

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134049.18 - phase: 0
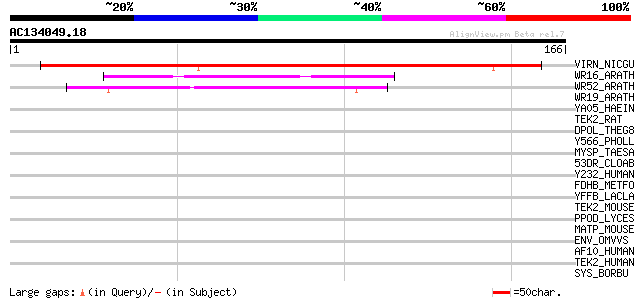
(166 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 143 2e-34
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 56 4e-08
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 47 2e-05
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 41 0.001
YA05_HAEIN (P44974) Hypothetical UPF0141 protein HI1005 32 0.87
TEK2_RAT (Q6AYM2) Tektin-2 (Testicular tektin) (Tektin-t) 31 1.1
DPOL_THEG8 (Q9HH84) DNA polymerase (EC 2.7.7.7) [Contains: Endon... 30 1.9
Y566_PHOLL (Q7N8Z3) Hypothetical UPF0246 protein plu0566 30 3.3
MYSP_TAESA (Q8T305) Paramyosin 30 3.3
53DR_CLOAB (Q97JQ5) Putative 5'(3')-deoxyribonucleotidase (EC 3.... 30 3.3
Y232_HUMAN (Q92628) Hypothetical protein KIAA0232 (Fragment) 29 4.3
FDHB_METFO (P06130) Formate dehydrogenase beta chain (EC 1.2.1.2) 29 4.3
YFFB_LACLA (Q48660) Hypothetical UPF0074 protein yffB 29 5.6
TEK2_MOUSE (Q922G7) Tektin-2 (Testicular tektin) (Tektin-t) 29 5.6
PPOD_LYCES (Q08306) Polyphenol oxidase D, chloroplast precursor ... 29 5.6
MATP_MOUSE (P58355) Membrane-associated transporter protein (AIM... 29 5.6
ENV_OMVVS (P16899) ENV polyprotein precursor (Coat polyprotein) 29 5.6
AF10_HUMAN (P55197) AF-10 protein 29 5.6
TEK2_HUMAN (Q9UIF3) Tektin-2 (Testicular tektin) (Tektin-t) (Tes... 28 7.3
SYS_BORBU (O51244) Seryl-tRNA synthetase (EC 6.1.1.11) (Serine--... 28 7.3
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 143 bits (360), Expect = 2e-34
Identities = 72/152 (47%), Positives = 107/152 (70%), Gaps = 2/152 (1%)
Query: 10 SSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVR 68
+SS+++ + ++VF+SFR EDTR FTSHL L I+T+ D+ L G I L +
Sbjct: 2 ASSSSSSRWSYDVFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIPGELCK 61
Query: 69 AIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYA 128
AIEE++ +++VFS+NYA S++CL+EL+KI+ECK + V+PIFYDVDP+ VRNQ+ S+A
Sbjct: 62 AIEESQFAIVVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQKESFA 121
Query: 129 EAFAKHEKNSEEKIK-VQEWRNGLMEAANYSG 159
+AF +HE ++ ++ +Q WR L EAAN G
Sbjct: 122 KAFEEHETKYKDDVEGIQRWRIALNEAANLKG 153
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 55.8 bits (133), Expect = 4e-08
Identities = 31/87 (35%), Positives = 52/87 (59%), Gaps = 6/87 (6%)
Query: 29 EDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEISTTLVRAIEEAELSVIVFSKNYAASK 88
E+ R +F SHL+ AL+R + D ++S D +S +E A +SV++ N S
Sbjct: 14 EEVRYSFVSHLSKALQRKGVN---DVFIDSDDSLSNESQSMVERARVSVMILPGNRTVS- 69
Query: 89 FCLDELMKILECKRMKGKMVVPIFYDV 115
LD+L+K+L+C++ K ++VVP+ Y V
Sbjct: 70 --LDKLVKVLDCQKNKDQVVVPVLYGV 94
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein
RRS1) (Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 47.4 bits (111), Expect = 2e-05
Identities = 34/98 (34%), Positives = 53/98 (53%), Gaps = 3/98 (3%)
Query: 18 EKHEVFISFRS-EDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEISTTLVRAIEEAELS 76
