

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133779.6 - phase: 0
(1061 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
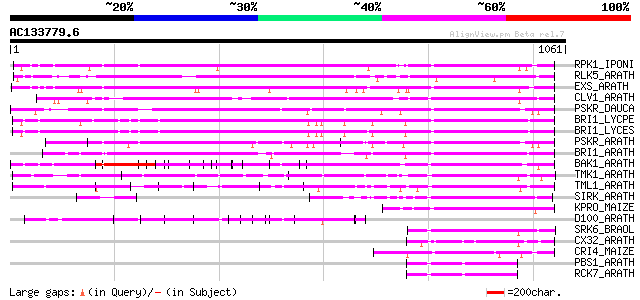
Score E
Sequences producing significant alignments: (bits) Value
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 543 e-154
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 486 e-136
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 483 e-136
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 442 e-123
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 427 e-118
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 418 e-116
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 418 e-116
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 399 e-110
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 385 e-106
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 245 5e-64
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 224 1e-57
TML1_ARATH (P33543) Putative kinase-like protein TMKL1 precursor 204 8e-52
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 173 3e-42
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 162 6e-39
D100_ARATH (Q00874) DNA-damage-repair/toleration protein DRT100 ... 154 9e-37
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 148 7e-35
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 147 1e-34
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 145 6e-34
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 144 2e-33
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 141 8e-33
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 543 bits (1399), Expect = e-154
Identities = 375/1115 (33%), Positives = 545/1115 (48%), Gaps = 112/1115 (10%)
Query: 8 IMILCVLPTLSVA----EDSEAKLALLKWKASFDNQSQSILSTWK-NTTNPCSKWRGIEC 62
++ LC ++ A D A L+L + S + I +W + + PCS W G+EC
Sbjct: 9 LLFLCSTSSIYAAFALNSDGAALLSLTRHWTSIPSD---ITQSWNASDSTPCS-WLGVEC 64
Query: 63 DKSNLISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLNF 122
D+ + T++L++ G+ G S +L + + N F+G+IP Q+GN S + ++
Sbjct: 65 DRRQFVDTLNLSSYGISGEFGP-EISHLKHLKKVVLSGNGFFGSIPSQLGNCSLLEHIDL 123
Query: 123 SKNPIIGSIPQEMYTLRSLKGLDFFFCTL------------------------SGEIDKS 158
S N G+IP + L++L+ L FF +L +G I +
Sbjct: 124 SSNSFTGNIPDTLGALQNLRNLSLFFNSLIGPFPESLLSIPHLETVYFTGNGLNGSIPSN 183
Query: 159 IGNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTYID 218
IGN++ L+ L L N FSG P+P +G + L+ L + +LVG++P + L NL Y+D
Sbjct: 184 IGNMSELTTLWLDDNQFSG-PVPSSLGNITTLQELYLNDNNLVGTLPVTLNNLENLVYLD 242
Query: 219 LSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIP 278
+ NN L G IP + +++ + +NN + G +P L N +SL ++ +LSG IP
Sbjct: 243 VRNNSLVGAIPLDFVSCKQIDTISLSNN-QFTGGLPPGLGNCTSLREFGAFSCALSGPIP 301
Query: 279 DSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYF 338
L LD L L N+ SG IP +G K++ L L+ N+L G IP +G L L+Y
Sbjct: 302 SCFGQLTKLDTLYLAGNHFSGRIPPELGKCKSMIDLQLQQNQLEGEIPGELGMLSQLQYL 361
Query: 339 SVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVG--- 395
+ NNL+G +P +I ++ L ++ N L G +P + + S + EN F G
Sbjct: 362 HLYTNNLSGEVPLSIWKIQSLQSLQLYQNNLSGELPVDMTELKQLVSLALYENHFTGVIP 421
Query: 396 ---------------------HLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERI 434
H+P +C+ LK L +N G VP+ L CS++ER+
Sbjct: 422 QDLGANSSLEVLDLTRNMFTGHIPPNLCSQKKLKRLLLGYNYLEGSVPSDLGGCSTLERL 481
Query: 435 RIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIP 494
+E N + G + DF NL + DLS N F G I P+ G ++ +S+ +SG IP
Sbjct: 482 ILEENNLRGGLP-DFVEKQNLLFFDLSGNNFTGPIPPSLGNLKNVTAIYLSSNQLSGSIP 540
Query: 495 LDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFTDSIPTEIGLLQRLEE 554
+ L KL L+LS N L G LP E L L L S+N SIP+ +G L L +
Sbjct: 541 PELGSLVKLEHLNLSHNILKGILPSE-LSNCHKLSELDASHNLLNGSIPSTLGSLTELTK 599
Query: 555 LDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFD-SALASIDLSGNRLNGNIP 613
L LG N SG IP + + KL L L N + G IP AL S++LS N+LNG +P
Sbjct: 600 LSLGENSFSGGIPTSLFQSNKLLNLQLGGNLLAGDIPPVGALQALRSLNLSSNKLNGQLP 659
Query: 614 TSLGFLVQLSMLNLSHNMLSGTIPSTFSM-SLDFVNISDNQLDGPLPEN-PAFLRAPFES 671
LG L L L++SHN LSGT+ ++ SL F+NIS N GP+P + FL + S
Sbjct: 660 IDLGKLKMLEELDVSHNNLSGTLRVLSTIQSLTFINISHNLFSGPVPPSLTKFLNSSPTS 719
Query: 672 FKNNKGLCGNITG----------LVPCATSQIHSRKSKNILQSVFIALGALILVLSGVGI 721
F N LC N L PC + + L I LGAL+ ++
Sbjct: 720 FSGNSDLCINCPADGLACPESSILRPCNMQSNTGKGGLSTLGIAMIVLGALLFIICLFLF 779
Query: 722 SMYVFFRRKKPNEEIQTEEEVQKGVLFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQG 781
S ++F KK +EI Q+G DG ++ ++EATEN +DKY+IG G+ G
Sbjct: 780 SAFLFLHCKKSVQEIAIS--AQEG--------DGSLL-NKVLEATENLNDKYVIGKGAHG 828
Query: 782 NVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSK 841
+YKA L V AVKKL + S S EIET+ ++HRN+IKL F +
Sbjct: 829 TIYKATLSPDKVYAVKKLVFTGIKN----GSVSMVREIETIGKVRHRNLIKLEEFWLRKE 884
Query: 842 FSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDIS 901
+ ++Y +ME GSL IL+ DW R N+ G A+ L+YLH DC P I+HRDI
Sbjct: 885 YGLILYTYMENGSLHDILHETNPPKPLDWSTRHNIAVGTAHGLAYLHFDCDPAIVHRDIK 944
Query: 902 SKNILLNLDYEAHVSDFGTAKFLKPDLHS--WTQFAGTFGYAAPELSQTMEVNEKCDVYS 959
NILL+ D E H+SDFG AK L S GT GY APE + T + + DVYS
Sbjct: 945 PMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAPENAFTTVKSRESDVYS 1004
Query: 960 FGVLALEIIIGKH--------PGDLISLFLSPST-----RPTANDMLLTEVLDQRPQKVI 1006
+GV+ LE+I K D++ S T + + LL E++D
Sbjct: 1005 YGVVLLELITRKKALDPSFNGETDIVGWVRSVWTQTGEIQKIVDPSLLDELID------- 1057
Query: 1007 KPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
+ E+V LA C + RPTM V K L
Sbjct: 1058 SSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQL 1092
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 486 bits (1250), Expect = e-136
Identities = 360/1063 (33%), Positives = 533/1063 (49%), Gaps = 131/1063 (12%)
Query: 7 IIMILCV----LPTLSVAEDSEAKLALLKWKASFDNQSQSILSTWK--NTTNPCSKWRGI 60
+I++LC+ LP+LS+ +D+ L + K + +QS LS+W N PC KW G+
Sbjct: 5 LILLLCLSSTYLPSLSLNQDATI---LRQAKLGLSDPAQS-LSSWSDNNDVTPC-KWLGV 59
Query: 61 ECDKSNLISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTL 120
CD ++ + ++DL++ L G FP+++ +L +++L
Sbjct: 60 SCDATSNVVSVDLSSFMLVGP--------FPSILC-----------------HLPSLHSL 94
Query: 121 NFSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGNNFSGGPI 180
+ N I GS+ + DF C NL LDL N G I
Sbjct: 95 SLYNNSINGSLSAD----------DFDTCH-------------NLISLDLS-ENLLVGSI 130
Query: 181 PPEIG-KLKKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLN 239
P + L L++L I+ +L +IP G L ++L+ NFLSG IP ++GN++ L
Sbjct: 131 PKSLPFNLPNLKFLEISGNNLSDTIPSSFGEFRKLESLNLAGNFLSGTIPASLGNVTTLK 190
Query: 240 QLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSG 299
+L A N IP L N++ L +++L +L G IP S+ L +L L L N L+G
Sbjct: 191 ELKLAYNLFSPSQIPSQLGNLTELQVLWLAGCNLVGPIPPSLSRLTSLVNLDLTFNQLTG 250
Query: 300 FIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQL 359
IPS I LK + + L NN SG +P S+G N+ L
Sbjct: 251 SIPSWITQLKTVEQIELFNNSFSGELPESMG------------------------NMTTL 286
Query: 360 IVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNRFTG 419
F+ + NKL G+IP+ L N+ N S + EN G LP + +L L F+NR TG
Sbjct: 287 KRFDASMNKLTGKIPDNL-NLLNLESLNLFENMLEGPLPESITRSKTLSELKLFNNRLTG 345
Query: 420 PVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDL 479
+P+ L + S ++ + + N+ G+I + L Y+ L DN F G IS N GK L
Sbjct: 346 VLPSQLGANSPLQYVDLSYNRFSGEIPANVCGEGKLEYLILIDNSFSGEISNNLGKCKSL 405
Query: 480 ETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFT 539
+SN +SG IP F GL +L L LS N TG +PK I+G K+L L+IS N F+
Sbjct: 406 TRVRLSNNKLSGQIPHGFWGLPRLSLLELSDNSFTGSIPKTIIGA-KNLSNLRISKNRFS 464
Query: 540 DSIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFDS--A 597
SIP EIG L + E+ N+ SG IP + +L +L L+LS+N++ G IP
Sbjct: 465 GSIPNEIGSLNGIIEISGAENDFSGEIPESLVKLKQLSRLDLSKNQLSGEIPRELRGWKN 524
Query: 598 LASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTF-SMSLDFVNISDNQLDG 656
L ++L+ N L+G IP +G L L+ L+LS N SG IP ++ L+ +N+S N L G
Sbjct: 525 LNELNLANNHLSGEIPKEVGILPVLNYLDLSSNQFSGEIPLELQNLKLNVLNLSYNHLSG 584
Query: 657 PLPENPAFLRAPF-ESFKNNKGLCGNITGLVPCATSQIHSRKSKNI-----LQSVFIALG 710
+P P + + F N GLC ++ GL T +SKNI L ++F+ G
Sbjct: 585 KIP--PLYANKIYAHDFIGNPGLCVDLDGLCRKIT------RSKNIGYVWILLTIFLLAG 636
Query: 711 ALILVLSGVGISMYVFFRRKKPNEEIQTEEEVQKGVLFSIWSHDGKMMFENIIEATENFD 770
+ +V GI M++ RK + T + S W K+ F E + D
Sbjct: 637 LVFVV----GIVMFIAKCRKLRALKSST-------LAASKWRSFHKLHFSEH-EIADCLD 684
Query: 771 DKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKS-----FTSEIETLTGI 825
+K +IG GS G VYK EL G VVAVKKL+ +SS S F +E+ETL I
Sbjct: 685 EKNVIGFGSSGKVYKVELRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAEVETLGTI 744
Query: 826 KHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQA-IAFDWEKRVNVVKGVANAL 884
+H++I++L CS LVY++M GSL +L+ +++ + W +R+ + A L
Sbjct: 745 RHKSIVRLWCCCSSGDCKLLVYEYMPNGSLADVLHGDRKGGVVLGWPERLRIALDAAEGL 804
Query: 885 SYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLK----PDLHSWTQFAGTFGY 940
SYLHHDC PPI+HRD+ S NILL+ DY A V+DFG AK + + + AG+ GY
Sbjct: 805 SYLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAMSGIAGSCGY 864
Query: 941 AAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPGD--LISLFLSPSTRPTANDMLLTEVL 998
APE T+ VNEK D+YSFGV+ LE++ GK P D L ++ + L V+
Sbjct: 865 IAPEYVYTLRVNEKSDIYSFGVVLLELVTGKQPTDSELGDKDMAKWVCTALDKCGLEPVI 924
Query: 999 DQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
D + K EE+ + + C + +P +RP+M +V ML
Sbjct: 925 DPKLDLKFK---EEISKVIHIGLLCTSPLPLNRPSMRKVVIML 964
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 483 bits (1244), Expect = e-136
Identities = 377/1211 (31%), Positives = 561/1211 (46%), Gaps = 197/1211 (16%)
Query: 3 LPTFIIMILCVLPTLSVAEDSEAKLALLKWKASFDNQSQSILSTWK--NTTNPCSKWRGI 60
L + + + ++ + S +L+ +K S +N S +LS+W ++ + C W G+
Sbjct: 4 LTALFLFLFFSFSSSAIVDLSSETTSLISFKRSLENPS--LLSSWNVSSSASHCD-WVGV 60
Query: 61 ECDKSNLISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTL 120
