

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133780.4 - phase: 1
(377 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
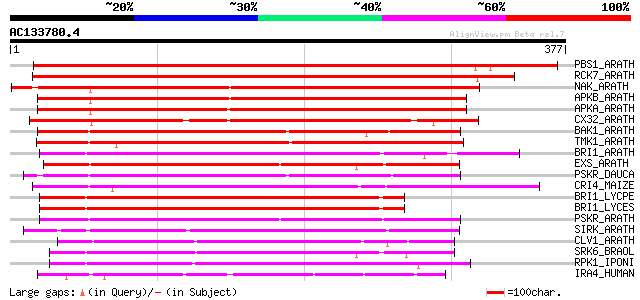
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 456 e-128
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 407 e-113
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 306 4e-83
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 295 1e-79
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 285 1e-76
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 272 9e-73
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 235 2e-61
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 227 4e-59
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 223 5e-58
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 223 6e-58
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 215 2e-55
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 213 6e-55
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 212 1e-54
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 211 2e-54
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 205 2e-52
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 196 8e-50
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 195 2e-49
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 193 5e-49
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 192 9e-49
IRA4_HUMAN (Q9NWZ3) Interleukin-1 receptor-associated kinase-4 (... 183 5e-46
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 456 bits (1173), Expect = e-128
Identities = 228/381 (59%), Positives = 280/381 (72%), Gaps = 25/381 (6%)
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
A F FRELA AT NF +TF+G+GGFG VYKG+L STGQ VAVK+LD G QG +EFLV
Sbjct: 71 AHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLV 130
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHHPNLV++IGYCA+GDQRLLVYE+MP+GSLE HLHDL PD E LDWN RM+I
Sbjct: 131 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKI 190
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRV 196
A GAA+GL +LH +A P VIYRD KSSNILLDEGF+PKLSDFGLAK GPTGD+S+V+TRV
Sbjct: 191 AAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRV 250
Query: 197 MGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAH-DKHLVDWARPLFR 255
MGT+GYCAPEYA TG+LT++SD+YSFGVV LELITGR+A D H +++LV WARPLF
Sbjct: 251 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFN 310
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYVP 315
D+ F KL DP L+G +P L AL +A MC++E RP D+V AL YL+++ Y P
Sbjct: 311 DRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTALSYLANQAYDP 370
Query: 316 -----------KASEIVSPGE-------------MEHDESPNETTMILVKDSLREQALAE 351
+ + +++ + E ++SP ET IL +D RE+A+AE
Sbjct: 371 SKDDSRRNRDERGARLITRNDDGGGSGSKFDLEGSEKEDSPRETARILNRDINRERAVAE 430
Query: 352 AKQWGETWREKRKQSGQNSPE 372
AK WGE+ REKR+QS Q + E
Sbjct: 431 AKMWGESLREKRRQSEQGTSE 451
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 407 bits (1046), Expect = e-113
Identities = 203/331 (61%), Positives = 247/331 (74%), Gaps = 3/331 (0%)
Query: 16 KAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFL 75
KAQ FTF+ELA AT NFR + F+G+GGFG V+KG + Q VA+K+LD G QG +EF+
Sbjct: 87 KAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFV 146
Query: 76 VEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMR 135
