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQAS 319
P++ P + P QQ GQ PPQQ Q QP FP QQ Y Q
Sbjct: 30 PQNPSQQQPQEQVPLVQQQQFPGQQQQFPPQQPYPQPQP----------FPSQQPYLQLQ 79
Query: 320 -----QRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQ 374
Q +PPQ Y Q Q + P+Q Y Q Y PQ P SQQQ Q QQ
Sbjct: 80 PFPQPQPFPPQLPYPQP-QSFPPQQPYPQQQPQYLQPQQP--ISQQQAQQQQQQQQQQQQ 136
Score = 50.1 bits (118), Expect = 1e-05
Identities = 44/121 (36%), Positives = 54/121 (44%), Gaps = 20/121 (16%)
Query: 272 PQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQA 331
PQN QQ Q + QQ GQQQ +FPPQQ Y Q Q +P QQ Y Q
Sbjct: 30 PQNPSQQQPQEQVPLVQQQQFPGQQQ----------QFPPQQPYPQP-QPFPSQQPYLQL 78
Query: 332 SQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQY-PPQQSFGPLGEGERRNYAPQ 390
P+ Q P PQ PQ++ QQ Y PQY PQQ P+ + + + Q
Sbjct: 79 QPFPQPQPFPPQLPY----PQ-PQSFPPQQPYPQQQPQYLQPQQ---PISQQQAQQQQQQ 130
Query: 391 Q 391
Q
Sbjct: 131 Q 131
Score = 37.7 bits (86), Expect = 0.067
Identities = 38/137 (27%), Positives = 55/137 (39%), Gaps = 35/137 (25%)
Query: 291 QNIGQQQPNIRIDSTSQR-FPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYS 349
QN QQQP ++ Q+ FP QQ Q++PPQQ Y Q
Sbjct: 31 QNPSQQQPQEQVPLVQQQQFPGQQ------QQFPPQQPYPQ------------------- 65
Query: 350 APQAPQNYSQQQTYGPASPQYPPQQSFGP-LGEGERRNYAPQQNFGPPGEVDRRNYAPQQ 408
PQ + QQ Y P +P Q F P L + +++ PQQ P + + PQQ
Sbjct: 66 ----PQPFPSQQPYLQLQP-FPQPQPFPPQLPYPQPQSFPPQQ---PYPQQQPQYLQPQQ 117
Query: 409 NFGPPRQGERRNYVPQQ 425
+ +++ QQ
Sbjct: 118 PISQQQAQQQQQQQQQQ 134
Score = 32.7 bits (73), Expect = 2.2
Identities = 26/93 (27%), Positives = 36/93 (37%), Gaps = 14/93 (15%)
Query: 277 PQQNHGQASH------IPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYP------- 323
P+Q+ QA H I QQ QQ+ ++ Q+ Q Q Q+ P
Sbjct: 190 PEQSQCQAIHNVAHAIIMHQQQQQQQEQKQQLQQQQQQQQQLQQQQQQQQQQPSSQVSFQ 249
Query: 324 -PQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQ 355
PQQ Y + + P Q QA + Q PQ
Sbjct: 250 QPQQQYPSSQVSFQPSQLNPQAQGSVQPQQLPQ 282
>SFPQ_HUMAN (P23246) Splicing factor, proline-and glutamine-rich
(Polypyrimidine tract-binding protein-associated
splicing factor) (PTB-associated splicing factor) (PSF)
(DNA-binding p52/p100 complex, 100 kDa subunit)
Length = 707
Score = 55.1 bits (131), Expect = 4e-07
Identities = 76/271 (28%), Positives = 90/271 (33%), Gaps = 38/271 (14%)
Query: 232 GRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQ 291
G GG D+R G NR M GP PP H Q PPQQ
Sbjct: 22 GGGRGGLHDFRSPPP-----GMGLNQNRGPMGPGPGQSGPKPPIPPPPPHQQQQQPPPQQ 76
Query: 292 NIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQ--SYDQASQGYLPKQNYGQAPQNYS 349
QQ P + PP Q Q PP Q S +QG P G AP S
Sbjct: 77 PPPQQPP-------PHQPPPHPQPHQQQQPPPPPQDSSKPVVAQGPGPAPGVGSAPPASS 129
Query: 350 A--PQAPQNYSQQQTYGPA-SPQYPPQQSFGPLG------EGERRNYAPQQNFG---PPG 397
+ P P GP +P PP + P G P Q G PP
Sbjct: 130 SAPPATPPTSGAPPGSGPGPTPTPPPAVTSAPPGAPPPTPPSSGVPTTPPQAGGPPPPPA 189
Query: 398 EVDRRNYAPQQNFGP--PRQGERRNYVPQQNFGPPGQGERGNSVPSEGGWDFKPSYMEEF 455
V P+Q GP P+ G+ GP G G S P G KP +
Sbjct: 190 AVPGPGPGPKQGPGPGGPKGGKMPG-------GPKPGGGPGLSTP---GGHPKPPHRGGG 239
