

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134049.1 - phase: 0
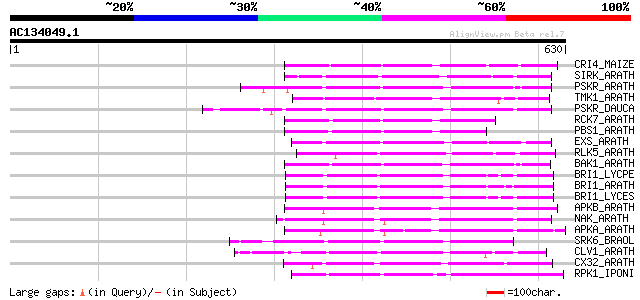
(630 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 213 1e-54
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 186 2e-46
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 186 2e-46
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 182 3e-45
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 179 1e-44
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 175 3e-43
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 174 8e-43
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 172 3e-42
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 169 2e-41
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 169 3e-41
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 167 8e-41
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 167 8e-41
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 166 1e-40
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 166 1e-40
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 165 4e-40
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 158 5e-38
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 157 6e-38
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 157 8e-38
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 157 1e-37
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 150 7e-36
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 213 bits (542), Expect = 1e-54
Identities = 126/309 (40%), Positives = 177/309 (56%), Gaps = 14/309 (4%)
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNE 372
F+Y+EL +T F ++G G F V+ G LRDG + AVK + + SSK F NE
Sbjct: 493 FSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKK-SSKEFHNE 551
Query: 373 ILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLE 432
+ +LS ++H +L+ L GYC D +LVY+++ +G+L +HLHG K + W R+
Sbjct: 552 LDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKK-RLNWARRVT 610
Query: 433 IAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSS 492
IA+Q A +EYLH PP++HRDI SSNI I++D +V DFGLS L S
Sbjct: 611 IAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSIL-------GPADS 663
Query: 493 GGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALA 552
G + P GT GYLDP+Y+R LT KSDVYSFGVVLLE++SG KA+D E +
Sbjct: 664 GTPLSELPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAID--MQFEEGNIV 721
Query: 553 DMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGEL 612
+ V I G + +LDPVL +D L+A+ +A +A +CV DRP +V L
Sbjct: 722 EWAVPLIKAGDIFAILDPVLSPPSD---LEALKKIASVACKCVRMRGKDRPSMDKVTTAL 778
Query: 613 KRVRSRISG 621
+ + + G
Sbjct: 779 EHALALLMG 787
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 186 bits (472), Expect = 2e-46
Identities = 110/303 (36%), Positives = 178/303 (58%), Gaps = 21/303 (6%)
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNE 372
F Y E+ TNNF+ R IG GGFG VY G + +G+ AVK L + +A K F E
Sbjct: 564 FKYSEVVNITNNFE--RVIGKGGFGKVYHGVI-NGEQVAVKVL---SEESAQGYKEFRAE 617
Query: 373 ILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLE 432
+ +L + H NL L GYC++ ++L+Y+Y+ N L ++L G +S ++++W+ RL+
Sbjct: 618 VDLLMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAGKRS----FILSWEERLK 673
Query: 433 IAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSS 492
I++ A +EYLH KPPIVHRD+ +NI + + ++ K+ DFGLSR ++ S Q
Sbjct: 674 ISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQ---- 729
Query: 493 GGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALA 552
+ T G+ GYLDP+Y+ + ++ EKSDVYS GVVLLE+I+G A+ + ++ + ++
Sbjct: 730 ---ISTVVAGSIGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKTEK-VHIS 785
Query: 553 DMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGEL 612
D V S + G ++ ++D L + + + ++E+A C RP +VV EL
Sbjct: 786 DHVRSILANGDIRGIVDQRL---RERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMEL 842
Query: 613 KRV 615
K++
Sbjct: 843 KQI 845
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 186 bits (472), Expect = 2e-46