VEVL LSL HPNLV +IG+CAEGDQRLLVYEYMP GSLE HLH L +PLDWNTRM+
Sbjct: 147 VEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMK 206
Query: 136 IAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATR 195
IA GAARGL YLH P VIYRDLK SNILL E + PKLSDFGLAK GP+GD+++V+TR
Sbjct: 207 IAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTR 266
Query: 196 VMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAH-DKHLVDWARPLF 254
VMGT+GYCAP+YA TG+LT +SDIYSFGVVLLELITGR+A D T+ D++LV WARPLF
Sbjct: 267 VMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPLF 326
Query: 255 RDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYV 314
+D+ NF K+VDP LQG YP+ GL AL ++ MC++E P +RP D+VLAL++L+S KY
Sbjct: 327 KDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLALNFLASSKYD 386
Query: 315 PK--ASEIVSPGEMEHDESPNETTMILVKDS 343
P +S D E LVK++
Sbjct: 387 PNSPSSSSGKNPSFHRDRDDEEKRPHLVKET 417
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 306 bits (785), Expect = 4e-83
Identities = 160/328 (48%), Positives = 218/328 (65%), Gaps = 15/328 (4%)
Query: 2 RSEATTAENTDISNKAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGS-------- 53
R+E +N ++ N F+ EL +AT+NFR ++ +G+GGFG V+KG +
Sbjct: 42 RTEGEILQNANLKN----FSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKP 97
Query: 54 -TGQAVAVKRLDTTGFQGEKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMG 112
TG +AVKRL+ GFQG +E+L E+ L L HPNLV +IGYC E + RLLVYE+M G
Sbjct: 98 GTGIVIAVKRLNQEGFQGHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRG 157
Query: 113 SLESHLHDLLPDNEPLDWNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFY 172
SLE+HL +PL WNTR+R+A+GAARGL +LH+ A+P VIYRD K+SNILLD +
Sbjct: 158 SLENHLFRRGTFYQPLSWNTRVRMALGAARGLAFLHN-AQPQVIYRDFKASNILLDSNYN 216
Query: 173 PKLSDFGLAKFGPTGDQSYVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITG 232
KLSDFGLA+ GP GD S+V+TRVMGT GY APEY TG L+++SD+YSFGVVLLEL++G
Sbjct: 217 AKLSDFGLARDGPMGDNSHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSG 276
Query: 233 RRAYDETRAHDKH-LVDWARPLFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLRED 291
RRA D+ + +H LVDWARP +K +++DP LQG Y ++ +A C+ D
Sbjct: 277 RRAIDKNQPVGEHNLVDWARPYLTNKRRLLRVMDPRLQGQYSLTRALKIAVLALDCISID 336
Query: 292 PRLRPSAGDIVLALDYLSSKKYVPKASE 319
+ RP+ +IV ++ L +K K +
Sbjct: 337 AKSRPTMNEIVKTMEELHIQKEASKEQQ 364
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 295 bits (755), Expect = 1e-79
Identities = 150/301 (49%), Positives = 208/301 (68%), Gaps = 11/301 (3%)
Query: 20 FTFRELATATKNFRDETFIGQGGFGTVYKGKLGS---------TGQAVAVKRLDTTGFQG 70
FTF EL AT+NFR ++ +G+GGFG+V+KG + TG +AVK+L+ G+QG
Sbjct: 57 FTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQG 116
Query: 71 EKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDW 130
+E+L EV L HPNLV +IGYC E + RLLVYE+MP GSLE+HL +PL W
Sbjct: 117 HQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQPLSW 176
Query: 131 NTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQS 190
R+++A+GAA+GL +LH+ AE SVIYRD K+SNILLD + KLSDFGLAK GPTGD+S
Sbjct: 177 TLRLKVALGAAKGLAFLHN-AETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTGDKS 235
Query: 191 YVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETR-AHDKHLVDW 249
+V+TR+MGT+GY APEY TG LT +SD+YS+GVVLLE+++GRRA D+ R ++ LV+W
Sbjct: 236 HVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQKLVEW 295
Query: 250 ARPLFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLS 309