C ++++ L +L L+G + SS NL L + N F G IPP+I NL + TL
Sbjct: 61 TCLLGR-VNSLSLPSLSLRGQIPK-EISSLKNLRELCLAGNQFSGKIPPEIWNLKHLQTL 118
Query: 121 NFSKNPIIG-------SIPQEMY------------------TLRSLKGLDFFFCTLSGEI 155
+ S N + G +PQ +Y +L +L LD +LSGEI
Sbjct: 119 DLSGNSLTGLLPRLLSELPQLLYLDLSDNHFSGSLPPSFFISLPALSSLDVSNNSLSGEI 178
Query: 156 DKSIGNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLT 215
IG L+NLS L +G N+FSG IP EIG + L+ A G +P+EI L +L
Sbjct: 179 PPEIGKLSNLSNLYMGLNSFSG-QIPSEIGNISLLKNFAAPSCFFNGPLPKEISKLKHLA 237
Query: 216 YIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSG 275
+DLS N L IP++ G + L+ L + +L G IP L N SL + L SLSG
Sbjct: 238 KLDLSYNPLKCSIPKSFGELHNLSILNLVS-AELIGLIPPELGNCKSLKSLMLSFNSLSG 296
Query: 276 SIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINL 335
+P + I L + N LSG +PS +G K L LLL NNR SG IP I + L
Sbjct: 297 PLPLELSE-IPLLTFSAERNQLSGSLPSWMGKWKVLDSLLLANNRFSGEIPHEIEDCPML 355
Query: 336 KYFSVQVNNLTGTIPATI------------GNLKQ------------------------- 358
K+ S+ N L+G+IP + GNL
Sbjct: 356 KHLSLASNLLSGSIPRELCGSGSLEAIDLSGNLLSGTIEEVFDGCSSLGELLLTNNQING 415
Query: 359 ----------LIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLK 408
L+ ++ SN G IP L+ TN F S N G+LP+++ SLK
Sbjct: 416 SIPEDLWKLPLMALDLDSNNFTGEIPKSLWKSTNLMEFTASYNRLEGYLPAEIGNAASLK 475
Query: 409 YLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGH 468
L N+ TG +P + +S+ + + N +G I + G +L +DL N G
Sbjct: 476 RLVLSDNQLTGEIPREIGKLTSLSVLNLNANMFQGKIPVELGDCTSLTTLDLGSNNLQGQ 535
Query: 469 ISPNWGKSLDLETFMISNTNISGGIPL------------DFIGLTKLGRLHLSSNQLTGK 516
I L+ ++S N+SG IP D L G LS N+L+G
Sbjct: 536 IPDKITALAQLQCLVLSYNNLSGSIPSKPSAYFHQIEMPDLSFLQHHGIFDLSYNRLSGP 595
Query: 517 LPKEILGGMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKL 576
+P+E LG L+ + +SNNH + IP + L L LDL GN L+G+IP E+ KL
Sbjct: 596 IPEE-LGECLVLVEISLSNNHLSGEIPASLSRLTNLTILDLSGNALTGSIPKEMGNSLKL 654
Query: 577 RMLNLSRNRIEGRIPSTFD--SALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSG 634
+ LNL+ N++ G IP +F +L ++L+ N+L+G +P SLG L +L+ ++LS N LSG
Sbjct: 655 QGLNLANNQLNGHIPESFGLLGSLVKLNLTKNKLDGPVPASLGNLKELTHMDLSFNNLSG 714
Query: 635 TIPSTFSM--------------------------SLDFVNISDNQLDGPLPE------NP 662
+ S S L+++++S+N L G +P N
Sbjct: 715 ELSSELSTMEKLVGLYIEQNKFTGEIPSELGNLTQLEYLDVSENLLSGEIPTKICGLPNL 774
Query: 663 AFLRAPFESFKN------------------NKGLCGNITGLVPCATSQIHSRKSKNILQS 704
FL + + NK LCG + G + +I K ++
Sbjct: 775 EFLNLAKNNLRGEVPSDGVCQDPSKALLSGNKELCGRVVG----SDCKIEGTKLRSAWGI 830
Query: 705 VFIALGALILVLSGV-GISMYVFFRRKKPNEEIQTEEE------VQKGVLFSIWSHDGK- 756
+ LG I+V V + + +R K ++ + EE V + + F S +
Sbjct: 831 AGLMLGFTIIVFVFVFSLRRWAMTKRVKQRDDPERMEESRLKGFVDQNLYFLSGSRSREP 890
Query: 757 -----MMFE---------NIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLV 802
MFE +I+EAT++F K +IG G G VYKA LP VAVKKL
Sbjct: 891 LSINIAMFEQPLLKVRLGDIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEA 950
Query: 803 RDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNE 862
+ + ++ F +E+ETL +KH N++ L G+CS S+ LVY++M GSLD L N+
Sbjct: 951 KTQ-----GNREFMAEMETLGKVKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQ 1005
Query: 863 KQAI-AFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTA 921
+ DW KR+ + G A L++LHH P IIHRDI + NILL+ D+E V+DFG A
Sbjct: 1006 TGMLEVLDWSKRLKIAVGAARGLAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLA 1065
Query: 922 KFLKP-DLHSWTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHP------- 973
+ + + H T AGTFGY PE Q+ K DVYSFGV+ LE++ GK P
Sbjct: 1066 RLISACESHVSTVIAGTFGYIPPEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKE 1125
Query: 974 ---GDLISLFLSPSTRPTANDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRS 1030
G+L+ + + A D++ P V + + + ++A CL + P
Sbjct: 1126 SEGGNLVGWAIQKINQGKAVDVI-------DPLLVSVALKNSQLRLLQIAMLCLAETPAK 1178
Query: 1031 RPTMDQVCKML 1041
RP M V K L
Sbjct: 1179 RPNMLDVLKAL 1189
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 442 bits (1137), Expect = e-123
Identities = 330/1020 (32%), Positives = 487/1020 (47%), Gaps = 102/1020 (10%)
Query: 52 NPCSKWRGIECDKSNLISTIDLANLGLKGTLHS------LTFSSFP-----NLITLNIYN 100
+PC + +E + S I GL +HS +FS +I+LN+
Sbjct: 20 SPCFAYTDMEVLLNLKSSMIGPKGHGLHDWIHSSSPDAHCSFSGVSCDDDARVISLNVSF 79
Query: 101 NHFYGTIPPQIGNLSRINTLNFSKNPIIGSIPQEMYTLRSLKGLDF-----FFCTLSGEI 155
+GTI P+IG L+ + L + N G +P EM +L SLK L+ T GEI
Sbjct: 80 TPLFGTISPEIGMLTHLVNLTLAANNFTGELPLEMKSLTSLKVLNISNNGNLTGTFPGEI 139
Query: 156 DKSIGNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLT 215
K+ + +L LD NNF+G +PPE+ +LKKL+
Sbjct: 140 LKA---MVDLEVLDTYNNNFNG-KLPPEMSELKKLK------------------------ 171
Query: 216 YIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYL-YNMSLS 274
Y+ NF SG IPE+ G++ L L N L G P L + +L +Y+ Y S +
Sbjct: 172 YLSFGGNFFSGEIPESYGDIQSLEYLGL-NGAGLSGKSPAFLSRLKNLREMYIGYYNSYT 230
Query: 275 GSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLIN 334
G +P L L++L + L+G IP+++ NLK+L L L N L+G IP + L++
Sbjct: 231 GGVPPEFGGLTKLEILDMASCTLTGEIPTSLSNLKHLHTLFLHINNLTGHIPPELSGLVS 290
Query: 335 LKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFV 394
LK + +N LTG IP + NL + + + N LYG+IP + + F V EN+F
Sbjct: 291 LKSLDLSINQLTGEIPQSFINLGNITLINLFRNNLYGQIPEAIGELPKLEVFEVWENNFT 350
Query: 395 GHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPN 454
LP+ + G+L L N TG +P L G ++E I
Sbjct: 351 LQLPANLGRNGNLIKLDVSDNHLTGLIPKDL----------CRGEKLEMLI--------- 391
Query: 455 LRYVDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLT 514
LS+N F G I GK L I ++G +P L + + L+ N +
Sbjct: 392 -----LSNNFFFGPIPEELGKCKSLTKIRIVKNLLNGTVPAGLFNLPLVTIIELTDNFFS 446
Query: 515 GKLPKEILGGMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIPNEVAELP 574
G+LP + G + +YL SNN F+ IP IG L+ L L N G IP E+ EL
Sbjct: 447 GELPVTMSGDVLDQIYL--SNNWFSGEIPPAIGNFPNLQTLFLDRNRFRGNIPREIFELK 504
Query: 575 KLRMLNLSRNRIEGRIPSTFD--SALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNML 632
L +N S N I G IP + S L S+DLS NR+NG IP + + L LN+S N L
Sbjct: 505 HLSRINTSANNITGGIPDSISRCSTLISVDLSRNRINGEIPKGINNVKNLGTLNISGNQL 564
Query: 633 SGTIPSTFS--MSLDFVNISDNQLDGPLPENPAFLRAPFESFKNNKGLCGNITGLVPCAT 690
+G+IP+ SL +++S N L G +P FL SF N LC P
Sbjct: 565 TGSIPTGIGNMTSLTTLDLSFNDLSGRVPLGGQFLVFNETSFAGNTYLCLPHRVSCPTRP 624
Query: 691 SQIHSRKSKNILQSVFIALGALILVLSGVGISMYVFFRRKKPNEEIQTEEEVQKGVLFSI 750
Q + I + + + + IS+ + KK N QK + + +
Sbjct: 625 GQTSDHNHTALFSPSRIVITVIAAITGLILISVAIRQMNKKKN---------QKSLAWKL 675
Query: 751 WSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFF 810
+ K+ F++ + E ++ +IG G G VY+ +P + VA+K+L +
Sbjct: 676 TAFQ-KLDFKSE-DVLECLKEENIIGKGGAGIVYRGSMPNNVDVAIKRLV----GRGTGR 729
Query: 811 SSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDW 870
S FT+EI+TL I+HR+I++L G+ ++ + L+Y++M GSL ++L+ K W
Sbjct: 730 SDHGFTAEIQTLGRIRHRHIVRLLGYVANKDTNLLLYEYMPNGSLGELLHGSKGG-HLQW 788
Query: 871 EKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPDLHS 930
E R V A L YLHHDCSP I+HRD+ S NILL+ D+EAHV+DFG AKFL S
Sbjct: 789 ETRHRVAVEAAKGLCYLHHDCSPLILHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAAS 848
Query: 931 --WTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHP-------GDLISLFL 981
+ AG++GY APE + T++V+EK DVYSFGV+ LE+I GK P D++
Sbjct: 849 ECMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIAGKKPVGEFGEGVDIVRWVR 908
Query: 982 SPSTRPTANDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
+ T V P+ P+ VI + K+A C+ + +RPTM +V ML
Sbjct: 909 NTEEEITQPSDAAIVVAIVDPRLTGYPL-TSVIHVFKIAMMCVEEEAAARPTMREVVHML 967
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 427 bits (1097), Expect = e-118
Identities = 332/1095 (30%), Positives = 513/1095 (46%), Gaps = 138/1095 (12%)
Query: 2 VLPTFIIMIL---CVLPTLSVAEDSEAKLALLKWKASFDNQSQSILSTWK-----NTTNP 53
VL ++I+IL CV + +++ LK F +S + WK + ++
Sbjct: 3 VLRVYVILILVGFCVQIVVVNSQNLTCNSNDLKALEGFMRGLESSIDGWKWNESSSFSSN 62
Query: 54 CSKWRGIECDKSNLISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGN 113
C W GI C S +LGL +
Sbjct: 63 CCDWVGISCKSS--------VSLGLD------------------------------DVNE 84
Query: 114 LSRINTLNFSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGN 173
R+ L + + G + + + L LK L+ +LSG I S+ NL+NL LDL N
Sbjct: 85 SGRVVELELGRRKLSGKLSESVAKLDQLKVLNLTHNSLSGSIAASLLNLSNLEVLDLSSN 144
Query: 174 NFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEI-GLLTNLTYIDLSNNFLSGVIPETI 232
+FSG + P + L LR L + + S G IP + L + IDL+ N+ G IP I
Sbjct: 145 DFSG--LFPSLINLPSLRVLNVYENSFHGLIPASLCNNLPRIREIDLAMNYFDGSIPVGI 202
Query: 233 GNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLAL 292
GN S + L A+N +LSGSIP + L NL VLAL
Sbjct: 203 GNCSSVEYLGLASN-------------------------NLSGSIPQELFQLSNLSVLAL 237
Query: 293 YMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPAT 352
N LSG + S +G L NL L + +N+ SG IP L L YFS Q N G +P +
Sbjct: 238 QNNRLSGALSSKLGKLSNLGRLDISSNKFSGKIPDVFLELNKLWYFSAQSNLFNGEMPRS 297
Query: 353 IGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSA 412
+ N + + + + +N L G+I +TN S ++ N F G +PS + LK ++
Sbjct: 298 LSNSRSISLLSLRNNTLSGQIYLNCSAMTNLTSLDLASNSFSGSIPSNLPNCLRLKTINF 357
Query: 413 FHNRFTGPVPTSLKSCSSIERIRIEGNQIE--GDIAEDFGVYPNLRYVDLSDNKFHGHIS 470
+F +P S K+ S+ + + I+ E NL+ + L+ N F
Sbjct: 358 AKIKFIAQIPESFKNFQSLTSLSFSNSSIQNISSALEILQHCQNLKTLVLTLN-FQKEEL 416
Query: 471 PNWG--KSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSL 528
P+ + +L+ +I++ + G +P L L LS NQL+G +P LG + SL
Sbjct: 417 PSVPSLQFKNLKVLIIASCQLRGTVPQWLSNSPSLQLLDLSWNQLSGTIP-PWLGSLNSL 475
Query: 529 LYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIP--------------NEVAELP 574
YL +SNN F IP + LQ L + E S P N+ + P
Sbjct: 476 FYLDLSNNTFIGEIPHSLTSLQSLVSKENAVEEPSPDFPFFKKKNTNAGGLQYNQPSSFP 535
Query: 575 KLRMLNLSRNRIEGRIPSTFDS--ALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNML 632
M++LS N + G I F L ++L N L+GNIP +L + L +L+LSHN L
Sbjct: 536 P--MIDLSYNSLNGSIWPEFGDLRQLHVLNLKNNNLSGNIPANLSGMTSLEVLDLSHNNL 593