EK E F+ E+ R +F SHL+ AL+R I + + ++ D + IE+A +S
Sbjct: 5 EKDEEFVCISCVEEVRYSFVSHLSEALRRKGINNVVVD-VDIDDLLFKESQAKIEKAGVS 63
Query: 77 VIVFSKNYAASKFCLDELMKILECKR-MKGKMVVPIFY 113
V+V N S+ LD+ K+LEC+R K + VV + Y
Sbjct: 64 VMVLPGNCDPSEVWLDKFAKVLECQRNNKDQAVVSVLY 101
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 40.8 bits (94), Expect = 0.001
Identities = 36/142 (25%), Positives = 59/142 (41%), Gaps = 24/142 (16%)
Query: 20 HEVFISFRSEDTRN-NFTSHLNGALKRLDIRTYIDNNLNSGDEISTTLVRAIEEAELSVI 78
++V I + D N +F SHL +L R I Y N V A+ + + +I
Sbjct: 668 YDVVIRYGRADISNEDFISHLRASLCRRGISVYEKFNE----------VDALPKCRVLII 717
Query: 79 VFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKNS 138
V + Y S L+ ILE + + ++V PIFY + P D +Y + + E
Sbjct: 718 VLTSTYVPSN-----LLNILEHQHTEDRVVYPIFYRLSPYDFVCNSKNYERFYLQDEP-- 770
Query: 139 EEKIKVQEWRNGLMEAANYSGW 160
++W+ L E G+
Sbjct: 771 ------KKWQAALKEITQMPGY 786
>YA05_HAEIN (P44974) Hypothetical UPF0141 protein HI1005
Length = 519
Score = 31.6 bits (70), Expect = 0.87
Identities = 25/109 (22%), Positives = 50/109 (44%), Gaps = 13/109 (11%)
Query: 26 FRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEISTTLVRAIEEAELSVIVFSKNYA 85
+R+E +T ++ K+ ++TY +N E T R + A+ ++ + Y
Sbjct: 266 YRAEKGETVYTDNIISLAKKAGVKTYWISNQGKIGEFDTIASRIGQSADETIFMKPLGYN 325
Query: 86 ASKFCLDELMKILECKRMK-----GKMVV-------PIFYDVDPTDVRN 122
+ K DE++ +L+ K +K K++V P F + P +V+N
Sbjct: 326 SKKVYDDEMLPVLD-KALKENISNSKLIVIHLIGSHPAFCERLPYEVKN 373
>TEK2_RAT (Q6AYM2) Tektin-2 (Testicular tektin) (Tektin-t)
Length = 430
Score = 31.2 bits (69), Expect = 1.1
Identities = 27/96 (28%), Positives = 41/96 (42%), Gaps = 3/96 (3%)
Query: 5 YSSSTSSSNNTPQEKHEVFISFR--SEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEI 62
YS NT +E E+ R ED R + L A RL+ RTY N D+
Sbjct: 276 YSELKWQEKNTLEEIAELQADIRRLEEDLRRKMMN-LKLAHTRLESRTYRPNVELCRDQT 334
Query: 63 STTLVRAIEEAELSVIVFSKNYAASKFCLDELMKIL 98
L+ + + E ++ + + A ++ LD L K L
Sbjct: 335 QYGLIDEVHQLEATISIMKQKLAQTQDALDALFKHL 370
>DPOL_THEG8 (Q9HH84) DNA polymerase (EC 2.7.7.7) [Contains:
Endonuclease PI-TspGE8I (EC 3.1.-.-) (Tsp-GE8 pol-1
intein); Endonuclease PI-TspGE8II (EC 3.1.-.-) (Tsp-GE8
pol-2 intein)]
Length = 1699
Score = 30.4 bits (67), Expect = 1.9
Identities = 21/84 (25%), Positives = 39/84 (46%), Gaps = 8/84 (9%)
Query: 39 LNGALKRLDIRTYIDNNLNSGDEISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKIL 98
+NG LK + I ++DN + G + +E + + I F++ +K + +K L
Sbjct: 503 VNGRLKLVRIGDFVDNTMKKGQPLENDGTEVLEVSGIEAISFNRK---TKIAEIKPVKAL 559
Query: 99 ECKRMKGKMVVPIFYDVDPTDVRN 122
R +GK+ YD+ + RN
Sbjct: 560 IRHRYRGKV-----YDIKLSSGRN 578
>Y566_PHOLL (Q7N8Z3) Hypothetical UPF0246 protein plu0566
Length = 258
Score = 29.6 bits (65), Expect = 3.3
Identities = 19/56 (33%), Positives = 31/56 (54%), Gaps = 8/56 (14%)
Query: 59 GDEISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYD 114
GD I+ L A+E+ +V+V N A+ DE K + K++ GK++ P+F D