ARPL +K +++D LQ Y + +A CL + +LRP+ ++V L+++
Sbjct: 296 ARPLLANKRKLFRVIDNRLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHLEHIQ 355
Query: 310 S 310
+
Sbjct: 356 T 356
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 285 bits (729), Expect = 1e-76
Identities = 146/301 (48%), Positives = 205/301 (67%), Gaps = 11/301 (3%)
Query: 20 FTFRELATATKNFRDETFIGQGGFGTVYKGKLGS---------TGQAVAVKRLDTTGFQG 70
F+F EL +AT+NFR ++ +G+GGFG V+KG + TG +AVK+L+ G+QG
Sbjct: 56 FSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQG 115
Query: 71 EKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDW 130
+E+L EV L H +LV +IGYC E + RLLVYE+MP GSLE+HL +PL W
Sbjct: 116 HQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQPLSW 175
Query: 131 NTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQS 190
R+++A+GAA+GL +LH +E VIYRD K+SNILLD + KLSDFGLAK GP GD+S
Sbjct: 176 KLRLKVALGAAKGLAFLH-SSETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKS 234
Query: 191 YVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETR-AHDKHLVDW 249
+V+TRVMGTHGY APEY TG LT +SD+YSFGVVLLEL++GRRA D+ R + +++LV+W
Sbjct: 235 HVSTRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEW 294
Query: 250 ARPLFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLS 309
A+P +K +++D LQ Y + ++ CL + +LRP+ ++V L+++
Sbjct: 295 AKPYLVNKRKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHLEHIQ 354
Query: 310 S 310
S
Sbjct: 355 S 355
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 272 bits (696), Expect = 9e-73
Identities = 144/318 (45%), Positives = 205/318 (64%), Gaps = 20/318 (6%)
Query: 14 SNKAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGST---------GQAVAVKRLD 64
S +++ F +L TATKNF+ ++ +GQGGFG VY+G + +T G VA+KRL+
Sbjct: 68 SPNLKVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLN 127
Query: 65 TTGFQGEKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPD 124
+ QG E+ EV L +L H NLV ++GYC E + LLVYE+MP GSLESHL
Sbjct: 128 SESVQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHL---FRR 184
Query: 125 NEPLDWNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFG 184
N+P W+ R++I +GAARGL +LH + VIYRD K+SNILLD + KLSDFGLAK G
Sbjct: 185 NDPFPWDLRIKIVIGAARGLAFLH-SLQREVIYRDFKASNILLDSNYDAKLSDFGLAKLG 243
Query: 185 PTGDQSYVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAH-D 243
P ++S+V TR+MGT+GY APEY TG L ++SD+++FGVVLLE++TG A++ R
Sbjct: 244 PADEKSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQ 303
Query: 244 KHLVDWARPLFRDKGNFRKLVDPHLQGHYPISGLRMALEMARM---CLREDPRLRPSAGD 300
+ LVDW RP +K ++++D ++G Y ++A EMAR+ C+ DP+ RP +
Sbjct: 304 ESLVDWLRPELSNKHRVKQIMDKGIKGQYT---TKVATEMARITLSCIEPDPKNRPHMKE 360
Query: 301 IVLALDYLSSKKYVPKAS 318
+V L+++ VP S
Sbjct: 361 VVEVLEHIQGLNVVPNRS 378
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 235 bits (599), Expect = 2e-61
Identities = 130/291 (44%), Positives = 182/291 (61%), Gaps = 7/291 (2%)
Query: 20 FTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQG-EKEFLVEV 78
F+ REL A+ NF ++ +G+GGFG VYKG+L G VAVKRL QG E +F EV
Sbjct: 277 FSLRELQVASDNFSNKNILGRGGFGKVYKGRLAD-GTLVAVKRLKEERTQGGELQFQTEV 335
Query: 79 LMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAV 138
M+S+ H NL+ + G+C +RLLVY YM GS+ S L + PLDW R RIA+
Sbjct: 336 EMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIAL 395
Query: 139 GAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVMG 198