Query: 633 SGTIPSTFSMSLDFV---NISDNQLDGPLPENPAFLRAPFESFKNNKGLCGNITGLVPCA 689
SG IP + + L F+ +++ N+L GP+P F P SF+ N+GLCG
Sbjct: 594 SGNIPPSL-VKLSFLSTFSVAYNKLSGPIPTGVQFQTFPNSSFEGNQGLCGEHASPCHIT 652
Query: 690 TSQIHS---RKSKNILQSVFIA----LGALILVLSGVGISMYVFFRRKKPNEEIQTEEEV 742
H + KNI + V +A LG + L+ + I + R + E+ +E+
Sbjct: 653 DQSPHGSAVKSKKNIRKIVAVAVGTGLGTVFLLTVTLLIILRTTSRGEVDPEKKADADEI 712
Query: 743 QKG----VLFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKK 798
+ G VLF + ++ ++I+++T +F+ +IG G G VYKA LP G VA+K+
Sbjct: 713 ELGSRSVVLFHNKDSNNELSLDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKR 772
Query: 799 LHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQI 858
L + + F +E+ETL+ +H N++ L G+C++ L+Y +M+ GSLD
Sbjct: 773 LSGDTGQ-----MDREFQAEVETLSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYW 827
Query: 859 LNNEKQA-IAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSD 917
L+ + + DW+ R+ + +G A L+YLH C P I+HRDI S NILL+ + AH++D
Sbjct: 828 LHEKVDGPPSLDWKTRLRIARGAAEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLAD 887
Query: 918 FGTAKFLKP-DLHSWTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPGDL 976
FG A+ + P D H T GT GY PE Q K DVYSFGV+ LE++ G+ P D+
Sbjct: 888 FGLARLILPYDTHVTTDLVGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDV 947
Query: 977 ISLFLSPSTRPTANDMLLTEVL----DQRPQKVIKPI------DEEVILIAKLAFSCLNQ 1026
+P + L++ VL ++R ++ P EE++L+ ++A CL +
Sbjct: 948 --------CKPRGSRDLISWVLQMKTEKRESEIFDPFIYDKDHAEEMLLVLEIACRCLGE 999
Query: 1027 VPRSRPTMDQVCKML 1041
P++RPT Q+ L
Sbjct: 1000 NPKTRPTTQQLVSWL 1014
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 418 bits (1074), Expect = e-116
Identities = 354/1163 (30%), Positives = 548/1163 (46%), Gaps = 151/1163 (12%)
Query: 6 FIIMILCVLPTLSVA-------EDSEAKLALLKWKASFDNQSQSILSTWKNTTNPCSKWR 58
F+++++ LP S A +DS+ LL +KA+ + ++L W ++T+PCS +
Sbjct: 20 FVLLLIFFLPPASPAASVNGLYKDSQQ---LLSFKAALP-PTPTLLQNWLSSTDPCS-FT 74
Query: 59 GIECDKSNLISTIDLANLGLKGTLHSLTFSSFP--NLITLNIYNNHFYGTIPPQIGNLSR 116
G+ C K++ +S+IDL+N L +T P NL +L + N + G++ +
Sbjct: 75 GVSC-KNSRVSSIDLSNTFLSVDFSLVTSYLLPLSNLESLVLKNANLSGSLTSAAKSQCG 133
Query: 117 I--NTLNFSKNPIIGSIPQ-----EMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLD 169
+ ++++ ++N I G I L+SL F E+ K G +L LD
Sbjct: 134 VTLDSIDLAENTISGPISDISSFGVCSNLKSLNLSKNFLDPPGKEMLK--GATFSLQVLD 191
Query: 170 LGGNNFSGGPIPPEIGKLK--KLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGV 227
L NN SG + P + + +L + +I L GSIP+ NL+Y+DLS N S V
Sbjct: 192 LSYNNISGFNLFPWVSSMGFVELEFFSIKGNKLAGSIPELD--FKNLSYLDLSANNFSTV 249
Query: 228 IPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINL 287
P + + S L L ++N K YG I SL + L+ + L N G +P +L
Sbjct: 250 FP-SFKDCSNLQHLDLSSN-KFYGDIGSSLSSCGKLSFLNLTNNQFVGLVPKLPSE--SL 305
Query: 288 DVLALYMNNLSGFIPSTIGNL-KNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLT 346
L L N+ G P+ + +L K + L L N SG +P S+G +L+ + NN +
Sbjct: 306 QYLYLRGNDFQGVYPNQLADLCKTVVELDLSYNNFSGMVPESLGECSSLELVDISNNNFS 365
Query: 347 GTIPA-TIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTG- 404
G +P T+ L + ++ NK G +P+ N+ + +S N+ G +PS +C
Sbjct: 366 GKLPVDTLLKLSNIKTMVLSFNKFVGGLPDSFSNLPKLETLDMSSNNLTGIIPSGICKDP 425
Query: 405 -GSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDN 463
+LK L +N F GP+P SL +CS + + + N + G I G L+ + L N
Sbjct: 426 MNNLKVLYLQNNLFKGPIPDSLSNCSQLVSLDLSFNYLTGSIPSSLGSLSKLKDLILWLN 485
Query: 464 KFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILG 523
+ G I LE ++ +++G IP TKL + LS+NQL+G++P LG
Sbjct: 486 QLSGEIPQELMYLQALENLILDFNDLTGPIPASLSNCTKLNWISLSNNQLSGEIPAS-LG 544
Query: 524 GMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIPNE---------VAELP 574
+ +L LK+ NN + +IP E+G Q L LDL N L+G+IP VA L
Sbjct: 545 RLSNLAILKLGNNSISGNIPAELGNCQSLIWLDLNTNFLNGSIPPPLFKQSGNIAVALLT 604
Query: 575 KLRMLNLSRN-----------------------RIEGRIPSTF--------------DSA 597
R + + + RI R P F + +
Sbjct: 605 GKRYVYIKNDGSKECHGAGNLLEFGGIRQEQLDRISTRHPCNFTRVYRGITQPTFNHNGS 664
Query: 598 LASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPS------------------- 638
+ +DLS N+L G+IP LG + LS+LNL HN LSG IP
Sbjct: 665 MIFLDLSYNKLEGSIPKELGAMYYLSILNLGHNDLSGMIPQQLGGLKNVAILDLSYNRFN 724
Query: 639 -------TFSMSLDFVNISDNQLDGPLPENPAFLRAPFESFKNNKGLCGNITGLVPCATS 691
T L +++S+N L G +PE+ F P F NN LCG L PC++
Sbjct: 725 GTIPNSLTSLTLLGEIDLSNNNLSGMIPESAPFDTFPDYRFANN-SLCGYPLPL-PCSSG 782
Query: 692 --------QIHSRKSKNILQSVFIALGALILVLSGVGISMYVFFRRKKPNE---EIQTEE 740
Q R+ ++ SV + L + + G+ I +R++ E E +
Sbjct: 783 PKSDANQHQKSHRRQASLAGSVAMGLLFSLFCIFGLIIVAIETKKRRRKKEAALEAYMDG 842
Query: 741 EVQKGVLFSIWSHDG-----------------KMMFENIIEATENFDDKYLIGVGSQGNV 783
S W K+ F +++EAT F + L+G G G+V
Sbjct: 843 HSHSATANSAWKFTSAREALSINLAAFEKPLRKLTFADLLEATNGFHNDSLVGSGGFGDV 902
Query: 784 YKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFS 843
YKA+L G VVA+KKL V + + FT+E+ET+ IKHRN++ L G+C +
Sbjct: 903 YKAQLKDGSVVAIKKLIHVSGQ-----GDREFTAEMETIGKIKHRNLVPLLGYCKVGEER 957
Query: 844 FLVYKFMEGGSLDQILNNEKQ-AIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISS 902
LVY++M+ GSL+ +L++ K+ I +W R + G A L++LHH+C P IIHRD+ S
Sbjct: 958 LLVYEYMKYGSLEDVLHDRKKTGIKLNWPARRKIAIGAARGLAFLHHNCIPHIIHRDMKS 1017
Query: 903 KNILLNLDYEAHVSDFGTAKFLKP-DLH-SWTQFAGTFGYAAPELSQTMEVNEKCDVYSF 960
N+LL+ + EA VSDFG A+ + D H S + AGT GY PE Q+ + K DVYS+
Sbjct: 1018 SNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSY 1077
Query: 961 GVLALEIIIGKHPGDLISL---FLSPSTRPTANDMLLTEVLDQRPQKVIKPIDEEVILIA 1017
GV+ LE++ GK P D L + A +T+V D+ K I+ E++
Sbjct: 1078 GVVLLELLTGKQPTDSADFGDNNLVGWVKLHAKGK-ITDVFDRELLKEDASIEIELLQHL 1136
Query: 1018 KLAFSCLNQVPRSRPTMDQVCKM 1040
K+A +CL+ RPTM QV M
Sbjct: 1137 KVACACLDDRHWKRPTMIQVMAM 1159
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 418 bits (1074), Expect = e-116
Identities = 350/1161 (30%), Positives = 547/1161 (46%), Gaps = 147/1161 (12%)
Query: 6 FIIMILCVLPTLSVA-------EDSEAKLALLKWKASFDNQSQSILSTWKNTTNPCSKWR 58
F+++++ LP S A +DS+ LL +KA+ + ++L W ++T PCS +
Sbjct: 20 FVLLLIFFLPPASPAASVNGLYKDSQQ---LLSFKAALP-PTPTLLQNWLSSTGPCS-FT 74
Query: 59 GIECDKSNLISTIDLANLGLKGTLHSLTFSSFP--NLITLNIYNNHFYGTIPPQIGNLSR 116
G+ C K++ +S+IDL+N L +T P NL +L + N + G++ +
Sbjct: 75 GVSC-KNSRVSSIDLSNTFLSVDFSLVTSYLLPLSNLESLVLKNANLSGSLTSAAKSQCG 133
Query: 117 I--NTLNFSKNPIIGSIPQ--EMYTLRSLKGLDFFFCTLSGEIDKSIGNLT-NLSYLDLG 171
+ ++++ ++N I G I +LK L+ L + + T +L LDL
Sbjct: 134 VTLDSIDLAENTISGPISDISSFGVCSNLKSLNLSKNFLDPPGKEMLKAATFSLQVLDLS 193
Query: 172 GNNFSGGPIPPEIGKLK--KLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIP 229
NN SG + P + + +L + ++ L GSIP+ NL+Y+DLS N S V P
Sbjct: 194 YNNISGFNLFPWVSSMGFVELEFFSLKGNKLAGSIPELD--FKNLSYLDLSANNFSTVFP 251
Query: 230 ETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDV 289
+ + S L L ++N K YG I SL + L+ + L N G +P +L
Sbjct: 252 -SFKDCSNLQHLDLSSN-KFYGDIGSSLSSCGKLSFLNLTNNQFVGLVPKLPSE--SLQY 307
Query: 290 LALYMNNLSGFIPSTIGNL-KNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGT 348
L L N+ G P+ + +L K + L L N SG +P S+G +L+ + NN +G
Sbjct: 308 LYLRGNDFQGVYPNQLADLCKTVVELDLSYNNFSGMVPESLGECSSLELVDISYNNFSGK 367
Query: 349 IPA-TIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTG--G 405
+P T+ L + ++ NK G +P+ N+ + +S N+ G +PS +C
Sbjct: 368 LPVDTLSKLSNIKTMVLSFNKFVGGLPDSFSNLLKLETLDMSSNNLTGVIPSGICKDPMN 427
Query: 406 SLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKF 465
+LK L +N F GP+P SL +CS + + + N + G I G L+ + L N+
Sbjct: 428 NLKVLYLQNNLFKGPIPDSLSNCSQLVSLDLSFNYLTGSIPSSLGSLSKLKDLILWLNQL 487
Query: 466 HGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGM 525
G I LE ++ +++G IP TKL + LS+NQL+G++P LG +
Sbjct: 488 SGEIPQELMYLQALENLILDFNDLTGPIPASLSNCTKLNWISLSNNQLSGEIPAS-LGRL 546
Query: 526 KSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIPNE---------VAELPKL 576
+L LK+ NN + +IP E+G Q L LDL N L+G+IP VA L
Sbjct: 547 SNLAILKLGNNSISGNIPAELGNCQSLIWLDLNTNFLNGSIPPPLFKQSGNIAVALLTGK 606
Query: 577 RMLNLSRN-----------------------RIEGRIPSTF--------------DSALA 599
R + + + RI R P F + ++
Sbjct: 607 RYVYIKNDGSKECHGAGNLLEFGGIRQEQLDRISTRHPCNFTRVYRGITQPTFNHNGSMI 666
Query: 600 SIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPS--------------------- 638
+DLS N+L G+IP LG + LS+LNL HN LSG IP
Sbjct: 667 FLDLSYNKLEGSIPKELGAMYYLSILNLGHNDLSGMIPQQLGGLKNVAILDLSYNRFNGT 726
Query: 639 -----TFSMSLDFVNISDNQLDGPLPENPAFLRAPFESFKNNKGLCGNITGLVPCATS-- 691
T L +++S+N L G +PE+ F P F NN LCG +PC++
Sbjct: 727 IPNSLTSLTLLGEIDLSNNNLSGMIPESAPFDTFPDYRFANN-SLCGYPLP-IPCSSGPK 784
Query: 692 ------QIHSRKSKNILQSVFIALGALILVLSGVGISMYVFFRRKKPNE---EIQTEEEV 742
Q R+ ++ SV + L + + G+ I +R++ E E +
Sbjct: 785 SDANQHQKSHRRQASLAGSVAMGLLFSLFCIFGLIIVAIETKKRRRKKEAALEAYMDGHS 844
Query: 743 QKGVLFSIWSHDG-----------------KMMFENIIEATENFDDKYLIGVGSQGNVYK 785
S W K+ F +++EAT F + L+G G G+VYK
Sbjct: 845 HSATANSAWKFTSAREALSINLAAFEKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYK 904
Query: 786 AELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFL 845
A+L G VVA+KKL V + + FT+E+ET+ IKHRN++ L G+C + L
Sbjct: 905 AQLKDGSVVAIKKLIHVSGQ-----GDREFTAEMETIGKIKHRNLVPLLGYCKVGEERLL 959
Query: 846 VYKFMEGGSLDQILNNEKQ-AIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKN 904
VY++M+ GSL+ +L++ K+ I +W R + G A L++LHH+C P IIHRD+ S N
Sbjct: 960 VYEYMKYGSLEDVLHDRKKIGIKLNWPARRKIAIGAARGLAFLHHNCIPHIIHRDMKSSN 1019
Query: 905 ILLNLDYEAHVSDFGTAKFLKP-DLH-SWTQFAGTFGYAAPELSQTMEVNEKCDVYSFGV 962
+LL+ + EA VSDFG A+ + D H S + AGT GY PE Q+ + K DVYS+GV
Sbjct: 1020 VLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGV 1079
Query: 963 LALEIIIGKHPGDLISL---FLSPSTRPTANDMLLTEVLDQRPQKVIKPIDEEVILIAKL 1019
+ LE++ GK P D L + A +T+V D+ K I+ E++ K+
Sbjct: 1080 VLLELLTGKQPTDSADFGDNNLVGWVKLHAKGK-ITDVFDRELLKEDASIEIELLQHLKV 1138
Query: 1020 AFSCLNQVPRSRPTMDQVCKM 1040
A +CL+ RPTM QV M
Sbjct: 1139 ACACLDDRHWKRPTMIQVMAM 1159
