

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133780.9 - phase: 0 /pseudo
(562 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
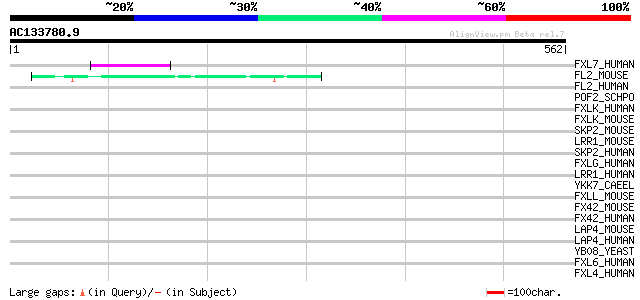
Score E
Sequences producing significant alignments: (bits) Value
FXL7_HUMAN (Q9UJT9) F-box/LRR-repeat protein 7 (F-box and leucin... 55 5e-07
FL2_MOUSE (Q8BH16) F-box/LRR-repeat protein 2 (F-box and leucine... 47 2e-04
FL2_HUMAN (Q9UKC9) F-box/LRR-repeat protein 2 (F-box and leucine... 44 0.001
POF2_SCHPO (O74783) F-box/LRR-repeat protein 2 (F-box and leucin... 42 0.006
FXLK_HUMAN (Q96IG2) F-box/LRR-repeat protein 20 (F-box and leuci... 41 0.009
FXLK_MOUSE (Q9CZV8) F-box/LRR-repeat protein 20 (F-box and leuci... 40 0.012
SKP2_MOUSE (Q9Z0Z3) S-phase kinase-associated protein 2 (F-box p... 40 0.016
LRR1_MOUSE (Q80VQ1) Leucine-rich repeat-containing protein 1 40 0.016
SKP2_HUMAN (Q13309) S-phase kinase-associated protein 2 (F-box p... 39 0.028
FXLG_HUMAN (Q8N461) F-box/LRR-repeat protein 16 (F-box and leuci... 39 0.028
LRR1_HUMAN (Q9BTT6) Leucine-rich repeat-containing protein 1 (LA... 39 0.036
YKK7_CAEEL (P34284) Hypothetical F-box/LRR-repeat protein C02F5.... 39 0.047
FXLL_MOUSE (Q8BFZ4) F-box/LRR-repeat protein 21 (F-box and leuci... 39 0.047
FX42_MOUSE (Q6PDJ6) F-box only protein 42 39 0.047
FX42_HUMAN (Q6P3S6) F-box only protein 42 39 0.047
LAP4_MOUSE (Q80U72) LAP4 protein (Scribble homolog protein) 37 0.18
LAP4_HUMAN (Q14160) LAP4 protein (Scribble homolog protein) (hSc... 37 0.18
YB08_YEAST (P38285) Hypothetical 62.7 kDa protein in CNS1-CDC28 ... 36 0.31
FXL6_HUMAN (Q8N531) F-box/LRR-repeat protein 6 (F-box and leucin... 36 0.31
FXL4_HUMAN (Q9UKA2) F-box/LRR-repeat protein 4 (F-box and leucin... 35 0.52
>FXL7_HUMAN (Q9UJT9) F-box/LRR-repeat protein 7 (F-box and
leucine-rich repeat protein 7) (F-box protein FBL6/FBL7)
Length = 491
Score = 55.1 bits (131), Expect = 5e-07
Identities = 36/82 (43%), Positives = 46/82 (55%), Gaps = 1/82 (1%)
Query: 83 GWGGFVYPWIEALAKNKVGLEELRL-KRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTD 141
G G +E LAKN L+ L + K +VSD LE L+ + N K L L SCE T
Sbjct: 385 GCEGITDHGVEYLAKNCTKLKSLDIGKCPLVSDTGLECLALNCFNLKRLSLKSCESITGQ 444
Query: 142 GLAAVAANCRSLRELDLQENEV 163
GL VAANC L+ L++Q+ EV
Sbjct: 445 GLQIVAANCFDLQTLNVQDCEV 466
Score = 35.0 bits (79), Expect = 0.52
Identities = 60/278 (21%), Positives = 106/278 (37%), Gaps = 23/278 (8%)
Query: 41 QRVFIGNCYSISPERL---VERFPDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAK 97
+R+ + CY+IS E + V P+L+ L + G SL + L
Sbjct: 215 RRLEVSGCYNISNEAVFDVVSLCPNLEHLDVSGCSKVTCISLTREA-----SIKLSPLHG 269
Query: 98 NKVGLEELRLKR-MVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLREL 156
++ + L + V+ DE L ++ L L C T +GL + C S++EL
Sbjct: 270 KQISIRYLDMTDCFVLEDEGLHTIAAHCTQLTHLYLRRCVRLTDEGLRYLVIYCASIKEL 329