Sbjct: 148 GDLITEKLNEALEQQGDNVLV---NLAS-----DEYFKSVNTKKLAGKIIKPVFLD 195
>MYSP_TAESA (Q8T305) Paramyosin
Length = 863
Score = 29.6 bits (65), Expect = 3.3
Identities = 15/34 (44%), Positives = 22/34 (64%)
Query: 66 LVRAIEEAELSVIVFSKNYAASKFCLDELMKILE 99
LVRA +E E V+ FSK AA + LD+L + ++
Sbjct: 199 LVRANQEYEAQVVTFSKTKAALESQLDDLKRAMD 232
>53DR_CLOAB (Q97JQ5) Putative 5'(3')-deoxyribonucleotidase (EC
3.1.3.-)
Length = 180
Score = 29.6 bits (65), Expect = 3.3
Identities = 16/65 (24%), Positives = 30/65 (45%), Gaps = 1/65 (1%)
Query: 83 NYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKNSEEKI 142
N+ KF I+ C KG + + I D +P ++ N +G+ A H K+ +
Sbjct: 103 NWIKEKFPFISYQNIIFCHN-KGLVHLDILIDDNPLNLENFKGNKILFDAHHNKSENRFV 161
Query: 143 KVQEW 147
+ ++W
Sbjct: 162 RARDW 166
>Y232_HUMAN (Q92628) Hypothetical protein KIAA0232 (Fragment)
Length = 1402
Score = 29.3 bits (64), Expect = 4.3
Identities = 26/106 (24%), Positives = 40/106 (37%), Gaps = 9/106 (8%)
Query: 3 WSYSSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEI 62
WS+S T S N T + E F +ED +N + R+ Y+D L +
Sbjct: 721 WSHSEETRSDNETLNIQFEESTQFNAED--------INYVVPRVS-SNYVDEELLDFLQD 771
Query: 63 STTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMV 108
T + E+ +VF K C +L + E K + V
Sbjct: 772 ETCQQNSRTLGEIPTLVFKKTSKLESVCGIQLEQKTENKNFETTQV 817
>FDHB_METFO (P06130) Formate dehydrogenase beta chain (EC 1.2.1.2)
Length = 399
Score = 29.3 bits (64), Expect = 4.3
Identities = 16/65 (24%), Positives = 32/65 (48%), Gaps = 1/65 (1%)
Query: 103 MKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKNSEEKIKVQEWRN-GLMEAANYSGWD 161
+KG+ ++ FY+VDP V + + + + E +E++I + E + G N +
Sbjct: 136 VKGRQMMEEFYEVDPDSVVKEEIAKGKLIVETEDGTEKEIPIDELEDEGFGRRTNCRRCE 195
Query: 162 CNVNR 166
N+ R
Sbjct: 196 VNIPR 200
>YFFB_LACLA (Q48660) Hypothetical UPF0074 protein yffB
Length = 156
Score = 28.9 bits (63), Expect = 5.6
Identities = 20/68 (29%), Positives = 29/68 (42%), Gaps = 7/68 (10%)
Query: 26 FRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEISTTLVRAIEEAELSVIVFSKNYA 85
F+S+ NF +G KR I + +D EI TLVR + L+ + Y
Sbjct: 90 FQSQGLLQNFIGSESGKAKRCAITSALD-------EIENTLVRTLSNVSLAQVADETQYN 142
Query: 86 ASKFCLDE 93
+ LDE
Sbjct: 143 YNLGYLDE 150
>TEK2_MOUSE (Q922G7) Tektin-2 (Testicular tektin) (Tektin-t)
Length = 430
Score = 28.9 bits (63), Expect = 5.6
Identities = 27/96 (28%), Positives = 40/96 (41%), Gaps = 3/96 (3%)
Query: 5 YSSSTSSSNNTPQEKHEVFISFR--SEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEI 62
YS NT +E E+ R ED R + L A RL+ RTY N D+
Sbjct: 276 YSELKWQEKNTLEEIAELQGDIRRLEEDLRRKMMN-LKLAHTRLESRTYRSNVELCRDQT 334
Query: 63 STTLVRAIEEAELSVIVFSKNYAASKFCLDELMKIL 98
L+ + + E ++ + A ++ LD L K L
Sbjct: 335 QYGLIDEVHQLEATINTMKQKLAQTQNALDALFKHL 370
>PPOD_LYCES (Q08306) Polyphenol oxidase D, chloroplast precursor (EC
1.10.3.1) (PPO) (Catechol oxidase)
Length = 591
Score = 28.9 bits (63), Expect = 5.6
Identities = 18/59 (30%), Positives = 30/59 (50%), Gaps = 5/59 (8%)