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 399 bits (1026), Expect = e-110
Identities = 305/968 (31%), Positives = 466/968 (47%), Gaps = 100/968 (10%)
Query: 149 CTLSGEIDKSIGNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEI 208
C +G I + N + L+LG SG + +GKL ++R L +++ + SIP I
Sbjct: 63 CNWTG-ITCNSNNTGRVIRLELGNKKLSG-KLSESLGKLDEIRVLNLSRNFIKDSIPLSI 120
Query: 209 GLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIP-HSLWNMSSLTLIY 267
L NL +DLS+N LSG IP +I N+ L ++N K G +P H N + + ++
Sbjct: 121 FNLKNLQTLDLSSNDLSGGIPTSI-NLPALQSFDLSSN-KFNGSLPSHICHNSTQIRVVK 178
Query: 268 LYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPA 327
L +G+ + L+ L L MN+L+G IP + +LK L LL ++ NRLSGS+
Sbjct: 179 LAVNYFAGNFTSGFGKCVLLEHLCLGMNDLTGNIPEDLFHLKRLNLLGIQENRLSGSLSR 238
Query: 328 SIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFV 387
I NL +L V N +G IP L QL F +N G IP L N +
Sbjct: 239 EIRNLSSLVRLDVSWNLFSGEIPDVFDELPQLKFFLGQTNGFIGGIPKSLANSPSLNLLN 298
Query: 388 VSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAE 447
+ N G L +L L NRF G +P +L C ++ + + N G + E
Sbjct: 299 LRNNSLSGRLMLNCTAMIALNSLDLGTNRFNGRLPENLPDCKRLKNVNLARNTFHGQVPE 358
Query: 448 DFGVYPNLRYVDLSDN-------------------------KFHGHISPNWGKSL---DL 479
F + +L Y LS++ FHG P+ SL L
Sbjct: 359 SFKNFESLSYFSLSNSSLANISSALGILQHCKNLTTLVLTLNFHGEALPD-DSSLHFEKL 417
Query: 480 ETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFT 539
+ +++N ++G +P +L L LS N+LTG +P I G K+L YL +SNN FT
Sbjct: 418 KVLVVANCRLTGSMPRWLSSSNELQLLDLSWNRLTGAIPSWI-GDFKALFYLDLSNNSFT 476
Query: 540 DSIPTEIGLLQRLEELDLGGNELSGTIP-----NEVAELPKLRM-------LNLSRNRIE 587
IP + L+ L ++ NE S P NE A + + L N +
Sbjct: 477 GEIPKSLTKLESLTSRNISVNEPSPDFPFFMKRNESARALQYNQIFGFPPTIELGHNNLS 536
Query: 588 GRIPSTFDS--ALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTFSM--S 643
G I F + L DL N L+G+IP+SL + L L+LS+N LSG+IP +
Sbjct: 537 GPIWEEFGNLKKLHVFDLKWNALSGSIPSSLSGMTSLEALDLSNNRLSGSIPVSLQQLSF 596
Query: 644 LDFVNISDNQLDGPLPENPAFLRAPFESFKNNKGLCGNITGLVPCATSQ-----IHSRKS 698
L +++ N L G +P F P SF++N LCG PC+ SR+S
Sbjct: 597 LSKFSVAYNNLSGVIPSGGQFQTFPNSSFESNH-LCGE--HRFPCSEGTESALIKRSRRS 653
Query: 699 K--NILQSVFIALGALILVLSGVGISMYVFFRRKKPNE---EIQTEEEVQKGVLFSI--- 750
+ +I ++ IA G++ L+ +S+ V R++ E EI+ E + + L I
Sbjct: 654 RGGDIGMAIGIAFGSVFLLTL---LSLIVLRARRRSGEVDPEIEESESMNRKELGEIGSK 710
Query: 751 -----WSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDE 805
S+D ++ +++++++T +FD +IG G G VYKA LP G VA+KKL +
Sbjct: 711 LVVLFQSNDKELSYDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQ 770
Query: 806 EMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQA 865
+ F +E+ETL+ +H N++ L GFC + L+Y +ME GSLD L+
Sbjct: 771 -----IEREFEAEVETLSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDG 825
Query: 866 IA-FDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFL 924
A W+ R+ + +G A L YLH C P I+HRDI S NILL+ ++ +H++DFG A+ +
Sbjct: 826 PALLKWKTRLRIAQGAAKGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLM 885
Query: 925 KP-DLHSWTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPGDLISLFLSP 983
P + H T GT GY PE Q K DVYSFGV+ LE++ K P D+
Sbjct: 886 SPYETHVSTDLVGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDM------- 938
Query: 984 STRPTANDMLLTEVL----DQRPQKVIKPI------DEEVILIAKLAFSCLNQVPRSRPT 1033
+P L++ V+ + R +V P+ D+E+ + ++A CL++ P+ RPT
Sbjct: 939 -CKPKGCRDLISWVVKMKHESRASEVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPT 997
Query: 1034 MDQVCKML 1041
Q+ L
Sbjct: 998 TQQLVSWL 1005
Score = 165 bits (418), Expect = 5e-40
Identities = 156/599 (26%), Positives = 256/599 (42%), Gaps = 54/599 (9%)
Query: 68 ISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLNFSKNPI 127
+ + DL++ G+L S + + + + N+F G G + L N +
Sbjct: 149 LQSFDLSSNKFNGSLPSHICHNSTQIRVVKLAVNYFAGNFTSGFGKCVLLEHLCLGMNDL 208
Query: 128 IGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGNNFSGGPIPPEIGKL 187
G+IP++++ L+ L L LSG + + I NL++L LD+ N FS G IP +L
Sbjct: 209 TGNIPEDLFHLKRLNLLGIQENRLSGSLSREIRNLSSLVRLDVSWNLFS-GEIPDVFDEL 267
Query: 188 KKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLS---------------------- 225
+L++ +G IP+ + +L ++L NN LS
Sbjct: 268 PQLKFFLGQTNGFIGGIPKSLANSPSLNLLNLRNNSLSGRLMLNCTAMIALNSLDLGTNR 327
Query: 226 --GVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLS--GSIPDSV 281
G +PE + + +L + A NT +G +P S N SL+ L N SL+ S +
Sbjct: 328 FNGRLPENLPDCKRLKNVNLARNT-FHGQVPESFKNFESLSYFSLSNSSLANISSALGIL 386
Query: 282 QNLINLDVLALYMNNLSGFIPSTIG-NLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSV 340
Q+ NL L L +N +P + + L +L++ N RL+GS+P + + L+ +
Sbjct: 387 QHCKNLTTLVLTLNFHGEALPDDSSLHFEKLKVLVVANCRLTGSMPRWLSSSNELQLLDL 446
Query: 341 QVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQ 400
N LTG IP+ IG+ K L ++++N G IP L + + S +S N+ P
Sbjct: 447 SWNRLTGAIPSWIGDFKALFYLDLSNNSFTGEIPKSLTKLESLTSRNISVNEPSPDFPFF 506
Query: 401 MCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDL 460
M S + L +N+ G PT I + N + G I E+FG L DL
Sbjct: 507 MKRNESARALQ--YNQIFGFPPT----------IELGHNNLSGPIWEEFGNLKKLHVFDL 554
Query: 461 SDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKE 520
N G I + LE +SN +SG IP+ L+ L + ++ N L+G +P
Sbjct: 555 KWNALSGSIPSSLSGMTSLEALDLSNNRLSGSIPVSLQQLSFLSKFSVAYNNLSGVIPSG 614
Query: 521 ILGGMKSLLYLKISNNH------FTDSIPTEIGLLQRLEELDLGGNELS-GTIPNEVAEL 573
G ++ +NH F S TE L++R G ++ G V L
Sbjct: 615 --GQFQTFPNSSFESNHLCGEHRFPCSEGTESALIKRSRRSRGGDIGMAIGIAFGSVFLL 672
Query: 574 PKLRMLNLSRNRIEGRIPSTFDSALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNML 632
L ++ L R G + D + + + G I + L L Q + LS++ L
Sbjct: 673 TLLSLIVLRARRRSGEV----DPEIEESESMNRKELGEIGSKLVVLFQSNDKELSYDDL 727
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 385 bits (988), Expect = e-106
Identities = 320/1036 (30%), Positives = 484/1036 (45%), Gaps = 111/1036 (10%)
Query: 64 KSNLISTIDLANLGLKGT--LHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLN 121
K N + +DL+ + G + + L L I N G + + + L+
Sbjct: 171 KLNSLEVLDLSANSISGANVVGWVLSDGCGELKHLAISGNKISGDV--DVSRCVNLEFLD 228
Query: 122 FSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGNNFSGGPIP 181
S N IP + +L+ LD LSG+ ++I T L L++ N F G PIP
Sbjct: 229 VSSNNFSTGIPF-LGDCSALQHLDISGNKLSGDFSRAISTCTELKLLNISSNQFVG-PIP 286
Query: 182 PEIGKLKKLRYLAITQGSLVGSIPQEI-GLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQ 240
P LK L+YL++ + G IP + G LT +DLS N G +P G+ S L
Sbjct: 287 PL--PLKSLQYLSLAENKFTGEIPDFLSGACDTLTGLDLSGNHFYGAVPPFFGSCSLLES 344
Query: 241 LMFANNTKLYGPIP-HSLWNMSSLTLIYLYNMSLSGSIPDSVQNL-INLDVLALYMNNLS 298
L ++N G +P +L M L ++ L SG +P+S+ NL +L L L NN S
Sbjct: 345 LALSSNN-FSGELPMDTLLKMRGLKVLDLSFNEFSGELPESLTNLSASLLTLDLSSNNFS 403
Query: 299 G-FIPSTIGNLKN-LTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNL 356
G +P+ N KN L L L+NN +G IP ++ N L + N L+GTIP+++G+L
Sbjct: 404 GPILPNLCQNPKNTLQELYLQNNGFTGKIPPTLSNCSELVSLHLSFNYLSGTIPSSLGSL 463
Query: 357 KQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNR 416
+L ++ N L G IP L + + ++ ND G +PS + +L ++S +NR
Sbjct: 464 SKLRDLKLWLNMLEGEIPQELMYVKTLETLILDFNDLTGEIPSGLSNCTNLNWISLSNNR 523
Query: 417 FTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKS 476
TG +P + ++ +++ N G+I + G +L ++DL+ N F+G I K
Sbjct: 524 LTGEIPKWIGRLENLAILKLSNNSFSGNIPAELGDCRSLIWLDLNTNLFNGTIPAAMFKQ 583
Query: 477 LDLETFMISNTNISGGIPLDFI-----------GLTKLGRLHLSSNQLTGK-LPKEILGG 524
SG I +FI G+ K H + N L + + E L
Sbjct: 584 -------------SGKIAANFIAGKRYVYIKNDGMKK--ECHGAGNLLEFQGIRSEQLNR 628
Query: 525 MKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRN 584
+ + I++ + + LD+ N LSG IP E+ +P L +LNL N
Sbjct: 629 LSTRNPCNITSRVYGGHTSPTFDNNGSMMFLDMSYNMLSGYIPKEIGSMPYLFILNLGHN 688
Query: 585 RIEGRIPSTFDS--ALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTFSM 642
I G IP L +DLS N+L+G IP ++ L L+ ++LS+N LSG
Sbjct: 689 DISGSIPDEVGDLRGLNILDLSSNKLDGRIPQAMSALTMLTEIDLSNNNLSG-------- 740
Query: 643 SLDFVNISDNQLDGPLPENPAFLRAPFESFKNNKGLCG---------NITGLVPCATSQI 693
P+PE F P F NN GLCG N G S
Sbjct: 741 --------------PIPEMGQFETFPPAKFLNNPGLCGYPLPRCDPSNADGYAHHQRS-- 784
Query: 694 HSRKSKNILQSVFIALGALILVLSG---VGISMYVFFRRKKPNEEIQTEEEVQKG---VL 747
H R+ ++ SV + L + + G VG M R+K+ E+ E G
Sbjct: 785 HGRRPASLAGSVAMGLLFSFVCIFGLILVGREMRKRRRKKEAELEMYAEGHGNSGDRTAN 844
Query: 748 FSIWSHDG-----------------KMMFENIIEATENFDDKYLIGVGSQGNVYKAELPT 790
+ W G K+ F ++++AT F + LIG G G+VYKA L
Sbjct: 845 NTNWKLTGVKEALSINLAAFEKPLRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKD 904
Query: 791 GLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFM 850
G VA+KKL V + + F +E+ET+ IKHRN++ L G+C LVY+FM
Sbjct: 905 GSAVAIKKLIHVSGQ-----GDREFMAEMETIGKIKHRNLVPLLGYCKVGDERLLVYEFM 959
Query: 851 EGGSLDQILNNEKQA-IAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNL 909
+ GSL+ +L++ K+A + +W R + G A L++LHH+CSP IIHRD+ S N+LL+
Sbjct: 960 KYGSLEDVLHDPKKAGVKLNWSTRRKIAIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDE 1019
Query: 910 DYEAHVSDFGTAKFLKP-DLH-SWTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEI 967
+ EA VSDFG A+ + D H S + AGT GY PE Q+ + K DVYS+GV+ LE+
Sbjct: 1020 NLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLEL 1079
Query: 968 IIGKHPGDLISL---FLSPSTRPTANDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCL 1024
+ GK P D L + A + +++V D K ++ E++ K+A +CL
Sbjct: 1080 LTGKRPTDSPDFGDNNLVGWVKQHAK-LRISDVFDPELMKEDPALEIELLQHLKVAVACL 1138
Query: 1025 NQVPRSRPTMDQVCKM 1040
+ RPTM QV M
Sbjct: 1139 DDRAWRRPTMVQVMAM 1154
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated receptor
kinase 1 precursor (EC 2.7.1.37) (BRI1-associated
receptor kinase 1) (Somatic embryogenesis receptor-like
kinase 3)
Length = 615
Score = 245 bits (625), Expect = 5e-64
Identities = 175/511 (34%), Positives = 257/511 (50%), Gaps = 44/511 (8%)
Query: 555 LDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFDSA--LASIDLSGNRLNGNI 612