Query: 157 DLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVS--RSPNLKSLRLN 214
+ + G ES +S+ D+ + + + S R N +
Sbjct: 330 SVSDCRFVSDFGLREIAKLESRLRYLSIAHCGRVTDVGIRYVAKYCSKLRYLNARGCEGI 389
Query: 215 RSVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAM--FKATILKCKSITSLSGFLEVA 272
V+ L + T+ L D+G D + A+ F L KS S++G
Sbjct: 390 TDHGVEYLAKNCTKLKSL-DIGKCPLVSDTGLECLALNCFNLKRLSLKSCESITG----- 443
Query: 273 PFSLAAIYPICQNLTSLNLSYAAGILGIELIKLI-RHC 309
L + C +L +LN+ + +E ++ + RHC
Sbjct: 444 -QGLQIVAANCFDLQTLNVQDCE--VSVEALRFVKRHC 478
>FL2_MOUSE (Q8BH16) F-box/LRR-repeat protein 2 (F-box and
leucine-rich repeat protein 2)
Length = 423
Score = 46.6 bits (109), Expect = 2e-04
Identities = 78/307 (25%), Positives = 117/307 (37%), Gaps = 46/307 (14%)
Query: 23 NSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPD-----LKSLTLKGKPHFADF 77
N L+L +W R++ F Q G R+VE L+ L+L+G D
Sbjct: 43 NILALDGSNWQRVDLFNFQTDVEG--------RVVENISKRCGGFLRKLSLRGCIGVGDS 94
Query: 78 SLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMV-VSDESLELLSRSFVNFKSLVLVSCE 136
SL + A+N +E L L ++D + LSR K L L SC
Sbjct: 95 SL-------------KTFAQNCRNIEHLNLNGCTKITDSTCYSLSRFCSKLKHLDLTSCV 141
Query: 137 GFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINL- 195
T L ++ CR+L L+L + +G + C L +L L+G L
Sbjct: 142 SVTNSSLKGISEGCRNLEYLNLSWCDQITKEG--IEALVRGCRGLKAL---LLRGCTQLE 196
Query: 196 -GALERLVSRSPNLKSLRLN--RSVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMF 252
AL+ + + L SL L + D + +I +L L + ++ A
Sbjct: 197 DEALKHIQNHCHELVSLNLQSCSRITDDGVVQICRGCHRLQALCLSG----CSNLTDASL 252
Query: 253 KATILKCKSITSLS----GFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRH 308
A L C + L L A F+L A C L ++L I L++L H
Sbjct: 253 TALGLNCPRLQVLEAARCSHLTDAGFTLLA--RNCHELEKMDLEECVLITDSTLVQLSIH 310
Query: 309 CGKLQRL 315
C KLQ L
Sbjct: 311 CPKLQAL 317
>FL2_HUMAN (Q9UKC9) F-box/LRR-repeat protein 2 (F-box and
leucine-rich repeat protein 2) (F-box protein FBL2/FBL3)
Length = 423
Score = 43.5 bits (101), Expect = 0.001
Identities = 79/307 (25%), Positives = 116/307 (37%), Gaps = 46/307 (14%)
Query: 23 NSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPD-----LKSLTLKGKPHFADF 77
N L+L +W RI+ F Q G R+VE L+ L+L+G D
Sbjct: 43 NILALDGSNWQRIDLFNFQIDVEG--------RVVENISKRCGGFLRKLSLRGCIGVGDS 94
Query: 78 SLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMV-VSDESLELLSRSFVNFKSLVLVSCE 136
SL + A+N +E L L ++D + LSR K L L SC
Sbjct: 95 SL-------------KTFAQNCRNIEHLNLNGCTKITDSTCYSLSRFCSKLKHLDLTSCV 141
Query: 137 GFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINL- 195
T L ++ CR+L L+L + G + C L +L L+G L
Sbjct: 142 SITNSSLKGISEGCRNLEYLNLSWCDQITKDG--IEALVRGCRGLKAL---LLRGCTQLE 196
Query: 196 -GALERLVSRSPNLKSLRLN--RSVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMF 252
AL+ + + L SL L + + + +I +L L + ++ A
Sbjct: 197 DEALKHIQNYCHELVSLNLQSCSRITDEGVVQICRGCHRLQALCLSG----CSNLTDASL 252