G+ARGL YLH +P +I+RD+K++NILLDE F + DFGLAK D ++V T V G
Sbjct: 396 GSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKD-THVTTAVRG 454
Query: 199 THGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRA---HDKHLVDWARPLFR 255
T G+ APEY +TGK + ++D++ +GV+LLELITG+RA+D R D L+DW + L +
Sbjct: 455 TIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLK 514
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALD 306
+K LVD LQG+Y + +++A +C + P RP ++V L+
Sbjct: 515 EK-KLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLE 564
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 227 bits (578), Expect = 4e-59
Identities = 121/296 (40%), Positives = 183/296 (60%), Gaps = 8/296 (2%)
Query: 19 IFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGE--KEFLV 76
+ + + L + T NF + +G GGFG VYKG+L G +AVKR++ G+ EF
Sbjct: 575 LISIQVLRSVTNNFSSDNILGSGGFGVVYKGELHD-GTKIAVKRMENGVIAGKGFAEFKS 633
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDN-EPLDWNTRMR 135
E+ +L+ + H +LV+++GYC +G+++LLVYEYMP G+L HL + + +PL W R+
Sbjct: 634 EIAVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLT 693
Query: 136 IAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATR 195
+A+ ARG+ YLH A S I+RDLK SNILL + K++DFGL + P G S + TR
Sbjct: 694 LALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGS-IETR 752
Query: 196 VMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDK-HLVDWARPLF 254
+ GT GY APEYA TG++T + D+YSFGV+L+ELITGR++ DE++ + HLV W + ++
Sbjct: 753 IAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMY 812
Query: 255 RDK-GNFRKLVDPHLQ-GHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYL 308
+K +F+K +D + ++ + E+A C +P RP G V L L
Sbjct: 813 INKEASFKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGHAVNILSSL 868
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 223 bits (569), Expect = 5e-58
Identities = 127/328 (38%), Positives = 186/328 (55%), Gaps = 11/328 (3%)
Query: 21 TFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLVEVLM 80
TF +L AT F +++ IG GGFG VYK L G AVA+K+L QG++EF+ E+
Sbjct: 872 TFADLLQATNGFHNDSLIGSGGFGDVYKAIL-KDGSAVAIKKLIHVSGQGDREFMAEMET 930
Query: 81 LSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGA 140
+ + H NLV ++GYC GD+RLLVYE+M GSLE LHD L+W+TR +IA+G+
Sbjct: 931 IGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIAIGS 990
Query: 141 ARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTH 200
ARGL +LHH P +I+RD+KSSN+LLDE ++SDFG+A+ D + + GT
Sbjct: 991 ARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTP 1050
Query: 201 GYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKHLVDWARPLFRDKGNF 260
GY PEY + + + + D+YS+GVVLLEL+TG+R D D +LV W + K
Sbjct: 1051 GYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSPDFGDNNLVGWVKQ--HAKLRI 1108
Query: 261 RKLVDPHLQGHYPISGLRMA--LEMARMCLREDPRLRPSAGDIVLALDYLSSKKYVPKAS 318
+ DP L P + + L++A CL + RP+ + ++ K + S
Sbjct: 1109 SDVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPT------MVQVMAMFKEIQAGS 1162
Query: 319 EIVSPGEMEHDESPNETTMILVKDSLRE 346
I S + E +T+ +V S++E
Sbjct: 1163 GIDSQSTIRSIEDGGFSTIEMVDMSIKE 1190
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 223 bits (568), Expect = 6e-58
Identities = 124/284 (43%), Positives = 178/284 (62%), Gaps = 5/284 (1%)
Query: 24 ELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLVEVLMLSL 83
++ AT +F + IG GGFGTVYK L + VAVK+L QG +EF+ E+ L