+DLG LSG + ++ +LP L+ L L N I G IP + L S+DL N L+G I
Sbjct: 73 VDLGNANLSGQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNNLSGPI 132
Query: 613 PTSLGFLVQLSMLNLSHNMLSGTIPSTFS--MSLDFVNISDNQLDGPLPENPAFLRAPFE 670
P++LG L +L L L++N LSG IP + + ++L +++S+N L G +P N +F
Sbjct: 133 PSTLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNNPLTGDIPVNGSFSLFTPI 192
Query: 671 SFKNNKGLCGNITGLVPCATSQIHSRKSKNILQSVFIALGALILVLSGVGISMYVFFRRK 730
SF N K + P + + S I ++ + A +L V ++RRK
Sbjct: 193 SFANTKLTPLPASPPPPISPTPPSPAGSNRITGAIAGGVAAGAALLFAVPAIALAWWRRK 252
Query: 731 KPNE---EIQTEE--EVQKGVLFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYK 785
KP + ++ EE EV G L + + A++NF +K ++G G G VYK
Sbjct: 253 KPQDHFFDVPAEEDPEVHLGQL-------KRFSLRELQVASDNFSNKNILGRGGFGKVYK 305
Query: 786 AELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFL 845
L G +VAVK+L EE + F +E+E ++ HRN+++L GFC L
Sbjct: 306 GRLADGTLVAVKRLK----EERTQGGELQFQTEVEMISMAVHRNLLRLRGFCMTPTERLL 361
Query: 846 VYKFMEGGSLDQILNNEKQAIA-FDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKN 904
VY +M GS+ L ++ DW KR + G A L+YLH C P IIHRD+ + N
Sbjct: 362 VYPYMANGSVASCLRERPESQPPLDWPKRQRIALGSARGLAYLHDHCDPKIIHRDVKAAN 421
Query: 905 ILLNLDYEAHVSDFGTAKFLK-PDLHSWTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVL 963
ILL+ ++EA V DFG AK + D H T GT G+ APE T + +EK DV+ +GV+
Sbjct: 422 ILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGVM 481
Query: 964 ALEIIIGKHPGDLISLFLSPSTRPTAND---MLLTEVLDQRPQKVIKPI----------D 1010
LE+I G+ DL L AND MLL V +K ++ + D
Sbjct: 482 LLELITGQRAFDLARL---------ANDDDVMLLDWVKGLLKEKKLEALVDVDLQGNYKD 532
Query: 1011 EEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
EEV + ++A C P RP M +V +ML
Sbjct: 533 EEVEQLIQVALLCTQSSPMERPKMSEVVRML 563
Score = 90.5 bits (223), Expect = 2e-17
Identities = 59/178 (33%), Positives = 96/178 (53%), Gaps = 5/178 (2%)
Query: 1 MVLPTFIIMILCVLPTLSVAEDSEAKLALLKWKASFDNQSQSILSTWKNT-TNPCSKWRG 59
+++P F +IL + L V+ ++E AL K S + ++ +L +W T PC+ W
Sbjct: 5 LMIPCFFWLILVLDLVLRVSGNAEGD-ALSALKNSLADPNK-VLQSWDATLVTPCT-WFH 61
Query: 60 IECDKSNLISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINT 119
+ C+ N ++ +DL N L G L + PNL L +Y+N+ GTIP Q+GNL+ + +
Sbjct: 62 VTCNSDNSVTRVDLGNANLSGQL-VMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVS 120
Query: 120 LNFSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGNNFSG 177
L+ N + G IP + L+ L+ L +LSGEI +S+ + L LDL N +G
Sbjct: 121 LDLYLNNLSGPIPSTLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNNPLTG 178
Score = 87.4 bits (215), Expect = 2e-16
Identities = 50/124 (40%), Positives = 70/124 (56%), Gaps = 1/124 (0%)
Query: 246 NTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTI 305
N L G + L + +L + LY+ +++G+IP+ + NL L L LY+NNLSG IPST+
Sbjct: 77 NANLSGQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNNLSGPIPSTL 136
Query: 306 GNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVA 365
G LK L L L NN LSG IP S+ ++ L+ + N LTG IP G+ A
Sbjct: 137 GRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNNPLTGDIPVN-GSFSLFTPISFA 195
Query: 366 SNKL 369
+ KL
Sbjct: 196 NTKL 199
Score = 84.0 bits (206), Expect = 2e-15
Identities = 49/127 (38%), Positives = 76/127 (59%), Gaps = 1/127 (0%)
Query: 261 SSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNR 320
+S+T + L N +LSG + + L NL L LY NN++G IP +GNL L L L N
Sbjct: 68 NSVTRVDLGNANLSGQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNN 127
Query: 321 LSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIP-NGLYN 379
LSG IP+++G L L++ + N+L+G IP ++ + L V ++++N L G IP NG ++
Sbjct: 128 LSGPIPSTLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNNPLTGDIPVNGSFS 187
Query: 380 ITNWYSF 386
+ SF
Sbjct: 188 LFTPISF 194
Score = 80.1 bits (196), Expect = 3e-14
Identities = 46/115 (40%), Positives = 70/115 (60%), Gaps = 2/115 (1%)
Query: 164 NLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNF 223
+++ +DLG N SG + ++G+L L+YL + ++ G+IP+++G LT L +DL N
Sbjct: 69 SVTRVDLGNANLSG-QLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNN 127
Query: 224 LSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIP 278
LSG IP T+G + KL L NN L G IP SL + +L ++ L N L+G IP
Sbjct: 128 LSGPIPSTLGRLKKLRFLRL-NNNSLSGEIPRSLTAVLTLQVLDLSNNPLTGDIP 181
Score = 76.6 bits (187), Expect = 3e-13
Identities = 46/131 (35%), Positives = 67/131 (51%), Gaps = 6/131 (4%)
Query: 296 NLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGN 355
NLSG + +G L NL L L +N ++G+IP +GNL L + +NNL+G IP+T+G
Sbjct: 79 NLSGQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNNLSGPIPSTLGR 138
Query: 356 LKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLS--AF 413
LK+L + +N L G IP L + +S N G +P GS + +F
Sbjct: 139 LKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNNPLTGDIP----VNGSFSLFTPISF 194
Query: 414 HNRFTGPVPTS 424
N P+P S
Sbjct: 195 ANTKLTPLPAS 205
Score = 70.1 bits (170), Expect = 3e-11
Identities = 38/121 (31%), Positives = 63/121 (51%)
Query: 304 TIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFE 363
T + ++T + L N LSG + +G L NL+Y + NN+TGTIP +GNL +L+ +
Sbjct: 63 TCNSDNSVTRVDLGNANLSGQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLD 122
Query: 364 VASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPT 423
+ N L G IP+ L + ++ N G +P + +L+ L +N TG +P
Sbjct: 123 LYLNNLSGPIPSTLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNNPLTGDIPV 182
Query: 424 S 424
+
Sbjct: 183 N 183
Score = 65.9 bits (159), Expect = 6e-10
Identities = 43/141 (30%), Positives = 66/141 (46%), Gaps = 25/141 (17%)
Query: 427 SCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMISN 486
S +S+ R+ + + G + G PNL+Y++L N
Sbjct: 66 SDNSVTRVDLGNANLSGQLVMQLGQLPNLQYLELYSN----------------------- 102
Query: 487 TNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFTDSIPTEI 546
NI+G IP LT+L L L N L+G +P LG +K L +L+++NN + IP +
Sbjct: 103 -NITGTIPEQLGNLTELVSLDLYLNNLSGPIP-STLGRLKKLRFLRLNNNSLSGEIPRSL 160
Query: 547 GLLQRLEELDLGGNELSGTIP 567
+ L+ LDL N L+G IP
Sbjct: 161 TAVLTLQVLDLSNNPLTGDIP 181
Score = 56.2 bits (134), Expect = 4e-07
Identities = 31/102 (30%), Positives = 53/102 (51%)
Query: 344 NLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCT 403
NL+G + +G L L E+ SN + G IP L N+T S + N+ G +PS +
Sbjct: 79 NLSGQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNNLSGPIPSTLGR 138
Query: 404 GGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDI 445
L++L +N +G +P SL + +++ + + N + GDI
Sbjct: 139 LKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNNPLTGDI 180
Score = 51.2 bits (121), Expect = 1e-05
Identities = 28/102 (27%), Positives = 50/102 (48%)
Query: 395 GHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPN 454
G L Q+ +L+YL + N TG +P L + + + + + N + G I G
Sbjct: 82 GQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNNLSGPIPSTLGRLKK 141
Query: 455 LRYVDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLD 496
LR++ L++N G I + L L+ +SN ++G IP++
Sbjct: 142 LRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNNPLTGDIPVN 183
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 224 bits (570), Expect = 1e-57
Identities = 228/849 (26%), Positives = 370/849 (42%), Gaps = 66/849 (7%)
Query: 214 LTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSL 273
+T I + ++ L G + + N+S+L +L N + GP+P SL ++SL ++ L N +
Sbjct: 66 VTRIQIGHSGLQGTLSPDLRNLSELERLELQWNN-ISGPVP-SLSGLASLQVLMLSNNNF 123
Query: 274 SGSIPDSVQNLINLDVLALYMNNLSGF-IPSTIGNLKNLTLLLLRNNRLSGSIPASIG-- 330
D Q L +L + + N + IP ++ N L + +SGS+P +G
Sbjct: 124 DSIPSDVFQGLTSLQSVEIDNNPFKSWEIPESLRNASALQNFSANSANVSGSLPGFLGPD 183
Query: 331 NLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSE 390
L + NNL G +P ++ Q+ + KL G I L N+T +
Sbjct: 184 EFPGLSILHLAFNNLEGELPMSLAG-SQVQSLWLNGQKLTGDI-TVLQNMTGLKEVWLHS 241
Query: 391 NDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFG 450
N F G LP L+ LS N FTGPVP SL S S++ + + N ++G +
Sbjct: 242 NKFSGPLPD-FSGLKELESLSLRDNSFTGPVPASLLSLESLKVVNLTNNHLQGPVP---- 296
Query: 451 VYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSS 510
V+ + VDL + +S +++ ++ I F +L +
Sbjct: 297 VFKSSVSVDLDKDSNSFCLSSPGECDPRVKSLLL--------IASSFDYPPRLAESWKGN 348
Query: 511 NQLTGKLPKEILGGMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIPNEV 570
+ T + G ++ + + T +I E G ++ L+ + LG N L+G IP E+
Sbjct: 349 DPCTNWIGIACSNG--NITVISLEKMELTGTISPEFGAIKSLQRIILGINNLTGMIPQEL 406
Query: 571 AELPKLRMLNLSRNRIEGRIPSTFDSALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHN 630
LP L+ L++S N++ G++P + + ++ +GN G +SL S S +
Sbjct: 407 TTLPNLKTLDVSSNKLFGKVPGFRSNVV--VNTNGNPDIGKDKSSLSSPGSSSPSGGSGS 464
Query: 631 MLSGTIPSTFSMSLDFVNISDNQLDGPLPENPAFLRAPFESFKNNKGLCGNITGLVPCAT 690
++G S F+ I + G L F +K + +G
Sbjct: 465 GINGDKDRRGMKSSTFIGIIVGSVLGGLLSIFLIGLLVFCWYKKRQK---RFSGSESSNA 521
Query: 691 SQIHSRKSKNILQSVFIALGALILVLSGVGISMYVFFRRKKPNEEIQTEEEVQKGVLFSI 750
+H R S + +SV I + + + G+ Y + + IQ E +L SI
Sbjct: 522 VVVHPRHSGSDNESVKITVAGSSVSVGGIS-DTYTLPGTSEVGDNIQMVEA--GNMLISI 578
Query: 751 WSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFF 810
+ + T NF ++G G G VYK EL G +AVK++ E
Sbjct: 579 ---------QVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRM------ENGVI 623
Query: 811 SSKSFT---SEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILN--NEKQA 865
+ K F SEI LT ++HR+++ L G+C LVY++M G+L + L +E+
Sbjct: 624 AGKGFAEFKSEIAVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGL 683
Query: 866 IAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLK 925
W++R+ + VA + YLH IHRD+ NILL D A V+DFG +
Sbjct: 684 KPLLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAP 743
Query: 926 PDLHSW-TQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGK------HPGDLIS 978
S T+ AGTFGY APE + T V K DVYSFGV+ +E+I G+ P + I
Sbjct: 744 EGKGSIETRIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIH 803
Query: 979 LFLSPSTRPTANDMLLTEVLDQRPQKVIKPIDEEVI----LIAKLAFSCLNQVPRSRPTM 1034