Query: 253 KATILKCKSITSLS----GFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRH 308
A L C + L L A F+L A C L ++L I LI+L H
Sbjct: 253 TALGLNCPRLQILEAARCSHLTDAGFTLLA--RNCHELEKMDLEECILITDSTLIQLSIH 310
Query: 309 CGKLQRL 315
C KLQ L
Sbjct: 311 CPKLQAL 317
>POF2_SCHPO (O74783) F-box/LRR-repeat protein 2 (F-box and
leucine-rich repeat protein 2) (F-box protein pof2)
Length = 463
Score = 41.6 bits (96), Expect = 0.006
Identities = 63/239 (26%), Positives = 97/239 (40%), Gaps = 45/239 (18%)
Query: 55 RLVERFPDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSD 114
RLV R LK L++ G + F F+ E A + L L + D
Sbjct: 218 RLVSRNRGLKELSMDGCTELSHFIT-------FLNLNCELDAMRALSLNNLP----DLKD 266
Query: 115 ESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQE-NEVEDHKGQWLSC 173
+EL++ F SL L C G T L ++ +SL L L E+ D Q C
Sbjct: 267 SDIELITCKFSKLNSLFLSKCIGLTDSSLLSLTKLSQSLTTLHLGHCYEITDIGVQ---C 323
Query: 174 FPESCTSLVSLNF-ACLK-GDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQ 231
+SC ++ ++F CL+ DI + A+ +L P L+ + L + + + L IL
Sbjct: 324 LLKSCKNITYIDFGGCLRLSDIAVSAIAKL----PYLQRVGLVKCICLTDLSVILLS--- 376
Query: 232 LMDLGIGSFFHDL-------------NSDAYAMFKATILKCKSITSLSGFL--EVAPFS 275
GSF +L S +Y M+ LK S+T ++ L E+ FS
Sbjct: 377 ------GSFSRNLERVHLSYCIGLTAKSVSYLMYNCKTLKHLSVTGINSILCTELRSFS 429
>FXLK_HUMAN (Q96IG2) F-box/LRR-repeat protein 20 (F-box and
leucine-rich repeat protein 20) (F-box/LRR-repeat
protein 2-like)
Length = 436
Score = 40.8 bits (94), Expect = 0.009
Identities = 73/310 (23%), Positives = 116/310 (36%), Gaps = 52/310 (16%)
Query: 23 NSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPD-----LKSLTLKGKPHFADF 77
N L+L +W RI+ F QR G R+VE L+ L+L+G D
Sbjct: 56 NVLALDGSNWQRIDLFDFQRDIEG--------RVVENISKRCGGFLRKLSLRGCLGVGDN 107
Query: 78 SLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMV-VSDESLELLSRSFVNFKSLVLVSCE 136
+L A+N +E L L +D + LS+ + L L SC
Sbjct: 108 AL-------------RTFAQNCRNIEVLNLNGCTKTTDATCTSLSKFCSKLRHLDLASCT 154
Query: 137 GFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLG 196
T L A++ C L +L++ + G + C L +L LKG L
Sbjct: 155 SITNMSLKALSEGCPLLEQLNISWCDQVTKDG--IQALVRGCGGLKAL---FLKGCTQLE 209
Query: 197 --ALERLVSRSPNLKSLRLNRSVPV--DALQRILTRAPQLMDLGIG-------SFFHDLN 245
AL+ + + P L +L L + + + L I +L L + + L
Sbjct: 210 DEALKYIGAHCPELVTLNLQTCLQITDEGLITICRGCHKLQSLCASGCSNITDAILNALG 269
Query: 246 SDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKL 305
+ + + +C +T + GF +A C L ++L I LI+L
Sbjct: 270 QNCPRLRILEVARCSQLTDV-GFTTLARN--------CHELEKMDLEECVQITDSTLIQL 320
Query: 306 IRHCGKLQRL 315
HC +LQ L
Sbjct: 321 SIHCPRLQVL 330
>FXLK_MOUSE (Q9CZV8) F-box/LRR-repeat protein 20 (F-box and
leucine-rich repeat protein 20) (F-box/LRR-repeat
protein 2-like)
Length = 436
Score = 40.4 bits (93), Expect = 0.012
Identities = 73/310 (23%), Positives = 116/310 (36%), Gaps = 52/310 (16%)