Query: 2 AWSYSSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGD 60
A S+S + +S+ TPQEK+ + E N + N R D+ +DNN+N+ +
Sbjct: 464 AVSFSINRPTSSRTPQEKNA-----QEEMLTFNSIRYDNRGYIRFDVFLNVDNNVNANE 517
>MATP_MOUSE (P58355) Membrane-associated transporter protein (AIM-1
protein) (Melanoma antigen AIM1) (Underwhite protein)
Length = 530
Score = 28.9 bits (63), Expect = 5.6
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 9/43 (20%)
Query: 116 DPTDVRNQRGSYAEAFAKHEKNSEEKIK---------VQEWRN 149
DP ++ +GS A HE S EK+K VQEW+N
Sbjct: 248 DPPSQQDPQGSSLSASGMHEYGSIEKVKNGGADTEQPVQEWKN 290
>ENV_OMVVS (P16899) ENV polyprotein precursor (Coat polyprotein)
Length = 990
Score = 28.9 bits (63), Expect = 5.6
Identities = 19/86 (22%), Positives = 35/86 (40%), Gaps = 1/86 (1%)
Query: 75 LSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKH 134
+ VI F CL ++ E + + +V+ D++ + GS +H
Sbjct: 847 VGVICFRLLMCVITMCLQAYRQVREIRYTRVTVVIEAPVDLEEKQREERDGSSGSENLEH 906
Query: 135 EKNSEEKIKVQEWRNGLMEAANYSGW 160
EK + + +Q WR ++A S W
Sbjct: 907 EKRTSPRSFIQIWR-ATVQAWKTSPW 931
>AF10_HUMAN (P55197) AF-10 protein
Length = 1027
Score = 28.9 bits (63), Expect = 5.6
Identities = 19/64 (29%), Positives = 29/64 (44%), Gaps = 4/64 (6%)
Query: 4 SYSSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEIS 63
S SS+ SS +PQ+ F+SF D RN+ SH + D+ + G
Sbjct: 359 SVKSSSGSSVQSPQD----FLSFTDSDLRNDSYSHSQQSSATKDVHKGESGSQEGGVNSF 414
Query: 64 TTLV 67
+TL+
Sbjct: 415 STLI 418
>TEK2_HUMAN (Q9UIF3) Tektin-2 (Testicular tektin) (Tektin-t)
(Testicular tektin B1-like protein) (Tektin-B1) (TEKTB1)
Length = 430
Score = 28.5 bits (62), Expect = 7.3
Identities = 27/96 (28%), Positives = 39/96 (40%), Gaps = 3/96 (3%)
Query: 5 YSSSTSSSNNTPQEKHEVFISFR--SEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEI 62
YS NT +E E+ R ED R S L + RL+ RTY N D+
Sbjct: 276 YSELKWQEKNTLEEIAELQEDIRHLEEDLRTKLLS-LKLSHTRLEARTYRPNVELCRDQA 334
Query: 63 STTLVRAIEEAELSVIVFSKNYAASKFCLDELMKIL 98
L + + E ++ + A ++ LD L K L
Sbjct: 335 QYGLTDEVHQLEATIAALKQKLAQAQDALDALCKHL 370
>SYS_BORBU (O51244) Seryl-tRNA synthetase (EC 6.1.1.11)
(Serine--tRNA ligase) (SerRS)
Length = 425
Score = 28.5 bits (62), Expect = 7.3
Identities = 12/37 (32%), Positives = 20/37 (53%)
Query: 5 YSSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNG 41
Y TS+SN T + + I ++ +D +N F +NG
Sbjct: 347 YGEVTSTSNCTDYQSRRLKIRYKDQDGQNKFAHMVNG 383
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.128 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,064,964
Number of Sequences: 164201
Number of extensions: 718885
Number of successful extensions: 1561
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 1548
Number of HSP's gapped (non-prelim): 24
length of query: 166
length of database: 59,974,054
effective HSP length: 102
effective length of query: 64
effective length of database: 43,225,552
effective search space: 2766435328
effective search space used: 2766435328
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC134049.18