L +S R N E ++ +DEE + +A+LA C + P RP M
Sbjct: 804 L-VSWFKRMYIN----KEASFKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDM 858
Query: 1035 DQVCKMLGA 1043
+L +
Sbjct: 859 GHAVNILSS 867
Score = 114 bits (285), Expect = 1e-24
Identities = 131/537 (24%), Positives = 227/537 (41%), Gaps = 67/537 (12%)
Query: 5 TFIIMILCVLPTLSVAE-DSEAKL-ALLKWKASFDNQSQSILSTWKNTTNPCSKWRGIEC 62
TF++ L LS+++ DS+ L A+L K S + S W + +PC KW I C
Sbjct: 6 TFLLFSFTFLLLLSLSKADSDGDLSAMLSLKKSLNPPSSF---GWSDP-DPC-KWTHIVC 60
Query: 63 DKSNLISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLNF 122
+ ++ I + + GL+GTL P + NLS + L
Sbjct: 61 TGTKRVTRIQIGHSGLQGTLS-------------------------PDLRNLSELERLEL 95
Query: 123 SKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGNNFSGGPIPP 182
N I G +P + L SL+ L LT+L +++ N F IP
Sbjct: 96 QWNNISGPVPS-LSGLASLQVLMLSNNNFDSIPSDVFQGLTSLQSVEIDNNPFKSWEIPE 154
Query: 183 EIGKLKKLRYLAITQGSLVGSIPQEIGL--LTNLTYIDLSNNFLSGVIPETIGNMSKLNQ 240
+ L+ + ++ GS+P +G L+ + L+ N L G +P ++ Q
Sbjct: 155 SLRNASALQNFSANSANVSGSLPGFLGPDEFPGLSILHLAFNNLEGELPMSLAGSQV--Q 212
Query: 241 LMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGF 300
++ N KL G I L NM+ L ++L++ SG +PD L L+ L+L N+ +G
Sbjct: 213 SLWLNGQKLTGDIT-VLQNMTGLKEVWLHSNKFSGPLPD-FSGLKELESLSLRDNSFTGP 270
Query: 301 IPSTIGNLKNLTLLLLRNNRLSGSIPASIGNL-INLKYFSVQVNNLTGTIPATIGNLKQL 359
+P+++ +L++L ++ L NN L G +P ++ ++L S N+ + P +
Sbjct: 271 VPASLLSLESLKVVNLTNNHLQGPVPVFKSSVSVDLDKDS---NSFCLSSPGECDPRVKS 327
Query: 360 IVFEVASNKLYGRIPN---GLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNR 416
++ +S R+ G TNW C+ G++ +S
Sbjct: 328 LLLIASSFDYPPRLAESWKGNDPCTNWIGIA--------------CSNGNITVISLEKME 373
Query: 417 FTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKS 476
TG + + S++RI + N + G I ++ PNL+ +D+S NK +GK
Sbjct: 374 LTGTISPEFGAIKSLQRIILGINNLTGMIPQELTTLPNLKTLDVSSNKL-------FGKV 426
Query: 477 LDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKI 533
+ ++ NTN + I D L+ G S +G + GMKS ++ I
Sbjct: 427 PGFRSNVVVNTNGNPDIGKDKSSLSSPGSSSPSGGSGSGINGDKDRRGMKSSTFIGI 483
Score = 40.4 bits (93), Expect = 0.025
Identities = 41/166 (24%), Positives = 70/166 (41%), Gaps = 31/166 (18%)
Query: 550 QRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPS---------------TF 594
+R+ + +G + L GT+ ++ L +L L L N I G +PS F
Sbjct: 64 KRVTRIQIGHSGLQGTLSPDLRNLSELERLELQWNNISGPVPSLSGLASLQVLMLSNNNF 123
Query: 595 DS----------ALASIDLSGNRLNG-NIPTSLGFLVQLSMLNLSHNMLSGTIPSTFS-- 641
DS +L S+++ N IP SL L + + +SG++P
Sbjct: 124 DSIPSDVFQGLTSLQSVEIDNNPFKSWEIPESLRNASALQNFSANSANVSGSLPGFLGPD 183
Query: 642 --MSLDFVNISDNQLDGPLPENPAFLRAPFESFKNNKGLCGNITGL 685
L ++++ N L+G LP + A + + N + L G+IT L
Sbjct: 184 EFPGLSILHLAFNNLEGELPMSLAGSQVQ-SLWLNGQKLTGDITVL 228
>TML1_ARATH (P33543) Putative kinase-like protein TMKL1 precursor
Length = 674
Score = 204 bits (520), Expect = 8e-52
Identities = 180/599 (30%), Positives = 285/599 (47%), Gaps = 66/599 (11%)
Query: 478 DLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNH 537
DL + +NT++ L + L L S LTG LP+EI G L + ++ N
Sbjct: 82 DLSSPQWTNTSLFNDSSLHLLSL------QLPSANLTGSLPREI-GEFSMLQSVFLNINS 134
Query: 538 FTDSIPTEIGLLQRLEELDLGGNELSGTIPNEVAEL-PKLRMLNLSRNRIEG-----RIP 591
+ SIP E+G L ++DL GN L+G +P + L KL + N + G +P
Sbjct: 135 LSGSIPLELGYTSSLSDVDLSGNALAGVLPPSIWNLCDKLVSFKIHGNNLSGVLPEPALP 194
Query: 592 STFDSALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTFS-MSLDFVNIS 650
++ L +DL GN+ +G P + + L+LS N+ G +P + L+ +N+S
Sbjct: 195 NSTCGNLQVLDLGGNKFSGEFPEFITRFKGVKSLDLSSNVFEGLVPEGLGVLELESLNLS 254
Query: 651 DNQLDGPLPENPAFLRAPF--ESFKNNK-GLCGNITGLVPCATSQIHSRKSKNILQSVFI 707
N G LP+ F + F ESF+ N LCG L PC S SR S + + I
Sbjct: 255 HNNFSGMLPD---FGESKFGAESFEGNSPSLCG--LPLKPCLGS---SRLSPGAVAGLVI 306
Query: 708 ALGALILVLSGVGISMYVFFRRKKPNEEIQTEEEVQKG----VLFSIWSHDGKMMFENII 763
L + +V++ + I + + KK I++E+++++G + +GK++ +
Sbjct: 307 GLMSGAVVVASLLIG---YLQNKKRKSSIESEDDLEEGDEEDEIGEKEGGEGKLV---VF 360
Query: 764 EATENFD-DKYLIGVG------SQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFT 816
+ EN D L G S G VYKA+L G +A++ L E + S
Sbjct: 361 QGGENLTLDDVLNATGQVMEKTSYGTVYKAKLSDGGNIALRLLR-----EGTCKDRSSCL 415
Query: 817 SEIETLTGIKHRNIIKLHGFCSHSKFS-FLVYKFMEGGSLDQILNNEK-QAIAFDWEKRV 874
I L I+H N++ L F + L+Y ++ SL +L+ K + A +W +R
Sbjct: 416 PVIRQLGRIRHENLVPLRAFYQGKRGEKLLIYDYLPNISLHDLLHESKPRKPALNWARRH 475
Query: 875 NVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFL-KPDLHSWTQ 933
+ G+A L+YLH PIIH +I SKN+L++ + A +++FG K + +
Sbjct: 476 KIALGIARGLAYLHTGQEVPIIHGNIRSKNVLVDDFFFARLTEFGLDKIMVQAVADEIVS 535
Query: 934 FAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPG----------DLISLFLSP 983
A + GY APEL + + N + DVY+FG+L LEI++GK PG DL SL +
Sbjct: 536 QAKSDGYKAPELHKMKKCNPRSDVYAFGILLLEILMGKKPGKSGRNGNEFVDLPSLVKAA 595
Query: 984 STRPTANDMLLTEVLDQRPQKVIK-PIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
T EV D K I+ P++E ++ KLA C V RP+M++V K L
Sbjct: 596 VLEETT-----MEVFDLEAMKGIRSPMEEGLVHALKLAMGCCAPVTTVRPSMEEVVKQL 649
Score = 92.0 bits (227), Expect = 7e-18
Identities = 75/254 (29%), Positives = 128/254 (49%), Gaps = 33/254 (12%)
Query: 6 FIIMILCVLPTLSVAEDSEAKLALLKWKASFDNQSQSILSTWKNTTNPCSKWRGIECDKS 65
F++++ C T S++ S+ KL L K K+S S+S+L + N++ P +WRG++ S
Sbjct: 15 FVLILHCHCGT-SLSGSSDVKLLLGKIKSSLQGNSESLLLSSWNSSVPVCQWRGVKWVFS 73
Query: 66 NLISTIDLANLGL-KGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLNFSK 124
N S + ++L + T SL S +L++L + + + G++P +IG S + ++ +
Sbjct: 74 N-GSPLQCSDLSSPQWTNTSLFNDSSLHLLSLQLPSANLTGSLPREIGEFSMLQSVFLNI 132
Query: 125 NPIIGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLT--------------------- 163
N + GSIP E+ SL +D L+G + SI NL
Sbjct: 133 NSLSGSIPLELGYTSSLSDVDLSGNALAGVLPPSIWNLCDKLVSFKIHGNNLSGVLPEPA 192
Query: 164 -------NLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTY 216
NL LDLGGN FS G P I + K ++ L ++ G +P+ +G+L L
Sbjct: 193 LPNSTCGNLQVLDLGGNKFS-GEFPEFITRFKGVKSLDLSSNVFEGLVPEGLGVL-ELES 250
Query: 217 IDLSNNFLSGVIPE 230
++LS+N SG++P+
Sbjct: 251 LNLSHNNFSGMLPD 264
Score = 82.8 bits (203), Expect = 4e-15
Identities = 81/281 (28%), Positives = 128/281 (44%), Gaps = 32/281 (11%)
Query: 285 INLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNN 344
++L L L NL+G +P IG L + L N LSGSIP +G +L + N
Sbjct: 99 LHLLSLQLPSANLTGSLPREIGEFSMLQSVFLNINSLSGSIPLELGYTSSLSDVDLSGNA 158
Query: 345 LTGTIPATIGNL-KQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCT 403
L G +P +I NL +L+ F++ N L G +P LP+ C
Sbjct: 159 LAGVLPPSIWNLCDKLVSFKIHGNNLSGVLPEPA-------------------LPNSTC- 198
Query: 404 GGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDN 463
G+L+ L N+F+G P + ++ + + N EG + E GV L ++LS N
Sbjct: 199 -GNLQVLDLGGNKFSGEFPEFITRFKGVKSLDLSSNVFEGLVPEGLGVL-ELESLNLSHN 256
Query: 464 KFHGHISPNWGKS-LDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEIL 522
F G + P++G+S E+F ++ ++ G+PL LG LS + G + +
Sbjct: 257 NFSGML-PDFGESKFGAESFEGNSPSLC-GLPLK----PCLGSSRLSPGAVAGLVIGLMS 310
Query: 523 GG--MKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNE 561
G + SLL + N SI +E L + EE ++G E
Sbjct: 311 GAVVVASLLIGYLQNKKRKSSIESEDDLEEGDEEDEIGEKE 351
Score = 74.3 bits (181), Expect = 2e-12
Identities = 57/191 (29%), Positives = 89/191 (45%), Gaps = 31/191 (16%)
Query: 164 NLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNF 223
+L L L N +G +P EIG+ L+ + + SL GSIP E+G ++L+ +DLS N
Sbjct: 100 HLLSLQLPSANLTGS-LPREIGEFSMLQSVFLNINSLSGSIPLELGYTSSLSDVDLSGNA 158
Query: 224 LSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNM-SSLTLIYLYNMSLSGSIPDSV- 281
L+GV+P S+WN+ L ++ +LSG +P+
Sbjct: 159 LAGVLPP-------------------------SIWNLCDKLVSFKIHGNNLSGVLPEPAL 193
Query: 282 --QNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFS 339
NL VL L N SG P I K + L L +N G +P +G ++ L+ +
Sbjct: 194 PNSTCGNLQVLDLGGNKFSGEFPEFITRFKGVKSLDLSSNVFEGLVPEGLG-VLELESLN 252
Query: 340 VQVNNLTGTIP 350
+ NN +G +P
Sbjct: 253 LSHNNFSGMLP 263
>SIRK_ARATH (O64483) Senescence-induced receptor-like serine/threonine
kinase precursor (FLG22-induced receptor-like kinase 1)
Length = 876
Score = 173 bits (438), Expect = 3e-42
Identities = 137/471 (29%), Positives = 226/471 (47%), Gaps = 53/471 (11%)
Query: 574 PKLRMLNLSRNRIEGRIPSTFDS--ALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNM 631
P++ LN+S + + G+I F + ++ +DLSGN L G IP L L L+ LN+ N
Sbjct: 414 PRVVSLNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNK 473
Query: 632 LSGTIPSTFSMSLDFVNISDNQLDGPLPENPAFLRAPFESFKNNKGLCGNITGLVPCATS 691
L+G +P + + +G L LR F N LC + + C+ +
Sbjct: 474 LTGIVPQ---------RLHERSKNGSLS-----LR-----FGRNPDLCLSDS----CSNT 510
Query: 692 QIHSRKSKNILQSVFIALGALILVLSGVGISMYVFFRRKKPNEEIQTEEEVQKGVLFSIW 751
+ +K+KN + +G ++++L+ + + FRR K ++ T E + G L +
Sbjct: 511 K---KKNKNGYIIPLVVVGIIVVLLTALAL-----FRRFKKKQQRGTLGE-RNGPLKTAK 561
Query: 752 SHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFS 811
+ + ++ T NF+ +IG G G VY + G VAVK L E S
Sbjct: 562 RY---FKYSEVVNITNNFER--VIGKGGFGKVYHGVI-NGEQVAVKVL-----SEESAQG 610
Query: 812 SKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWE 871
K F +E++ L + H N+ L G+C+ L+Y++M +L L K++ WE
Sbjct: 611 YKEFRAEVDLLMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAG-KRSFILSWE 669
Query: 872 KRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPDLHSW 931
+R+ + A L YLH+ C PPI+HRD+ NILLN +A ++DFG ++ +
Sbjct: 670 ERLKISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQ 729
Query: 932 --TQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGK---HPGDLISLFLSPSTR 986
T AG+ GY PE T ++NEK DVYS GV+ LE+I G+ + +S R
Sbjct: 730 ISTVVAGSIGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKTEKVHISDHVR 789
Query: 987 PTANDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQV 1037
+ + ++DQR ++ + ++++A +C RPTM QV
Sbjct: 790 SILANGDIRGIVDQRLRE--RYDVGSAWKMSEIALACTEHTSAQRPTMSQV 838
Score = 46.2 bits (108), Expect = 5e-04