Sbjct: 909 DIVEATDHFSKKNIIGDGGFGTVYKACLPGE-KTVAVKKLSEAKTQGNREFMAEMETLGK 967
Query: 84 LHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGAARG 143
+ HPNLVS++GYC+ +++LLVYEYM GSL+ L + E LDW+ R++IAVGAARG
Sbjct: 968 VKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARG 1027
Query: 144 LNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTHGYC 203
L +LHH P +I+RD+K+SNILLD F PK++DFGLA+ + +S+V+T + GT GY
Sbjct: 1028 LAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARL-ISACESHVSTVIAGTFGYI 1086
Query: 204 APEYATTGKLTMRSDIYSFGVVLLELITGRR--AYDETRAHDKHLVDWARPLFRDKGNFR 261
PEY + + T + D+YSFGV+LLEL+TG+ D + +LV WA ++G
Sbjct: 1087 PPEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKI-NQGKAV 1145
Query: 262 KLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLAL 305
++DP L + L++A +CL E P RP+ D++ AL
Sbjct: 1146 DVIDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKAL 1189
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 215 bits (547), Expect = 2e-55
Identities = 123/298 (41%), Positives = 172/298 (57%), Gaps = 7/298 (2%)
Query: 10 NTDISNKAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQ 69
N D +N+ + ++ +T +F IG GGFG VYK L G VA+KRL Q
Sbjct: 724 NKDSNNELSL---DDILKSTSSFNQANIIGCGGFGLVYKATLPD-GTKVAIKRLSGDTGQ 779
Query: 70 GEKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLD 129
++EF EV LS HPNLV ++GYC + +LL+Y YM GSL+ LH+ + LD
Sbjct: 780 MDREFQAEVETLSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLD 839
Query: 130 WNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQ 189
W TR+RIA GAA GL YLH EP +++RD+KSSNILL + F L+DFGLA+ D
Sbjct: 840 WKTRLRIARGAAEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYD- 898
Query: 190 SYVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAH-DKHLVD 248
++V T ++GT GY PEY T + D+YSFGVVLLEL+TGRR D + + L+
Sbjct: 899 THVTTDLVGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLIS 958
Query: 249 WARPLFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALD 306
W + +K ++ DP + + + LE+A CL E+P+ RP+ +V L+
Sbjct: 959 WVLQMKTEKRE-SEIFDPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLE 1015
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 213 bits (542), Expect = 6e-55
Identities = 134/350 (38%), Positives = 204/350 (58%), Gaps = 8/350 (2%)
Query: 16 KAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGF--QGEKE 73
+AQ F++ EL AT F +++ +G+G F V+KG L G VAVKR + KE
Sbjct: 489 RAQEFSYEELEQATGGFSEDSQVGKGSFSCVFKGILRD-GTVVAVKRAIKASDVKKSSKE 547
Query: 74 FLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPD-NEPLDWNT 132
F E+ +LS L+H +L++++GYC +G +RLLVYE+M GSL HLH P+ + L+W
Sbjct: 548 FHNELDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWAR 607
Query: 133 RMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYV 192
R+ IAV AARG+ YLH A P VI+RD+KSSNIL+DE +++DFGL+ GP + +
Sbjct: 608 RVTIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPL 667
Query: 193 ATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKHLVDWARP 252
+ GT GY PEY LT +SD+YSFGVVLLE+++GR+A D + + ++V+WA P
Sbjct: 668 SELPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAID-MQFEEGNIVEWAVP 726
Query: 253 LFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDY-LSSK 311
L + G+ ++DP L + L+ +A C+R + RPS + AL++ L+
Sbjct: 727 LIK-AGDIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEHALALL 785
Query: 312 KYVPKASEIVSPGEMEHDES-PNETTMILVKDSLREQALAEAKQWGETWR 360