Query: 23 NSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPD-----LKSLTLKGKPHFADF 77
N L+L +W RI+ F QR G R+VE L+ L+L+G D
Sbjct: 56 NVLALDGSNWQRIDLFDFQRDIEG--------RVVENISKRCGGFLRKLSLRGCLGVGDN 107
Query: 78 SLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMV-VSDESLELLSRSFVNFKSLVLVSCE 136
+L A+N +E L L +D + LS+ + L L SC
Sbjct: 108 AL-------------RTFAQNCRNIEVLSLNGCTKTTDATCTSLSKFCSKLRHLDLASCT 154
Query: 137 GFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLG 196
T L A++ C L +L++ + G + C L +L LKG L
Sbjct: 155 SITNMSLKALSEGCPLLEQLNISWCDQVTKDG--IQALVRGCGGLKAL---FLKGCTQLE 209
Query: 197 --ALERLVSRSPNLKSLRLNRSVPV--DALQRILTRAPQLMDLGIG-------SFFHDLN 245
AL+ + + P L +L L + + + L I +L L + + L
Sbjct: 210 DEALKYIGAHCPELVTLNLQTCLQITDEGLITICRGCHKLQSLCASGCSNITDAILNALG 269
Query: 246 SDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKL 305
+ + + +C +T + GF +A C L ++L I LI+L
Sbjct: 270 QNCPRLRILEVARCSQLTDV-GFTTLARN--------CHELEKMDLEECVQITDSTLIQL 320
Query: 306 IRHCGKLQRL 315
HC +LQ L
Sbjct: 321 SIHCPRLQVL 330
>SKP2_MOUSE (Q9Z0Z3) S-phase kinase-associated protein 2 (F-box
protein Skp2) (Cyclin A/CDK2-associated protein p45)
(F-box/WD-40 protein 1) (FWD1)
Length = 424
Score = 40.0 bits (92), Expect = 0.016
Identities = 32/118 (27%), Positives = 59/118 (49%), Gaps = 6/118 (5%)
Query: 102 LEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQ-- 159
L+ L L+ + +SD ++ L+++ N L L C GF+ +A + ++C L EL+L
Sbjct: 208 LQNLSLEGLQLSDPIVKTLAQN-ENLVRLNLCGCSGFSESAVATLLSSCSRLDELNLSWC 266
Query: 160 ENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSV 217
+ E H ++ P + T LN + + ++ L ++ R PNL L L+ S+
Sbjct: 267 FDFTEKHVQAAVAHLPNTIT---QLNLSGYRKNLQKTDLCTIIKRCPNLIRLDLSDSI 321
>LRR1_MOUSE (Q80VQ1) Leucine-rich repeat-containing protein 1
Length = 524
Score = 40.0 bits (92), Expect = 0.016
Identities = 37/136 (27%), Positives = 62/136 (45%), Gaps = 12/136 (8%)
Query: 105 LRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVE 164
++L+++ +SD ++ L NF LV + + A C++L +V
Sbjct: 59 VKLRKLGLSDNEIQRLPPEIANFMQLVELDVSRNDIPEIPESIAFCKAL--------QVA 110
Query: 165 DHKGQWLSCFPESCTSLVSLNFACLK-GDINLGALERLVSRSPNLKSLRLNRSVPVDALQ 223
D G L+ PES L N CL DI+L +L + NL SL L ++ + L
Sbjct: 111 DFSGNPLTRLPESFPEL--QNLTCLSVNDISLQSLPENIGNLYNLASLELRENL-LTYLP 167
Query: 224 RILTRAPQLMDLGIGS 239
LT+ +L +L +G+
Sbjct: 168 DSLTQLRRLEELDLGN 183
>SKP2_HUMAN (Q13309) S-phase kinase-associated protein 2 (F-box
protein Skp2) (Cyclin A/CDK2-associated protein p45)
(p45skp2) (F-box/LRR-repeat protein 1)
Length = 424
Score = 39.3 bits (90), Expect = 0.028
Identities = 35/118 (29%), Positives = 57/118 (47%), Gaps = 6/118 (5%)
Query: 102 LEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQ-- 159
L+ L L+ + +SD + L+++ N L L C GF+ L + ++C L EL+L
Sbjct: 208 LQNLSLEGLRLSDPIVNTLAKNS-NLVRLNLSGCSGFSEFALQTLLSSCSRLDELNLSWC 266