Query: 456 EQGQKGNNHAEEQKESQQRFPPPGPGNFTGE 486
E +H ++ Q PP GPG + E
Sbjct: 240 EPRGGRQHHPPYHQQHHQGPPPGGPGGRSEE 270
>PRPH_MESAU (P06680) Acidic proline-rich protein HP43A precursor
Length = 183
Score = 55.1 bits (131), Expect = 4e-07
Identities = 41/149 (27%), Positives = 57/149 (37%), Gaps = 9/149 (6%)
Query: 273 QNFPPQQNHGQASHIPPQQNIGQQQPNI--RIDSTSQRFPPQQSYDQASQRYPPQQSYDQ 330
Q P Q H + PPQ + ++ + D + PPQQ D R P +
Sbjct: 31 QGQDPGQLHQRPGQFPPQPSASDEEGDDDGEEDGNAPEGPPQQGGDHQKPRPPKPGNQQG 90
Query: 331 ASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQ 390
Q +QN P N P + Q + P + + PPQQ G+ + R P
Sbjct: 91 PPQQEGQQQNRPPKPGNQEGPPQQEGQQQNRPPKPGNQEGPPQQE----GQQQNRPPKPG 146
Query: 391 QNFGPP---GEVDRRNYAPQQNFGPPRQG 416
GPP G+ R P GPP+QG
Sbjct: 147 NQEGPPQQEGQQQNRPPKPGNQEGPPQQG 175
Score = 50.1 bits (118), Expect = 1e-05
Identities = 55/193 (28%), Positives = 79/193 (40%), Gaps = 52/193 (26%)
Query: 303 DSTSQRFPPQQSYDQASQ------RYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQN 356
DS SQ +Q Q ++PPQ S +G + G AP+ PQ +
Sbjct: 20 DSISQLSEEEQQGQDPGQLHQRPGQFPPQPSASD-EEGDDDGEEDGNAPEG--PPQQGGD 76
Query: 357 YSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQ- 415
+ + + P + Q PPQQ EG+++N P+ P GPP+Q
Sbjct: 77 HQKPRPPKPGNQQGPPQQ------EGQQQNRPPK---------------PGNQEGPPQQE 115
Query: 416 GERRNYVPQ--QNFGPPGQ-GERGNSVPSEGGWDFKPSYMEEFEQGQKGNNHAEEQKESQ 472
G+++N P+ GPP Q G++ N P KP GN Q+E Q
Sbjct: 116 GQQQNRPPKPGNQEGPPQQEGQQQNRPP-------KP-----------GNQEGPPQQEGQ 157
Query: 473 QRFPPPGPGNFTG 485
Q+ PP PGN G
Sbjct: 158 QQNRPPKPGNQEG 170
Score = 40.0 bits (92), Expect = 0.014
Identities = 46/176 (26%), Positives = 68/176 (38%), Gaps = 27/176 (15%)
Query: 173 TTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDP--VNKQYGGDQYIEGQIIPRPPPVQ 230
T Y + ++EEE + + PG + P + + + G D E P PP Q
Sbjct: 16 TIYEDSISQLSEEEQQGQD--PGQLHQRPGQFPPQPSASDEEGDDDGEEDGNAPEGPPQQ 73
Query: 231 FGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQ------NFPPQ------ 278
G + R N Q P + N P+ G GPPQ N PP+
Sbjct: 74 GGDHQKPRPPKPGNQQGPPQQEGQQQNRP---PKPGNQEGPPQQEGQQQNRPPKPGNQEG 130
Query: 279 --QNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQAS 332
Q GQ + PP+ G Q+ + + Q PP+ +Q PPQQ ++ S
Sbjct: 131 PPQQEGQQQNRPPKP--GNQEGPPQQEGQQQNRPPK----PGNQEGPPQQGSEEQS 180
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.133 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 69,458,683
Number of Sequences: 164201
Number of extensions: 3576306
Number of successful extensions: 10951
Number of sequences better than 10.0: 497
Number of HSP's better than 10.0 without gapping: 72
Number of HSP's successfully gapped in prelim test: 449
Number of HSP's that attempted gapping in prelim test: 7683
Number of HSP's gapped (non-prelim): 1745
length of query: 488
length of database: 59,974,054
effective HSP length: 114
effective length of query: 374
effective length of database: 41,255,140
effective search space: 15429422360
effective search space used: 15429422360
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC134322.18