P + + P E+ S ++ + + S E LA+ + G +R
Sbjct: 786 MGSPCIEQPILPTEVVLGSSRMHKVSQMSSNHSCSENELADGEDQGIGYR 835
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 212 bits (540), Expect = 1e-54
Identities = 108/248 (43%), Positives = 151/248 (60%), Gaps = 3/248 (1%)
Query: 21 TFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLVEVLM 80
TF +L AT F +++ +G GGFG VYK +L G VA+K+L QG++EF E+
Sbjct: 877 TFADLLEATNGFHNDSLVGSGGFGDVYKAQL-KDGSVVAIKKLIHVSGQGDREFTAEMET 935
Query: 81 LSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGA 140
+ + H NLV ++GYC G++RLLVYEYM GSLE LHD L+W R +IA+GA
Sbjct: 936 IGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARRKIAIGA 995
Query: 141 ARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTH 200
ARGL +LHH P +I+RD+KSSN+LLDE ++SDFG+A+ D + + GT
Sbjct: 996 ARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTP 1055
Query: 201 GYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKHLVDWARPLFRDKGNF 260
GY PEY + + + + D+YS+GVVLLEL+TG++ D D +LV W + KG
Sbjct: 1056 GYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDNNLVGWVK--LHAKGKI 1113
Query: 261 RKLVDPHL 268
+ D L
Sbjct: 1114 TDVFDREL 1121
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 211 bits (538), Expect = 2e-54
Identities = 108/248 (43%), Positives = 151/248 (60%), Gaps = 3/248 (1%)
Query: 21 TFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLVEVLM 80
TF +L AT F +++ +G GGFG VYK +L G VA+K+L QG++EF E+
Sbjct: 877 TFADLLEATNGFHNDSLVGSGGFGDVYKAQL-KDGSVVAIKKLIHVSGQGDREFTAEMET 935
Query: 81 LSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGA 140
+ + H NLV ++GYC G++RLLVYEYM GSLE LHD L+W R +IA+GA
Sbjct: 936 IGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARRKIAIGA 995
Query: 141 ARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTH 200
ARGL +LHH P +I+RD+KSSN+LLDE ++SDFG+A+ D + + GT
Sbjct: 996 ARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTP 1055
Query: 201 GYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKHLVDWARPLFRDKGNF 260
GY PEY + + + + D+YS+GVVLLEL+TG++ D D +LV W + KG
Sbjct: 1056 GYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDNNLVGWVK--LHAKGKI 1113
Query: 261 RKLVDPHL 268
+ D L
Sbjct: 1114 TDVFDREL 1121
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 205 bits (521), Expect = 2e-52
Identities = 118/287 (41%), Positives = 169/287 (58%), Gaps = 4/287 (1%)
Query: 21 TFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLVEVLM 80
++ +L +T +F IG GGFG VYK L G+ VA+K+L Q E+EF EV
Sbjct: 723 SYDDLLDSTNSFDQANIIGCGGFGMVYKATLPD-GKKVAIKKLSGDCGQIEREFEAEVET 781
Query: 81 LSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGA 140
LS HPNLV + G+C + RLL+Y YM GSL+ LH+ L W TR+RIA GA
Sbjct: 782 LSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGA 841
Query: 141 ARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTH 200
A+GL YLH +P +++RD+KSSNILLDE F L+DFGLA+ + +++V+T ++GT
Sbjct: 842 AKGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARL-MSPYETHVSTDLVGTL 900
Query: 201 GYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHD-KHLVDWARPLFRDKGN 259
GY PEY T + D+YSFGVVLLEL+T +R D + + L+ W + + +
Sbjct: 901 GYIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKM-KHESR 959
Query: 260 FRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALD 306
++ DP + + LE+A +CL E+P+ RP+ +V LD
Sbjct: 960 ASEVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWLD 1006