Query: 160 ENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSV 217
+ E H ++ E+ T LN + + ++ L LV R PNL L L+ SV
Sbjct: 267 FDFTEKHVQVAVAHVSETIT---QLNLSGYRKNLQKSDLSTLVRRCPNLVHLDLSDSV 321
>FXLG_HUMAN (Q8N461) F-box/LRR-repeat protein 16 (F-box and
leucine-rich repeat protein 16) (cg14134)
Length = 479
Score = 39.3 bits (90), Expect = 0.028
Identities = 39/163 (23%), Positives = 69/163 (41%), Gaps = 20/163 (12%)
Query: 6 DEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKS 65
DE I + + S ++ L+ VCK+W R+ + + + G + + L P
Sbjct: 98 DEKILNGLFWYFSACEKCVLAQVCKAWRRV--LYQPKFWAGLTPVLHAKELYNVLPG--- 152
Query: 66 LTLKGKPHFADF-SLVPHGWGGF---------VYPWIEALAKNKVGLEELRLKRMVVSDE 115
G+ F + G+ GF + +I+ A +K G++ + LKR ++D
Sbjct: 153 ----GEKEFVNLQGFAARGFEGFCLVGVSDLDICEFIDNYALSKKGVKAMSLKRSTITDA 208
Query: 116 SLELLSRSFVNFKSLVLVSCEGFTTDGL-AAVAANCRSLRELD 157
LE++ L L C FT GL ++++A SL D
Sbjct: 209 GLEVMLEQMQGVVRLELSGCNDFTEAGLWSSLSARITSLSVSD 251
Score = 32.7 bits (73), Expect = 2.6
Identities = 17/36 (47%), Positives = 19/36 (52%)
Query: 123 SFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDL 158
S N +L L C T DG+ VA N R LR LDL
Sbjct: 318 SLPNLTALSLSGCSKVTDDGVELVAENLRKLRSLDL 353
>LRR1_HUMAN (Q9BTT6) Leucine-rich repeat-containing protein 1 (LAP
and no PDZ protein) (LANO adapter protein)
Length = 524
Score = 38.9 bits (89), Expect = 0.036
Identities = 36/136 (26%), Positives = 62/136 (45%), Gaps = 12/136 (8%)
Query: 105 LRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVE 164
++L+++ +SD ++ L NF LV + + + C++L +V
Sbjct: 59 VKLRKLGLSDNEIQRLPPEIANFMQLVELDVSRNEIPEIPESISFCKAL--------QVA 110
Query: 165 DHKGQWLSCFPESCTSLVSLNFACLK-GDINLGALERLVSRSPNLKSLRLNRSVPVDALQ 223
D G L+ PES L N CL DI+L +L + NL SL L ++ + L
Sbjct: 111 DFSGNPLTRLPESFPEL--QNLTCLSVNDISLQSLPENIGNLYNLASLELRENL-LTYLP 167
Query: 224 RILTRAPQLMDLGIGS 239
LT+ +L +L +G+
Sbjct: 168 DSLTQLRRLEELDLGN 183
>YKK7_CAEEL (P34284) Hypothetical F-box/LRR-repeat protein C02F5.7
in chromosome III
Length = 461
Score = 38.5 bits (88), Expect = 0.047
Identities = 27/98 (27%), Positives = 46/98 (46%), Gaps = 5/98 (5%)
Query: 92 IEALAKNKVGLEELRLKRM-VVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANC 150
++ +A LE L + +SD SL L + N K L L C +G +A C
Sbjct: 271 VQNIANGATALEYLCMSNCNQISDRSLVSLGQHSHNLKVLELSGCTLLGDNGFIPLARGC 330
Query: 151 RSLRELDLQE-NEVEDHKGQWLSCFPESCTSLVSLNFA 187
R L LD+++ + + DH ++ +CT+L L+ +
Sbjct: 331 RQLERLDMEDCSLISDHT---INSLANNCTALRELSLS 365
Score = 31.2 bits (69), Expect = 7.5
Identities = 51/227 (22%), Positives = 91/227 (39%), Gaps = 49/227 (21%)
Query: 112 VSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWL 171
+ D ++++ + + +L+L CEG T + +V A+ ++++L+L L
Sbjct: 214 IQDRGVQIILSNCKSLDTLILRGCEGLTENVFGSVEAHMGAIKKLNL------------L 261
Query: 172 SCF-------PESCTSLVSLNFACLK--GDINLGALERLVSRSPNLKSLRL--------N 214
CF +L + C+ I+ +L L S NLK L L N