Identities = 129/364 (35%), Positives = 188/364 (51%), Gaps = 27/364 (7%)
Query: 263 AIVIAFTSLILFLSVVISILRSRK----VNTTVEEDPTAVFLHNHRNANLLPPVF----- 313
AI IAF S+ L + + +LR+R+ V+ +EE + + L +F
Sbjct: 661 AIGIAFGSVFLLTLLSLIVLRARRRSGEVDPEIEESESMNRKELGEIGSKLVVLFQSNDK 720
Query: 314 --TYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCN 371
+YD+L STN+FD IG GGFG VY L DGK A+K L + + F
Sbjct: 721 ELSYDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKL---SGDCGQIEREFEA 777
Query: 372 EILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRL 431
E+ LS HPNLV L G+C +L+Y Y+ NG+L LH ++ W+TRL
Sbjct: 778 EVETLSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPA--LLKWKTRL 835
Query: 432 EIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTS 491
IA A + YLH P I+HRDI SSNI ++++ + DFGL+RL+ E++ +T
Sbjct: 836 RIAQGAAKGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHVSTD 895
Query: 492 SGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMAL 551
GT GY+ P+Y ++ T K DVYSFGVVLLEL++ + VD C+ K L
Sbjct: 896 L--------VGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDL 947
Query: 552 ADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGE 611
VV H + EV DP++ +ND + V E+A C++ + RP ++++V
Sbjct: 948 ISWVVKMKHESRASEVFDPLI-YSKEND--KEMFRVLEIACLCLSENPKQRPTTQQLVSW 1004
Query: 612 LKRV 615
L V
Sbjct: 1005 LDDV 1008
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 182 bits (461), Expect = 3e-45
Identities = 115/295 (38%), Positives = 157/295 (52%), Gaps = 18/295 (6%)
Query: 322 TNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILILSSIDH 381
TNNF +G GGFG VY G L DG AVK + + A F +EI +L+ + H
Sbjct: 585 TNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRME-NGVIAGKGFAEFKSEIAVLTKVRH 643
Query: 382 PNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTALAM 441
+LV L GYC D +LVY+Y+P GTL+ HL S+ + W+ RL +A+ A +
Sbjct: 644 RHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLF-EWSEEGLKPLLWKQRLTLALDVARGV 702
Query: 442 EYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWTGPQ 501
EYLH +HRD+ SNI + DMR KV DFGL RL G + T
Sbjct: 703 EYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRL--------APEGKGSIETRIA 754
Query: 502 GTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALAD----MVVS 557
GT GYL P+Y + R+T K DVYSFGV+L+ELI+G K++D + + + L M ++
Sbjct: 755 GTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMYIN 814
Query: 558 RIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGEL 612
+ K+ +D +DL D + L +V VAELA C A + RPD V L
Sbjct: 815 K--EASFKKAIDTTIDL--DEETLASVHTVAELAGHCCAREPYQRPDMGHAVNIL 865
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 179 bits (455), Expect = 1e-44
Identities = 130/410 (31%), Positives = 195/410 (46%), Gaps = 44/410 (10%)
Query: 220 NNGFCGFNSSDTKKQFVCYHFHSKSTLSPPWIHKMKPSKIAVFAIVIAFTSLILFLSVVI 279
N G CG ++S H +S K KI A+ ++ L ++
Sbjct: 638 NQGLCGEHASPC-------HITDQSPHGSAVKSKKNIRKIVAVAVGTGLGTVFLLTVTLL 690
Query: 280 SILRSRKVNTTVEEDP--------------TAVFLHNHRNANLLPPVFTYDELNISTNNF 325
ILR+ + E DP + V HN + N L + D++ ST++F
Sbjct: 691 IILRT---TSRGEVDPEKKADADEIELGSRSVVLFHNKDSNNEL----SLDDILKSTSSF 743
Query: 326 DPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILILSSIDHPNLV 385
+ IG GGFG VY L DG A+K L + + F E+ LS HPNLV
Sbjct: 744 NQANIIGCGGFGLVYKATLPDGTKVAIKRL---SGDTGQMDREFQAEVETLSRAQHPNLV 800
Query: 386 KLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTALAMEYLH 445
L GYC+ +L+Y Y+ NG+L LH + W+TRL IA A + YLH
Sbjct: 801 HLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPS--LDWKTRLRIARGAAEGLAYLH 858
Query: 446 FSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWTGPQGTPG 505
S +P I+HRDI SSNI + + DFGL+RL++ +++ TT GT G
Sbjct: 859 QSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYDTHVTTDL--------VGTLG 910
Query: 506 YLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMVVSRIHTGQLK 565
Y+ P+Y ++ T K DVYSFGVVLLEL++G + +D C+ + L V+ +