Identities = 32/114 (28%), Positives = 49/114 (42%), Gaps = 25/114 (21%)
Query: 128 IGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGNNFSGGPIPPEIGKL 187
I I + T + L+ F L G+ID + NLT++ LDL GN
Sbjct: 403 IDCIQSDNTTNPRVVSLNISFSELRGQIDPAFSNLTSIRKLDLSGN-------------- 448
Query: 188 KKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQL 241
+L G IP + L NLT +++ N L+G++P+ + SK L
Sbjct: 449 -----------TLTGEIPAFLANLPNLTELNVEGNKLTGIVPQRLHERSKNGSL 491
Score = 43.9 bits (102), Expect = 0.002
Identities = 24/61 (39%), Positives = 33/61 (53%)
Query: 290 LALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTI 349
L + + L G I NL ++ L L N L+G IPA + NL NL +V+ N LTG +
Sbjct: 419 LNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKLTGIV 478
Query: 350 P 350
P
Sbjct: 479 P 479
Score = 43.5 bits (101), Expect = 0.003
Identities = 22/60 (36%), Positives = 31/60 (51%)
Query: 319 NRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLY 378
+ L G I + NL +++ + N LTG IPA + NL L V NKL G +P L+
Sbjct: 424 SELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKLTGIVPQRLH 483
Score = 43.1 bits (100), Expect = 0.004
Identities = 23/61 (37%), Positives = 32/61 (51%)
Query: 531 LKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRI 590
L IS + I L + +LDL GN L+G IP +A LP L LN+ N++ G +
Sbjct: 419 LNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKLTGIV 478
Query: 591 P 591
P
Sbjct: 479 P 479
Score = 41.6 bits (96), Expect = 0.011
Identities = 23/62 (37%), Positives = 35/62 (56%)
Query: 116 RINTLNFSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGNNF 175
R+ +LN S + + G I L S++ LD TL+GEI + NL NL+ L++ GN
Sbjct: 415 RVVSLNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKL 474
Query: 176 SG 177
+G
Sbjct: 475 TG 476
Score = 41.6 bits (96), Expect = 0.011
Identities = 22/54 (40%), Positives = 32/54 (58%)
Query: 273 LSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIP 326
L G I + NL ++ L L N L+G IP+ + NL NLT L + N+L+G +P
Sbjct: 426 LRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKLTGIVP 479
Score = 40.8 bits (94), Expect = 0.019
Identities = 25/80 (31%), Positives = 42/80 (52%), Gaps = 1/80 (1%)
Query: 506 LHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGT 565
L++S ++L G++ + S+ L +S N T IP + L L EL++ GN+L+G
Sbjct: 419 LNISFSELRGQIDPAF-SNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKLTGI 477
Query: 566 IPNEVAELPKLRMLNLSRNR 585
+P + E K L+L R
Sbjct: 478 VPQRLHERSKNGSLSLRFGR 497
Score = 39.7 bits (91), Expect = 0.043
Identities = 33/94 (35%), Positives = 45/94 (47%), Gaps = 6/94 (6%)
Query: 193 LAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGP 252
L I+ L G I LT++ +DLS N L+G IP + N+ L +L N KL G
Sbjct: 419 LNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGN-KLTGI 477
Query: 253 IPHSLWNMS---SLTLIYLYNMSLSGSIPDSVQN 283
+P L S SL+L + N L + DS N
Sbjct: 478 VPQRLHERSKNGSLSLRFGRNPDL--CLSDSCSN 509
Score = 35.4 bits (80), Expect = 0.82
Identities = 23/74 (31%), Positives = 37/74 (49%)
Query: 247 TKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIG 306
++L G I + N++S+ + L +L+G IP + NL NL L + N L+G +P +
Sbjct: 424 SELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKLTGIVPQRLH 483
Query: 307 NLKNLTLLLLRNNR 320
L LR R
Sbjct: 484 ERSKNGSLSLRFGR 497
Score = 34.3 bits (77), Expect = 1.8
Identities = 30/91 (32%), Positives = 46/91 (49%), Gaps = 14/91 (15%)
Query: 49 NTTNP--------CSKWRG-IECDKSNLIST--IDLANLGLKGTLHSLTFSSFPNLITLN 97
NTTNP S+ RG I+ SNL S +DL+ L G + + ++ PNL LN
Sbjct: 410 NTTNPRVVSLNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAF-LANLPNLTELN 468
Query: 98 IYNNHFYGTIPPQIGNLSRINTLN--FSKNP 126
+ N G +P ++ S+ +L+ F +NP
Sbjct: 469 VEGNKLTGIVPQRLHERSKNGSLSLRFGRNP 499
Score = 32.7 bits (73), Expect = 5.3
Identities = 25/106 (23%), Positives = 45/106 (41%), Gaps = 11/106 (10%)
Query: 358 QLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNRF 417
+++ ++ ++L G+I N+T+ +S N G +P+ + +L L+ N+
Sbjct: 415 RVVSLNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKL 474
Query: 418 TGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDN 463
TG VP L S G ++ FG P+L D N
Sbjct: 475 TGIVPQRLHERSK-----------NGSLSLRFGRNPDLCLSDSCSN 509
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precursor
(EC 2.7.1.37)
Length = 817
Score = 162 bits (409), Expect = 6e-39
Identities = 105/341 (30%), Positives = 169/341 (48%), Gaps = 24/341 (7%)
Query: 713 ILVLSGVGISMYVFFRRKKPNEEIQTEEEVQKGVLFSIWSHDGKMMFENIIEATENFDDK 772
++ +S + + + +R+ E+ E+ K + S+ + + +++AT F K
Sbjct: 484 VVEVSFISFAWFFVLKRELRPSELWASEKGYKAMT----SNFRRYSYRELVKATRKF--K 537
Query: 773 YLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIK 832
+G G G VYK L VAVKKL VR + F +E+ + I H N+++
Sbjct: 538 VELGRGESGTVYKGVLEDDRHVAVKKLENVRQ------GKEVFQAELSVIGRINHMNLVR 591
Query: 833 LHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVVKGVANALSYLHHDCS 892
+ GFCS LV +++E GSL IL +E I DWE R N+ GVA L+YLHH+C
Sbjct: 592 IWGFCSEGSHRLLVSEYVENGSLANILFSEGGNILLDWEGRFNIALGVAKGLAYLHHECL 651
Query: 893 PPIIHRDISSKNILLNLDYEAHVSDFGTAKFLK--PDLHSWTQFAGTFGYAAPELSQTME 950
+IH D+ +NILL+ +E ++DFG K L + + GT GY APE ++
Sbjct: 652 EWVIHCDVKPENILLDQAFEPKITDFGLVKLLNRGGSTQNVSHVRGTLGYIAPEWVSSLP 711
Query: 951 VNEKCDVYSFGVLALEIIIGKHPGDLISLFLSPSTRPTANDMLLTEVLDQRPQ------- 1003
+ K DVYS+GV+ LE++ G +L+ + +L+ L+ Q
Sbjct: 712 ITAKVDVYSYGVVLLELLTGTRVSELVGGTDEVHSMLRKLVRMLSAKLEGEEQSWIDGYL 771
Query: 1004 --KVIKPID-EEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
K+ +P++ + + KLA SCL + RPTM+ + L
Sbjct: 772 DSKLNRPVNYVQARTLIKLAVSCLEEDRSKRPTMEHAVQTL 812
>D100_ARATH (Q00874) DNA-damage-repair/toleration protein DRT100
precursor
Length = 372
Score = 154 bits (390), Expect = 9e-37
Identities = 108/324 (33%), Positives = 163/324 (49%), Gaps = 14/324 (4%)
Query: 28 ALLKWKASFDNQSQSILSTWKNTTNPCSKWRGIECDKSNLISTIDLANLGLKGTLHSLTF 87
AL +K+S + I +TW T+ C +W GI CD + T ++ L+G F
Sbjct: 34 ALNAFKSSLSEPNLGIFNTWSENTDCCKEWYGISCDPDSGRVT----DISLRGESEDAIF 89
Query: 88 SSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLNFSK-NPIIGSIPQEMYTLRSLKGLDF 146
+ + G+I P + +L+ + +L + I G IP + +L SL+ LD
Sbjct: 90 QKAGR-------SGYMSGSIDPAVCDLTALTSLVLADWKGITGEIPPCITSLASLRILDL 142
Query: 147 FFCTLSGEIDKSIGNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQ 206
++GEI IG L+ L+ L+L N S G IP + L +L++L +T+ + G IP
Sbjct: 143 AGNKITGEIPAEIGKLSKLAVLNLAENQMS-GEIPASLTSLIELKHLELTENGITGVIPA 201
Query: 207 EIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLI 266
+ G L L+ + L N L+G IPE+I M +L L + N + GPIP + NM L+L+
Sbjct: 202 DFGSLKMLSRVLLGRNELTGSIPESISGMERLADLDLSKN-HIEGPIPEWMGNMKVLSLL 260
Query: 267 YLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIP 326
L SL+G IP S+ + LDV L N L G IP G+ L L L +N LSG IP
Sbjct: 261 NLDCNSLTGPIPGSLLSNSGLDVANLSRNALEGTIPDVFGSKTYLVSLDLSHNSLSGRIP 320
Query: 327 ASIGNLINLKYFSVQVNNLTGTIP 350
S+ + + + + N L G IP
Sbjct: 321 DSLSSAKFVGHLDISHNKLCGRIP 344
Score = 147 bits (371), Expect = 1e-34
Identities = 92/246 (37%), Positives = 135/246 (54%), Gaps = 6/246 (2%)
Query: 418 TGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSL 477
TG +P + S +S+ + + GN+I G+I + G L ++L++N+ G I + +
Sbjct: 124 TGEIPPCITSLASLRILDLAGNKITGEIPAEIGKLSKLAVLNLAENQMSGEIPASLTSLI 183
Query: 478 DLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNH 537
+L+ ++ I+G IP DF L L R+ L N+LTG +P+ I GM+ L L +S NH
Sbjct: 184 ELKHLELTENGITGVIPADFGSLKMLSRVLLGRNELTGSIPESI-SGMERLADLDLSKNH 242
Query: 538 FTDSIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFDSA 597
IP +G ++ L L+L N L+G IP + L + NLSRN +EG IP F S
Sbjct: 243 IEGPIPEWMGNMKVLSLLNLDCNSLTGPIPGSLLSNSGLDVANLSRNALEGTIPDVFGSK 302
Query: 598 --LASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTFSMS-LDFVNISDNQ- 653
L S+DLS N L+G IP SL + L++SHN L G IP+ F L+ + SDNQ
Sbjct: 303 TYLVSLDLSHNSLSGRIPDSLSSAKFVGHLDISHNKLCGRIPTGFPFDHLEATSFSDNQC 362
Query: 654 -LDGPL 658
GPL
Sbjct: 363 LCGGPL 368
Score = 133 bits (335), Expect = 2e-30
Identities = 87/244 (35%), Positives = 123/244 (49%), Gaps = 5/244 (2%)
Query: 441 IEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGL 500
I G+I +LR +DL+ NK G I GK L ++ +SG IP L
Sbjct: 123 ITGEIPPCITSLASLRILDLAGNKITGEIPAEIGKLSKLAVLNLAENQMSGEIPASLTSL 182
Query: 501 TKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGN 560
+L L L+ N +TG +P + G +K L + + N T SIP I ++RL +LDL N
Sbjct: 183 IELKHLELTENGITGVIPADF-GSLKMLSRVLLGRNELTGSIPESISGMERLADLDLSKN 241
Query: 561 ELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTF--DSALASIDLSGNRLNGNIPTSLGF 618
+ G IP + + L +LNL N + G IP + +S L +LS N L G IP G
Sbjct: 242 HIEGPIPEWMGNMKVLSLLNLDCNSLTGPIPGSLLSNSGLDVANLSRNALEGTIPDVFGS 301
Query: 619 LVQLSMLNLSHNMLSGTIPSTFSMS--LDFVNISDNQLDGPLPENPAFLRAPFESFKNNK 676
L L+LSHN LSG IP + S + + ++IS N+L G +P F SF +N+
Sbjct: 302 KTYLVSLDLSHNSLSGRIPDSLSSAKFVGHLDISHNKLCGRIPTGFPFDHLEATSFSDNQ 361
Query: 677 GLCG 680
LCG
Sbjct: 362 CLCG 365
Score = 132 bits (332), Expect = 5e-30
Identities = 85/251 (33%), Positives = 133/251 (52%), Gaps = 7/251 (2%)
Query: 251 GPIPHSLWNMSSLTLIYLYNMS-LSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLK 309
G I ++ ++++LT + L + ++G IP + +L +L +L L N ++G IP+ IG L
Sbjct: 100 GSIDPAVCDLTALTSLVLADWKGITGEIPPCITSLASLRILDLAGNKITGEIPAEIGKLS 159
Query: 310 NLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKL 369
L +L L N++SG IPAS+ +LI LK+ + N +TG IPA G+LK L + N+L
Sbjct: 160 KLAVLNLAENQMSGEIPASLTSLIELKHLELTENGITGVIPADFGSLKMLSRVLLGRNEL 219
Query: 370 YGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFH---NRFTGPVPTSLK 426
G IP + + +S+N G +P M G++K LS + N TGP+P SL
Sbjct: 220 TGSIPESISGMERLADLDLSKNHIEGPIPEWM---GNMKVLSLLNLDCNSLTGPIPGSLL 276
Query: 427 SCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMISN 486
S S ++ + N +EG I + FG L +DLS N G I + + + IS+
Sbjct: 277 SNSGLDVANLSRNALEGTIPDVFGSKTYLVSLDLSHNSLSGRIPDSLSSAKFVGHLDISH 336
Query: 487 TNISGGIPLDF 497