Sbjct: 262 QCFQLTDITVQNIANGATALEYLCMSNCNQISDRSLVSLGQHSHNLKVLELSGCTLLGDN 321
Query: 215 RSVPVDALQRILTRAPQLMDLGIGSFFHDLNS---DAYAMFKATILKCKSIT--SLSGFL 269
+P+ R L R + D + S H +NS + A+ + ++ C+ IT S+
Sbjct: 322 GFIPLARGCRQLERL-DMEDCSLIS-DHTINSLANNCTALRELSLSHCELITDESIQNLA 379
Query: 270 EVAPFSLAAI-YPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQRL 315
+L + C LT LS+ +RHC L+R+
Sbjct: 380 SKHRETLNVLELDNCPQLTDSTLSH------------LRHCKALKRI 414
>FXLL_MOUSE (Q8BFZ4) F-box/LRR-repeat protein 21 (F-box and
leucine-rich repeat protein 21) (F-box/LRR-repeat
protein 3B) (F-box and leucine-rich repeat protein 3B)
Length = 434
Score = 38.5 bits (88), Expect = 0.047
Identities = 57/221 (25%), Positives = 90/221 (39%), Gaps = 29/221 (13%)
Query: 98 NKVGLEELRLKRMVVSDESLELL-SRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLREL 156
N L ++++ V D SL++L + + + L + SC ++DG+ VA +C+ LREL
Sbjct: 176 NSKSLSSIKIEDTPVDDPSLKILVANNSDTLRLLKMSSCPHVSSDGILCVADHCQGLREL 235
Query: 157 DLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRS 216
L + D LS S +N L+ D+ VS +P +
Sbjct: 236 ALNYYILSDEILLALS-------SETHVNLEHLRIDV--------VSENPGQIKFHSIKK 280
Query: 217 VPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSL 276
DAL + R +M +F + A FK +T L V+ L
Sbjct: 281 RSWDALIKHSPRVNVVM------YFFLYEEEFEAFFKEE----TPVTHLYFGRSVSRAIL 330
Query: 277 AAIYPICQNLTSLNLSYAAGILGI--ELIKLIRHCGKLQRL 315
I C L L + A G+L + ELI++ +HC L L
Sbjct: 331 GRIGLNCPRLIEL-VVCANGLLPLDSELIRIAKHCKNLTSL 370
>FX42_MOUSE (Q6PDJ6) F-box only protein 42
Length = 717
Score = 38.5 bits (88), Expect = 0.047
Identities = 13/46 (28%), Positives = 28/46 (60%)
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIG 46
M+ P+EV+E++ ++ + + + +LVCK WYR+ + + + G
Sbjct: 47 MSELPEEVLEYILSFLSPYQEHKTAALVCKQWYRLIKGVAHQCYHG 92
>FX42_HUMAN (Q6P3S6) F-box only protein 42
Length = 717
Score = 38.5 bits (88), Expect = 0.047
Identities = 13/46 (28%), Positives = 28/46 (60%)
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIG 46
M+ P+EV+E++ ++ + + + +LVCK WYR+ + + + G
Sbjct: 47 MSELPEEVLEYILSFLSPYQEHKTAALVCKQWYRLIKGVAHQCYHG 92
>LAP4_MOUSE (Q80U72) LAP4 protein (Scribble homolog protein)
Length = 1612
Score = 36.6 bits (83), Expect = 0.18
Identities = 38/134 (28%), Positives = 57/134 (42%), Gaps = 12/134 (8%)
Query: 105 LRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVE 164
L L+++ +SD ++ L NF LV + + C++L E+
Sbjct: 59 LNLRKLGLSDNEIQRLPPEVANFMQLVELDVSRNDIPEIPESIKFCKAL--------EIA 110
Query: 165 DHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSV--PVDAL 222
D G LS P+ T L SL L D++L AL V NL +L L ++ + A
Sbjct: 111 DFSGNPLSRLPDGFTQLRSLAHLAL-NDVSLQALPGDVGNLANLVTLELRENLLKSLPAS 169
Query: 223 QRILTRAPQLMDLG 236
L + QL DLG
Sbjct: 170 LSFLVKLEQL-DLG 182
>LAP4_HUMAN (Q14160) LAP4 protein (Scribble homolog protein)
(hScrib)
Length = 1630
Score = 36.6 bits (83), Expect = 0.18