Sbjct: 911 YIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRES 970
Query: 566 EVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELKRV 615
E+ DP + D D + + V E+A RC+ + RP ++++V L+ +
Sbjct: 971 EIFDPFI---YDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLENI 1017
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 175 bits (444), Expect = 3e-43
Identities = 98/240 (40%), Positives = 145/240 (59%), Gaps = 13/240 (5%)
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRD-GKLAAVKHLHRHNHTAAFSSKSFCN 371
FT+ EL +T NF +G+GGFG V+ G + ++ A+K L R+ + F
Sbjct: 91 FTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGI---REFVV 147
Query: 372 EILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRL 431
E+L LS DHPNLVKL G+C++ +LVY+Y+P G+L +HLH S +K + W TR+
Sbjct: 148 EVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKK--PLDWNTRM 205
Query: 432 EIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTS 491
+IA A +EYLH + PP+++RD+ SNI + +D + K+ DFGL+++ +
Sbjct: 206 KIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKV-------GPSG 258
Query: 492 SGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMAL 551
V T GT GY PDY + +LT KSD+YSFGVVLLELI+G KA+D + +++ L
Sbjct: 259 DKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNL 318
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 174 bits (440), Expect = 8e-43
Identities = 97/230 (42%), Positives = 137/230 (59%), Gaps = 13/230 (5%)
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRD-GKLAAVKHLHRHNHTAAFSSKSFCN 371
F + EL +T NF P +G+GGFG VY G L G++ AVK L R+ ++ F
Sbjct: 74 FAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQG---NREFLV 130
Query: 372 EILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRL 431
E+L+LS + HPNLV L GYC+D +LVY+++P G+L +HLH ++ + W R+
Sbjct: 131 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEA--LDWNMRM 188
Query: 432 EIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTS 491
+IA A +E+LH PP+++RD SSNI +++ K+ DFGL++L T
Sbjct: 189 KIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKL-------GPTG 241
Query: 492 SGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVD 541
V T GT GY P+Y + +LT KSDVYSFGVV LELI+G KA+D
Sbjct: 242 DKSHVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAID 291
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 172 bits (435), Expect = 3e-42
Identities = 108/298 (36%), Positives = 162/298 (54%), Gaps = 21/298 (7%)
Query: 321 STNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILILSSID 380
+T++F K IGDGGFG+VY L K AVK L + ++ F E+ L +
Sbjct: 913 ATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKL---SEAKTQGNREFMAEMETLGKVK 969
Query: 381 HPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTALA 440
HPNLV L GYCS +LVY+Y+ NG+L L + ++ W RL+IA+ A
Sbjct: 970 HPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLE--VLDWSKRLKIAVGAARG 1027
Query: 441 MEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWTGP 500
+ +LH P I+HRDI +SNI ++ D KV DFGL+RL+ ES+ +T
Sbjct: 1028 LAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVSTVIA------- 1080
Query: 501 QGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAV--DYCRDKREMALADMVVSR 558
GT GY+ P+Y +S R T K DVYSFGV+LLEL++G + D+ ++ L + +
Sbjct: 1081 -GTFGYIPPEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDF-KESEGGNLVGWAIQK 1138
Query: 559 IHTGQLKEVLDPVL-DLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELKRV 615
I+ G+ +V+DP+L + N L + ++A C+A RP+ +V+ LK +
Sbjct: 1139 INQGKAVDVIDPLLVSVALKNSQL----RLLQIAMLCLAETPAKRPNMLDVLKALKEI 1192
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 169 bits (428), Expect = 2e-41
Identities = 108/301 (35%), Positives = 162/301 (52%), Gaps = 20/301 (6%)
Query: 326 DPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRH--NHTAAFSSKS-----FCNEILILSS 378
D K IG G G VY LR G++ AVK L++ +SS S F E+ L +
Sbjct: 684 DEKNVIGFGSSGKVYKVELRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAEVETLGT 743
Query: 379 IDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTA 438