+ G IP F
Sbjct: 337 NKLCGRIPTGF 347
Score = 127 bits (318), Expect = 2e-28
Identities = 89/268 (33%), Positives = 136/268 (50%), Gaps = 3/268 (1%)
Query: 198 GSLVGSIPQEIGLLTNLTYIDLSN-NFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHS 256
G + GSI + LT LT + L++ ++G IP I +++ L L A N K+ G IP
Sbjct: 96 GYMSGSIDPAVCDLTALTSLVLADWKGITGEIPPCITSLASLRILDLAGN-KITGEIPAE 154
Query: 257 LWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLL 316
+ +S L ++ L +SG IP S+ +LI L L L N ++G IP+ G+LK L+ +LL
Sbjct: 155 IGKLSKLAVLNLAENQMSGEIPASLTSLIELKHLELTENGITGVIPADFGSLKMLSRVLL 214
Query: 317 RNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNG 376
N L+GSIP SI + L + N++ G IP +GN+K L + + N L G IP
Sbjct: 215 GRNELTGSIPESISGMERLADLDLSKNHIEGPIPEWMGNMKVLSLLNLDCNSLTGPIPGS 274
Query: 377 LYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRI 436
L + + +S N G +P + L L HN +G +P SL S + + I
Sbjct: 275 LLSNSGLDVANLSRNALEGTIPDVFGSKTYLVSLDLSHNSLSGRIPDSLSSAKFVGHLDI 334
Query: 437 EGNQIEGDIAEDFGVYPNLRYVDLSDNK 464
N++ G I F + +L SDN+
Sbjct: 335 SHNKLCGRIPTGF-PFDHLEATSFSDNQ 361
Score = 112 bits (279), Expect = 7e-24
Identities = 82/252 (32%), Positives = 124/252 (48%), Gaps = 8/252 (3%)
Query: 297 LSGFIPSTIGNLKNLTLLLLRNNR-LSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGN 355
+SG I + +L LT L+L + + ++G IP I +L +L+ + N +TG IPA IG
Sbjct: 98 MSGSIDPAVCDLTALTSLVLADWKGITGEIPPCITSLASLRILDLAGNKITGEIPAEIGK 157
Query: 356 LKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAF-- 413
L +L V +A N++ G IP L ++ ++EN G +P+ GSLK LS
Sbjct: 158 LSKLAVLNLAENQMSGEIPASLTSLIELKHLELTENGITGVIPADF---GSLKMLSRVLL 214
Query: 414 -HNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPN 472
N TG +P S+ + + + N IEG I E G L ++L N G I +
Sbjct: 215 GRNELTGSIPESISGMERLADLDLSKNHIEGPIPEWMGNMKVLSLLNLDCNSLTGPIPGS 274
Query: 473 WGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLK 532
+ L+ +S + G IP F T L L LS N L+G++P + L K + +L
Sbjct: 275 LLSNSGLDVANLSRNALEGTIPDVFGSKTYLVSLDLSHNSLSGRIP-DSLSSAKFVGHLD 333
Query: 533 ISNNHFTDSIPT 544
IS+N IPT
Sbjct: 334 ISHNKLCGRIPT 345
Score = 95.5 bits (236), Expect = 7e-19
Identities = 70/201 (34%), Positives = 99/201 (48%), Gaps = 30/201 (14%)
Query: 489 ISGGIPLDFIGLTKLGRLHLSS-NQLTGKLPKEILGGMKSLLYLKISNNHFTDSIPTEIG 547
+SG I LT L L L+ +TG++P I + SL L ++ N T IP EIG
Sbjct: 98 MSGSIDPAVCDLTALTSLVLADWKGITGEIPPCITS-LASLRILDLAGNKITGEIPAEIG 156
Query: 548 LLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFDS----------- 596
L +L L+L N++SG IP + L +L+ L L+ N I G IP+ F S
Sbjct: 157 KLSKLAVLNLAENQMSGEIPASLTSLIELKHLELTENGITGVIPADFGSLKMLSRVLLGR 216
Query: 597 ---------------ALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTF- 640
LA +DLS N + G IP +G + LS+LNL N L+G IP +
Sbjct: 217 NELTGSIPESISGMERLADLDLSKNHIEGPIPEWMGNMKVLSLLNLDCNSLTGPIPGSLL 276
Query: 641 -SMSLDFVNISDNQLDGPLPE 660
+ LD N+S N L+G +P+
Sbjct: 277 SNSGLDVANLSRNALEGTIPD 297
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase receptor
precursor (EC 2.7.1.37) (S-receptor kinase) (SRK)
Length = 849
Score = 148 bits (374), Expect = 7e-35
Identities = 94/298 (31%), Positives = 152/298 (50%), Gaps = 22/298 (7%)
Query: 760 ENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEI 819
E +++ATENF +G G G VYK L G +AVK+L + S + F +E+
Sbjct: 519 ETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRL-----SKTSVQGTDEFMNEV 573
Query: 820 ETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVVKG 879
+ ++H N++++ G C L+Y+++E SLD L + + +W +R ++ G
Sbjct: 574 TLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSKLNWNERFDITNG 633
Query: 880 VANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPD--LHSWTQFAGT 937
VA L YLH D IIHRD+ NILL+ + +SDFG A+ + D + + GT
Sbjct: 634 VARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANTMKVVGT 693
Query: 938 FGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGK---------HPGDLISLFLSPSTRPT 988
+GY +PE + +EK DV+SFGV+ LEI+ GK + DL+S S
Sbjct: 694 YGYMSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSRWKEGR 753
Query: 989 A---NDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKMLGA 1043
A D ++ + L +P + +P +EV+ ++ C+ ++ RP M V M G+
Sbjct: 754 ALEIVDPVIVDSLSSQP-SIFQP--QEVLKCIQIGLLCVQELAEHRPAMSSVVWMFGS 808
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 147 bits (371), Expect = 1e-34
Identities = 103/305 (33%), Positives = 163/305 (52%), Gaps = 37/305 (12%)
Query: 759 FENIIEATENFDDKYLIGVGSQGNVYK----------AELPTGLVVAVKKLHLVRDEEMS 808
F ++ AT+NF ++G G G VY+ + + +G++VA+K+L+ E +
Sbjct: 76 FLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLN---SESVQ 132
Query: 809 FFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAF 868
F+ + SE+ L + HRN++KL G+C K LVY+FM GSL+ L F
Sbjct: 133 GFAE--WRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLFRRNDP--F 188
Query: 869 DWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPD- 927
W+ R+ +V G A L++LH +I+RD + NILL+ +Y+A +SDFG AK D
Sbjct: 189 PWDLRIKIVIGAARGLAFLH-SLQREVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADE 247
Query: 928 -LHSWTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIG------KHPGDLISL- 979
H T+ GT+GYAAPE T + K DV++FGV+ LEI+ G K P SL
Sbjct: 248 KSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQESLV 307
Query: 980 -FLSPSTRPTANDMLLTEVLDQ--RPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQ 1036
+L P +N + +++D+ + Q K E +A++ SC+ P++RP M +
Sbjct: 308 DWLRPE---LSNKHRVKQIMDKGIKGQYTTKVATE----MARITLSCIEPDPKNRPHMKE 360
Query: 1037 VCKML 1041
V ++L
Sbjct: 361 VVEVL 365
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 145 bits (366), Expect = 6e-34
Identities = 119/378 (31%), Positives = 180/378 (47%), Gaps = 44/378 (11%)
Query: 695 SRKSKNILQSVFIA--LGALILVLSGVGISMYVFFRRKKPNEEIQTEE-EVQKGVLFSIW 751
SRK +F+A + A++LVLS V ++ ++ R K + + E + K +S
Sbjct: 414 SRKLMAFQMRIFVAEIVFAVVLVLS-VSVTTCLYVRHKLRHCQCSNRELRLAKSTAYSFR 472
Query: 752 SHDGKMM---------------FENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAV 796
+ K+ +E + +AT F + +G GS V+K L G VVAV
Sbjct: 473 KDNMKIQPDMEDLKIRRAQEFSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAV 532
Query: 797 KKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLD 856
K+ D + S SK F +E++ L+ + H +++ L G+C LVY+FM GSL
Sbjct: 533 KRAIKASDVKKS---SKEFHNELDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLY 589
Query: 857 QILNNEKQAIA--FDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAH 914
Q L+ + + +W +RV + A + YLH PP+IHRDI S NIL++ D+ A
Sbjct: 590 QHLHGKDPNLKKRLNWARRVTIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNAR 649
Query: 915 VSDFGTAKFLKPDLHSWTQF----AGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIG 970
V+DFG + D S T AGT GY PE + + K DVYSFGV+ LEI+ G
Sbjct: 650 VADFGLSILGPAD--SGTPLSELPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSG 707
Query: 971 KHPGDL------ISLFLSPSTRPTANDMLLTEVLDQRPQKVIKPIDEEVI-LIAKLAFSC 1023
+ D+ I + P + +L VL P D E + IA +A C
Sbjct: 708 RKAIDMQFEEGNIVEWAVPLIKAGDIFAILDPVLS-------PPSDLEALKKIASVACKC 760
Query: 1024 LNQVPRSRPTMDQVCKML 1041
+ + RP+MD+V L
Sbjct: 761 VRMRGKDRPSMDKVTTAL 778
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 144 bits (362), Expect = 2e-33
Identities = 85/219 (38%), Positives = 122/219 (54%), Gaps = 13/219 (5%)
Query: 759 FENIIEATENFDDKYLIGVGSQGNVYKAELP-TGLVVAVKKLHLVRDEEMSFFSSKSFTS 817
F + AT NF +G G G VYK L TG VVAVK+L + ++ F
Sbjct: 76 FRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQL-----DRNGLQGNREFLV 130
Query: 818 EIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNN---EKQAIAFDWEKRV 874
E+ L+ + H N++ L G+C+ LVY+FM GSL+ L++ +K+A+ DW R+
Sbjct: 131 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEAL--DWNMRM 188
Query: 875 NVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKF--LKPDLHSWT 932
+ G A L +LH +PP+I+RD S NILL+ + +SDFG AK H T
Sbjct: 189 KIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVST 248
Query: 933 QFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGK 971
+ GT+GY APE + T ++ K DVYSFGV+ LE+I G+
Sbjct: 249 RVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGR 287
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 141 bits (356), Expect = 8e-33
Identities = 82/219 (37%), Positives = 121/219 (54%), Gaps = 13/219 (5%)
Query: 759 FENIIEATENFDDKYLIGVGSQGNVYKAELPT-GLVVAVKKLHLVRDEEMSFFSSKSFTS 817
F+ + EAT NF +G G G V+K + VVA+K+L + + F
Sbjct: 93 FQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQL-----DRNGVQGIREFVV 147
Query: 818 EIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQ---ILNNEKQAIAFDWEKRV 874
E+ TL+ H N++KL GFC+ LVY++M GSL+ +L + K+ + DW R+
Sbjct: 148 EVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPL--DWNTRM 205
Query: 875 NVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKF--LKPDLHSWT 932
+ G A L YLH +PP+I+RD+ NILL DY+ +SDFG AK H T
Sbjct: 206 KIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVST 265
Query: 933 QFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGK 971
+ GT+GY AP+ + T ++ K D+YSFGV+ LE+I G+
Sbjct: 266 RVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGR 304
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.138 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 125,836,353
Number of Sequences: 164201
Number of extensions: 5563451
Number of successful extensions: 21682
Number of sequences better than 10.0: 1873
Number of HSP's better than 10.0 without gapping: 848
Number of HSP's successfully gapped in prelim test: 1025
Number of HSP's that attempted gapping in prelim test: 13457
Number of HSP's gapped (non-prelim): 3782
length of query: 1061
length of database: 59,974,054
effective HSP length: 121
effective length of query: 940
effective length of database: 40,105,733
effective search space: 37699389020
effective search space used: 37699389020
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC133779.6