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 196 bits (498), Expect = 8e-50
Identities = 110/296 (37%), Positives = 167/296 (56%), Gaps = 7/296 (2%)
Query: 10 NTDISNKAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQ 69
N + + F + E+ T NF E IG+GGFG VY G + G+ VAVK L Q
Sbjct: 554 NGPLKTAKRYFKYSEVVNITNNF--ERVIGKGGFGKVYHGVIN--GEQVAVKVLSEESAQ 609
Query: 70 GEKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLD 129
G KEF EV +L +HH NL S++GYC E + +L+YEYM +L +L + L
Sbjct: 610 GYKEFRAEVDLLMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAG--KRSFILS 667
Query: 130 WNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQ 189
W R++I++ AA+GL YLH+ +P +++RD+K +NILL+E K++DFGL++
Sbjct: 668 WEERLKISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGS 727
Query: 190 SYVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKHLVDW 249
++T V G+ GY PEY +T ++ +SD+YS GVVLLE+ITG+ A ++ H+ D
Sbjct: 728 GQISTVVAGSIGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKTEKVHISDH 787
Query: 250 ARPLFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLAL 305
R + + G+ R +VD L+ Y + E+A C RP+ +V+ L
Sbjct: 788 VRSILAN-GDIRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMEL 842
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 195 bits (495), Expect = 2e-49
Identities = 114/277 (41%), Positives = 162/277 (58%), Gaps = 12/277 (4%)
Query: 33 RDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGF-QGEKEFLVEVLMLSLLHHPNLVS 91
++E IG+GG G VY+G + + VA+KRL G + + F E+ L + H ++V
Sbjct: 693 KEENIIGKGGAGIVYRGSMPNNVD-VAIKRLVGRGTGRSDHGFTAEIQTLGRIRHRHIVR 751
Query: 92 MIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGAARGLNYLHHEA 151
++GY A D LL+YEYMP GSL LH + L W TR R+AV AA+GL YLHH+
Sbjct: 752 LLGYVANKDTNLLLYEYMPNGSLGELLHGSKGGH--LQWETRHRVAVEAAKGLCYLHHDC 809
Query: 152 EPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTHGYCAPEYATTG 211
P +++RD+KS+NILLD F ++DFGLAKF G S + + G++GY APEYA T
Sbjct: 810 SPLILHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPEYAYTL 869
Query: 212 KLTMRSDIYSFGVVLLELITGRRAYDETRAHDKHLVDWARPLFR------DKGNFRKLVD 265
K+ +SD+YSFGVVLLELI G++ E +V W R D +VD
Sbjct: 870 KVDEKSDVYSFGVVLLELIAGKKPVGEF-GEGVDIVRWVRNTEEEITQPSDAAIVVAIVD 928
Query: 266 PHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIV 302
P L G YP++ + ++A MC+ E+ RP+ ++V
Sbjct: 929 PRLTG-YPLTSVIHVFKIAMMCVEEEAAARPTMREVV 964
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 193 bits (491), Expect = 5e-49
Identities = 109/285 (38%), Positives = 171/285 (59%), Gaps = 15/285 (5%)
Query: 28 ATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLVEVLMLSLLHHP 87
AT+NF +GQGGFG VYKG+L G+ +AVKRL T QG EF+ EV +++ L H
Sbjct: 524 ATENFSSCNKLGQGGFGIVYKGRL-LDGKEIAVKRLSKTSVQGTDEFMNEVTLIARLQHI 582
Query: 88 NLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGAARGLNYL 147
NLV ++G C EGD+++L+YEY+ SL+S+L ++ L+WN R I G ARGL YL
Sbjct: 583 NLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSK-LNWNERFDITNGVARGLLYL 641
Query: 148 HHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTHGYCAPEY 207
H ++ +I+RDLK SNILLD+ PK+SDFG+A+ + +V+GT+GY +PEY
Sbjct: 642 HQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANTMKVVGTYGYMSPEY 701
Query: 208 ATTGKLTMRSDIYSFGVVLLELITGRR---AYDETRAHDKHLVDWARPLFRDKGNFRKLV 264
A G + +SD++SFGV++LE+++G++ Y+ +D W+R +G ++V