I H ++V+L CS +LVY+Y+PNG+LA+ LHG + + G ++ W RL IA+ A
Sbjct: 744 IRHKSIVRLWCCCSSGDCKLLVYEYMPNGSLADVLHGDR--KGGVVLGWPERLRIALDAA 801
Query: 439 LAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWT 498
+ YLH PPIVHRD+ SSNI ++ D KV DFG++++ + S + G
Sbjct: 802 EGLSYLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAMSGIA-- 859
Query: 499 GPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMVVSR 558
G+ GY+ P+Y + R+ EKSD+YSFGVVLLEL++G + D ++M A V +
Sbjct: 860 ---GSCGYIAPEYVYTLRVNEKSDIYSFGVVLLELVTGKQPTDSELGDKDM--AKWVCTA 914
Query: 559 IHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELKRVRSR 618
+ L+ V+DP LDL + + V + C + +RP ++VV L+ V
Sbjct: 915 LDKCGLEPVIDPKLDL----KFKEEISKVIHIGLLCTSPLPLNRPSMRKVVIMLQEVSGA 970
Query: 619 I 619
+
Sbjct: 971 V 971
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 169 bits (427), Expect = 3e-41
Identities = 103/303 (33%), Positives = 167/303 (54%), Gaps = 17/303 (5%)
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNE 372
F+ EL ++++NF K +G GGFG VY G L DG L AVK L F E
Sbjct: 277 FSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGG--ELQFQTE 334
Query: 373 ILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLE 432
+ ++S H NL++L G+C P +LVY Y+ NG++A L + + W R
Sbjct: 335 VEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPP--LDWPKRQR 392
Query: 433 IAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSS 492
IA+ +A + YLH P I+HRD+ ++NI ++++ VGDFGL++L+ ++++ TT+
Sbjct: 393 IALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTA- 451
Query: 493 GGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCR--DKREMA 550
+GT G++ P+Y + + +EK+DV+ +GV+LLELI+G +A D R + ++
Sbjct: 452 -------VRGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVM 504
Query: 551 LADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVG 610
L D V + +L+ ++D L GN D + V + ++A C S +RP EVV
Sbjct: 505 LLDWVKGLLKEKKLEALVDVDLQ-GNYKD--EEVEQLIQVALLCTQSSPMERPKMSEVVR 561
Query: 611 ELK 613
L+
Sbjct: 562 MLE 564
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 167 bits (423), Expect = 8e-41
Identities = 104/307 (33%), Positives = 165/307 (52%), Gaps = 21/307 (6%)
Query: 314 TYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEI 373
T+ +L +TN F +G GGFG VY L+DG + A+K L H + + F E+
Sbjct: 877 TFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLI---HVSGQGDREFTAEM 933
Query: 374 LILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEI 433
+ I H NLV L GYC +LVY+Y+ G+L + LH K + G + W R +I
Sbjct: 934 ETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRK--KTGIKLNWPARRKI 991
Query: 434 AIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSG 493
AI A + +LH + P I+HRD+ SSN+ +++++ +V DFG++RL+ +++ + S+
Sbjct: 992 AIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVST- 1050
Query: 494 GFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALAD 553
GTPGY+ P+Y++SFR + K DVYS+GVVLLEL++G + D D + L
Sbjct: 1051 ------LAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVG 1103
Query: 554 MVVSRIHT-GQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKD--DRPDSKEVVG 610
V ++H G++ +V D L DA + + L C D RP +V+
Sbjct: 1104 WV--KLHAKGKITDVFDREL---LKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMA 1158
Query: 611 ELKRVRS 617
K +++
Sbjct: 1159 MFKEIQA 1165
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 167 bits (423), Expect = 8e-41
Identities = 103/305 (33%), Positives = 168/305 (54%), Gaps = 17/305 (5%)
Query: 314 TYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEI 373
T+ +L +TN F IG GGFG VY L+DG A+K L H + + F E+
Sbjct: 872 TFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLI---HVSGQGDREFMAEM 928
Query: 374 LILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEI 433
+ I H NLV L GYC +LVY+++ G+L + LH K + G + W TR +I
Sbjct: 929 ETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPK--KAGVKLNWSTRRKI 986
Query: 434 AIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSG 493
AI +A + +LH + P I+HRD+ SSN+ +++++ +V DFG++RL+ +++ + S+