Identities = 38/134 (28%), Positives = 57/134 (42%), Gaps = 12/134 (8%)
Query: 105 LRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVE 164
L L+++ +SD ++ L NF LV + + C++L E+
Sbjct: 59 LNLRKLGLSDNEIQRLPPEVANFMQLVELDVSRNDIPEIPESIKFCKAL--------EIA 110
Query: 165 DHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSV--PVDAL 222
D G LS P+ T L SL L D++L AL V NL +L L ++ + A
Sbjct: 111 DFSGNPLSRLPDGFTQLRSLAHLAL-NDVSLQALPGDVGNLANLVTLELRENLLKSLPAS 169
Query: 223 QRILTRAPQLMDLG 236
L + QL DLG
Sbjct: 170 LSFLVKLEQL-DLG 182
>YB08_YEAST (P38285) Hypothetical 62.7 kDa protein in CNS1-CDC28
intergenic region
Length = 549
Score = 35.8 bits (81), Expect = 0.31
Identities = 16/47 (34%), Positives = 26/47 (55%)
Query: 112 VSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDL 158
+ D L LS+S N K LVL +C+ + G+ +A NC L+ ++
Sbjct: 349 IDDNFLLRLSQSIPNLKHLVLRACDNVSDSGVVCIALNCPKLKTFNI 395
>FXL6_HUMAN (Q8N531) F-box/LRR-repeat protein 6 (F-box and
leucine-rich repeat protein 6) (F-box protein FBL6)
Length = 539
Score = 35.8 bits (81), Expect = 0.31
Identities = 40/132 (30%), Positives = 54/132 (40%), Gaps = 23/132 (17%)
Query: 117 LELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVED----------- 165
L+L+ L L C G T D L +A C L LDLQ + VE
Sbjct: 212 LKLVGECCPRLTFLKLSGCHGVTADALVMLAKACCQLHSLDLQHSMVESTAVVSFLEEAG 271
Query: 166 --HKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQ 223
+ WL+ ++ L +L +C L LE VS N S+ L +PV+ALQ
Sbjct: 272 SRMRKLWLTYSSQTTAILGALLGSCCP---QLQVLE--VSTGINRNSIPL--QLPVEALQ 324
Query: 224 RILTRAPQLMDL 235
+ PQL L
Sbjct: 325 K---GCPQLQVL 333
>FXL4_HUMAN (Q9UKA2) F-box/LRR-repeat protein 4 (F-box and
leucine-rich repeat protein 4) (F-box protein FBL4/FBL5)
Length = 621
Score = 35.0 bits (79), Expect = 0.52
Identities = 37/114 (32%), Positives = 49/114 (42%), Gaps = 32/114 (28%)
Query: 128 KSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSLNFA 187
++L L C+ T +G+A +A+ C L ELDL W C +L S
Sbjct: 482 RTLDLWRCKNITENGIAELASGCPLLEELDL----------GW-------CPTLQS---- 520
Query: 188 CLKGDINLGALERLVSRSPNLKSLRL--NRSV---PVDALQRILTRAPQLMDLG 236
+ G RL + PNL+ L L NRSV +D L TR QL LG
Sbjct: 521 ------STGCFTRLAHQLPNLQKLFLTANRSVCDTDIDELACNCTRLQQLDILG 568
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.340 0.146 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 58,346,893
Number of Sequences: 164201
Number of extensions: 2169887
Number of successful extensions: 7218
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 7097
Number of HSP's gapped (non-prelim): 126
length of query: 562
length of database: 59,974,054
effective HSP length: 115
effective length of query: 447
effective length of database: 41,090,939
effective search space: 18367649733
effective search space used: 18367649733
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 68 (30.8 bits)
Medicago: description of AC133780.9