Sbjct: 702 AMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSR---WKEGRALEIV 758
Query: 265 DPHL-------QGHYPISGLRMALEMARMCLREDPRLRPSAGDIV 302
DP + + + +++ +C++E RP+ +V
Sbjct: 759 DPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVV 803
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 192 bits (489), Expect = 9e-49
Identities = 113/291 (38%), Positives = 162/291 (54%), Gaps = 7/291 (2%)
Query: 28 ATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQ-GEKEFLVEVLMLSLLHH 86
AT+N D+ IG+G GT+YK L S + AVK+L TG + G + E+ + + H
Sbjct: 812 ATENLNDKYVIGKGAHGTIYKATL-SPDKVYAVKKLVFTGIKNGSVSMVREIETIGKVRH 870
Query: 87 PNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGAARGLNY 146
NL+ + + + L++Y YM GSL LH+ P +PLDW+TR IAVG A GL Y
Sbjct: 871 RNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPP-KPLDWSTRHNIAVGTAHGLAY 929
Query: 147 LHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTHGYCAPE 206
LH + +P++++RD+K NILLD P +SDFG+AK S + V GT GY APE
Sbjct: 930 LHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAPE 989
Query: 207 YATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKHLVDWARPLFRDKGNFRKLVDP 266
A T + SD+YS+GVVLLELIT ++A D + + +V W R ++ G +K+VDP
Sbjct: 990 NAFTTVKSRESDVYSYGVVLLELITRKKALDPSFNGETDIVGWVRSVWTQTGEIQKIVDP 1049
Query: 267 HLQGHYPISG----LRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKY 313
L S + AL +A C ++ RP+ D+V L S + Y
Sbjct: 1050 SLLDELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQLTRWSIRSY 1100
>IRA4_HUMAN (Q9NWZ3) Interleukin-1 receptor-associated kinase-4 (EC
2.7.1.37) (IRAK-4) (NY-REN-64 antigen)
Length = 460
Score = 183 bits (465), Expect = 5e-46
Identities = 117/287 (40%), Positives = 164/287 (56%), Gaps = 19/287 (6%)
Query: 20 FTFRELATATKNFRDETF------IGQGGFGTVYKGKLGSTGQAVAVKRL----DTTGFQ 69
F+F EL T NF + +G+GGFG VYKG + +T VAVK+L D T +
Sbjct: 168 FSFYELKNVTNNFDERPISVGGNKMGEGGFGVVYKGYVNNT--TVAVKKLAAMVDITTEE 225
Query: 70 GEKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLD 129
+++F E+ +++ H NLV ++G+ ++GD LVY YMP GSL L L PL
Sbjct: 226 LKQQFDQEIKVMAKCQHENLVELLGFSSDGDDLCLVYVYMPNGSLLDRL-SCLDGTPPLS 284
Query: 130 WNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQ 189
W+ R +IA GAA G+N+LH E I+RD+KS+NILLDE F K+SDFGLA+ Q
Sbjct: 285 WHMRCKIAQGAANGINFLH---ENHHIHRDIKSANILLDEAFTAKISDFGLARASEKFAQ 341
Query: 190 SYVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKHLVDW 249
+ + +R++GT Y APE A G++T +SDIYSFGVVLLE+ITG A DE R + L+D
Sbjct: 342 TVMTSRIVGTTAYMAPE-ALRGEITPKSDIYSFGVVLLEIITGLPAVDEHR-EPQLLLDI 399
Query: 250 ARPLFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRP 296
+ ++ +D + S + +A CL E RP
Sbjct: 400 KEEIEDEEKTIEDYIDKKMNDADSTS-VEAMYSVASQCLHEKKNKRP 445
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,593,847
Number of Sequences: 164201
Number of extensions: 2009771
Number of successful extensions: 8574
Number of sequences better than 10.0: 1630
Number of HSP's better than 10.0 without gapping: 1169
Number of HSP's successfully gapped in prelim test: 461
Number of HSP's that attempted gapping in prelim test: 4407
Number of HSP's gapped (non-prelim): 1875
length of query: 377
length of database: 59,974,054
effective HSP length: 112
effective length of query: 265
effective length of database: 41,583,542
effective search space: 11019638630
effective search space used: 11019638630
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC133780.4