Sbjct: 987 AIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVST- 1045
Query: 494 GFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALAD 553
GTPGY+ P+Y++SFR + K DVYS+GVVLLEL++G + D D + L
Sbjct: 1046 ------LAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTD-SPDFGDNNLVG 1098
Query: 554 MVVSRIHTGQLKEVLDPVLDLGNDNDALD-AVGAVAELAFRCVASDKDDRPDSKEVVGEL 612
V ++ +V DP +L ++ AL+ + ++A C+ RP +V+
Sbjct: 1099 WVKQHAKL-RISDVFDP--ELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVMAMF 1155
Query: 613 KRVRS 617
K +++
Sbjct: 1156 KEIQA 1160
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 166 bits (421), Expect = 1e-40
Identities = 104/307 (33%), Positives = 165/307 (52%), Gaps = 21/307 (6%)
Query: 314 TYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEI 373
T+ +L +TN F +G GGFG VY L+DG + A+K L H + + F E+
Sbjct: 877 TFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLI---HVSGQGDREFTAEM 933
Query: 374 LILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEI 433
+ I H NLV L GYC +LVY+Y+ G+L + LH K + G + W R +I
Sbjct: 934 ETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRK--KIGIKLNWPARRKI 991
Query: 434 AIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSG 493
AI A + +LH + P I+HRD+ SSN+ +++++ +V DFG++RL+ +++ + S+
Sbjct: 992 AIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVST- 1050
Query: 494 GFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALAD 553
GTPGY+ P+Y++SFR + K DVYS+GVVLLEL++G + D D + L
Sbjct: 1051 ------LAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVG 1103
Query: 554 MVVSRIHT-GQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKD--DRPDSKEVVG 610
V ++H G++ +V D L DA + + L C D RP +V+
Sbjct: 1104 WV--KLHAKGKITDVFDREL---LKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMA 1158
Query: 611 ELKRVRS 617
K +++
Sbjct: 1159 MFKEIQA 1165
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 166 bits (421), Expect = 1e-40
Identities = 108/320 (33%), Positives = 168/320 (51%), Gaps = 25/320 (7%)
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKH-------LHRHNHTAAFS 365
FT+ EL +T NF P +G+GGFGSV+ G + + L A K + + N
Sbjct: 57 FTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQG 116
Query: 366 SKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYM- 424
+ + E+ L HPNLVKL GYC + +LVY+++P G+L HL +R Y
Sbjct: 117 HQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLF----RRGSYFQ 172
Query: 425 -MTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVL 483
++W RL++A+ A + +LH + + +++RD +SNI ++ + K+ DFGL++
Sbjct: 173 PLSWTLRLKVALGAAKGLAFLH-NAETSVIYRDFKTSNILLDSEYNAKLSDFGLAK---- 227
Query: 484 QESNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYC 543
+ T V T GT GY P+Y + LT KSDVYS+GVVLLE++SG +AVD
Sbjct: 228 ---DGPTGDKSHVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKN 284
Query: 544 RDKREMALADMVVSRI-HTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDR 602
R E L + + + +L V+D L D +++ VA LA RC+ + R
Sbjct: 285 RPPGEQKLVEWARPLLANKRKLFRVIDNRL---QDQYSMEEACKVATLALRCLTFEIKLR 341
Query: 603 PDSKEVVGELKRVRSRISGG 622
P+ EVV L+ +++ G
Sbjct: 342 PNMNEVVSHLEHIQTLNEAG 361
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 165 bits (417), Expect = 4e-40
Identities = 110/323 (34%), Positives = 167/323 (51%), Gaps = 29/323 (8%)
Query: 304 RNANLLPPVFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKH-------LH 356
+NANL F+ EL +T NF P +G+GGFG V+ G + + LA K +
Sbjct: 49 QNANLKN--FSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVK 106
Query: 357 RHNHTAAFSSKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGS 416
R N + + EI L +DHPNLVKL GYC + +LVY+++ G+L HL
Sbjct: 107 RLNQEGFQGHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLF-- 164
Query: 417 KSKRKGYM---MTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVG 473
R+G ++W TR+ +A+ A + +LH + +P +++RD +SNI ++ + K+
Sbjct: 165 ---RRGTFYQPLSWNTRVRMALGAARGLAFLH-NAQPQVIYRDFKASNILLDSNYNAKLS 220
Query: 474 DFGLSRLLVLQESNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLEL 533
DFGL+R + V T GT GY P+Y + L+ KSDVYSFGVVLLEL
Sbjct: 221 DFGLAR-------DGPMGDNSHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLEL 273
Query: 534 ISGLKAVDYCRDKREMALADMVVSRI-HTGQLKEVLDPVLDLGNDNDALDAVGAVAELAF 592
+SG +A+D + E L D + + +L V+DP L +L +A LA
Sbjct: 274 LSGRRAIDKNQPVGEHNLVDWARPYLTNKRRLLRVMDPRL---QGQYSLTRALKIAVLAL 330
Query: 593 RCVASDKDDRPDSKEVVGELKRV 615
C++ D RP E+V ++ +
Sbjct: 331 DCISIDAKSRPTMNEIVKTMEEL 353
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 158 bits (399), Expect = 5e-38
Identities = 107/329 (32%), Positives = 172/329 (51%), Gaps = 28/329 (8%)
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAA-------VKHLHRHNHTAAFS 365
F++ EL +T NF P +G+GGFG V+ G + + L A V + + N
Sbjct: 56 FSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQG 115
Query: 366 SKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYM- 424
+ + E+ L H +LVKL GYC + +LVY+++P G+L HL R+G
Sbjct: 116 HQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLF-----RRGLYF 170
Query: 425 --MTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLV 482
++W+ RL++A+ A + +LH S + +++RD +SNI ++ + K+ DFGL++
Sbjct: 171 QPLSWKLRLKVALGAAKGLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSDFGLAK--- 226
Query: 483 LQESNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDY 542
+ V T GT GY P+Y + LT KSDVYSFGVVLLEL+SG +AVD
Sbjct: 227 ----DGPIGDKSHVSTRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDK 282
Query: 543 CRDKREMALADMVVS-RIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDD 601
R E L + ++ ++ V+D L D +++ VA L+ RC+ ++
Sbjct: 283 NRPSGERNLVEWAKPYLVNKRKIFRVIDNRL---QDQYSMEEACKVATLSLRCLTTEIKL 339
Query: 602 RPDSKEVVGELKRVRSRISGGITRSMSTT 630
RP+ EVV L+ ++S ++ I +M T
Sbjct: 340 RPNMSEVVSHLEHIQS-LNAAIGGNMDKT 367
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 157 bits (398), Expect = 6e-38
Identities = 106/323 (32%), Positives = 164/323 (49%), Gaps = 26/323 (8%)
Query: 250 WIHKMKPSKIAVFAIVIAFTSLILFLSVVISILRSRKVNTTVEEDPTAVFLHNHRNANLL 309
W K K +K + AI IA T L + +L S++ F ++ L
Sbjct: 466 WKRKQKRAKAS--AISIANTQRNQNLPMNEMVLSSKRE-----------FSGEYKFEELE 512
Query: 310 PPVFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSF 369
P+ + + +T NF K+G GGFG VY G L DGK AVK L + T+ + F
Sbjct: 513 LPLIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSK---TSVQGTDEF 569
Query: 370 CNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQT 429
NE+ +++ + H NLV++ G C + +L+Y+Y+ N +L +L G + K + W
Sbjct: 570 MNEVTLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSK---LNWNE 626
Query: 430 RLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQT 489
R +I A + YLH + I+HRD+ SNI ++K+M K+ DFG++R+ E+
Sbjct: 627 RFDITNGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEAN 686
Query: 490 TSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREM 549
T GT GY+ P+Y +EKSDV+SFGV++LE++SG K + E
Sbjct: 687 TMK-------VVGTYGYMSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYEN 739
Query: 550 ALADMVVSRIHTGQLKEVLDPVL 572
L V SR G+ E++DPV+
Sbjct: 740 DLLSYVWSRWKEGRALEIVDPVI 762
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 157 bits (397), Expect = 8e-38
Identities = 122/361 (33%), Positives = 183/361 (49%), Gaps = 41/361 (11%)
Query: 256 PSKIAVFAIVIAFTSLILFLSVVISILRSRKVNTTVEEDPTAVFLHNHRNANLLPPVFTY 315
PS+I V ++ A T LIL +SV I + +K ++ TA + ++ ++L +
Sbjct: 638 PSRI-VITVIAAITGLIL-ISVAIRQMNKKKNQKSLAWKLTAFQKLDFKSEDVLECL--- 692
Query: 316 DELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILI 375
E NI IG GG G VY G++ + A+K L S F EI
Sbjct: 693 KEENI----------IGKGGAGIVYRGSMPNNVDVAIKRLVGRG--TGRSDHGFTAEIQT 740
Query: 376 LSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAI 435
L I H ++V+L GY ++ +L+Y+Y+PNG+L E LHGSK G + W+TR +A+
Sbjct: 741 LGRIRHRHIVRLLGYVANKDTNLLLYEYMPNGSLGELLHGSK----GGHLQWETRHRVAV 796
Query: 436 QTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGF 495
+ A + YLH P I+HRD+ S+NI ++ D V DFGL++ LV +++ SS
Sbjct: 797 EAAKGLCYLHHDCSPLILHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSS--- 853
Query: 496 VWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLK-------AVDYCRDKRE 548
G+ GY+ P+Y + ++ EKSDVYSFGVVLLELI+G K VD R R
Sbjct: 854 ----IAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIAGKKPVGEFGEGVDIVRWVRN 909
Query: 549 MALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEV 608
+ + + ++DP L L +V V ++A CV + RP +EV
Sbjct: 910 --TEEEITQPSDAAIVVAIVDPRL----TGYPLTSVIHVFKIAMMCVEEEAAARPTMREV 963
Query: 609 V 609
V
Sbjct: 964 V 964
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 157 bits (396), Expect = 1e-37
Identities = 100/316 (31%), Positives = 161/316 (50%), Gaps = 30/316 (9%)
Query: 312 VFTYDELNISTNNFDPKRKIGDGGFGSVYLG----------NLRDGKLAAVKHLHRHNHT 361
V+ + +L +T NF P +G GGFG VY G + G + A+K L N
Sbjct: 73 VYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRL---NSE 129
Query: 362 AAFSSKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRK 421
+ + +E+ L + H NLVKL GYC + + L+LVY+++P G+L HL R+
Sbjct: 130 SVQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLF-----RR 184
Query: 422 GYMMTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLL 481
W R++I I A + +LH S++ +++RD +SNI ++ + K+ DFGL++L
Sbjct: 185 NDPFPWDLRIKIVIGAARGLAFLH-SLQREVIYRDFKASNILLDSNYDAKLSDFGLAKLG 243
Query: 482 VLQESNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVD 541
E + T T GT GY P+Y + L KSDV++FGVVLLE+++GL A +
Sbjct: 244 PADEKSHVT-------TRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHN 296
Query: 542 YCRDKREMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGA-VAELAFRCVASDKD 600
R + + +L D + + K + ++D G V +A + C+ D
Sbjct: 297 TKRPRGQESLVDWLRPELSN---KHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPK 353
Query: 601 DRPDSKEVVGELKRVR 616
+RP KEVV L+ ++
Sbjct: 354 NRPHMKEVVEVLEHIQ 369
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 150 bits (380), Expect = 7e-36
Identities = 102/309 (33%), Positives = 158/309 (51%), Gaps = 13/309 (4%)
Query: 321 STNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILILSSID 380
+T N + K IG G G++Y L K+ AVK L S S EI + +
Sbjct: 812 ATENLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTG--IKNGSVSMVREIETIGKVR 869
Query: 381 HPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTALA 440
H NL+KL + +++Y Y+ NG+L + LH + + + W TR IA+ TA
Sbjct: 870 HRNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPPKP---LDWSTRHNIAVGTAHG 926
Query: 441 MEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWTGP 500
+ YLHF P IVHRDI NI ++ D+ + DFG+++LL + + T+ V
Sbjct: 927 LAYLHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLL---DQSATSIPSNTV---- 979
Query: 501 QGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMVVSRIH 560
QGT GY+ P+ + + +SDVYS+GVVLLELI+ KA+D + + +
Sbjct: 980 QGTIGYMAPENAFTTVKSRESDVYSYGVVLLELITRKKALDPSFNGETDIVGWVRSVWTQ 1039
Query: 561 TGQLKEVLDP-VLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELKRVRSRI 619
TG++++++DP +LD D+ ++ V LA RC + D RP ++VV +L R R
Sbjct: 1040 TGEIQKIVDPSLLDELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQLTRWSIRS 1099
Query: 620 SGGITRSMS 628
R+ S
Sbjct: 1100 YSSSVRNKS 1108
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.135 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 77,444,562
Number of Sequences: 164201
Number of extensions: 3403515
Number of successful extensions: 13609
Number of sequences better than 10.0: 1619
Number of HSP's better than 10.0 without gapping: 994
Number of HSP's successfully gapped in prelim test: 626
Number of HSP's that attempted gapping in prelim test: 9561
Number of HSP's gapped (non-prelim): 2139
length of query: 630
length of database: 59,974,054
effective HSP length: 116
effective length of query: 514
effective length of database: 40,926,738
effective search space: 21036343332
effective search space used: 21036343332
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC134049.1


