

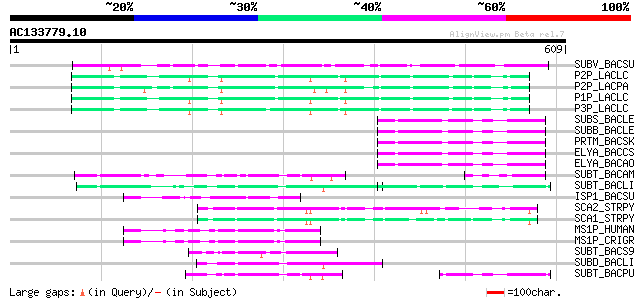
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133779.10 + phase: 2 /pseudo
(609 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SUBV_BACSU (P29141) Minor extracellular protease vpr precursor (... 115 4e-25
P2P_LACLC (P15293) PII-type proteinase precursor (EC 3.4.21.96) ... 82 4e-15
P2P_LACPA (Q02470) PII-type proteinase precursor (EC 3.4.21.96) ... 80 2e-14
P1P_LACLC (P16271) PI-type proteinase precursor (EC 3.4.21.-) (W... 79 3e-14
P3P_LACLC (P15292) PIII-type proteinase precursor (EC 3.4.21.96)... 77 1e-13
SUBS_BACLE (P29600) Subtilisin Savinase (EC 3.4.21.62) (Alkaline... 59 3e-08
SUBB_BACLE (P29599) Subtilisin BL (EC 3.4.21.62) (Alkaline prote... 59 3e-08
PRTM_BACSK (Q99405) M-protease (EC 3.4.21.-) 59 3e-08
ELYA_BACCS (P41362) Alkaline protease precursor (EC 3.4.21.-) 59 3e-08
ELYA_BACAO (P27693) Alkaline protease precursor (EC 3.4.21.-) 59 3e-08
SUBT_BACAM (P00782) Subtilisin BPN' precursor (EC 3.4.21.62) (Su... 58 6e-08
SUBT_BACLI (P00780) Subtilisin Carlsberg precursor (EC 3.4.21.62) 57 1e-07
ISP1_BACSU (P11018) Major intracellular serine protease precurso... 57 2e-07
SCA2_STRPY (P58099) C5a peptidase precursor (EC 3.4.21.-) (SCP) 54 1e-06
SCA1_STRPY (P15926) C5a peptidase precursor (EC 3.4.21.-) (SCP) 54 2e-06
MS1P_HUMAN (Q14703) Membrane-bound transcription factor site-1 p... 52 3e-06
MS1P_CRIGR (Q9Z2A8) Membrane-bound transcription factor site-1 p... 52 3e-06
SUBT_BACS9 (P28842) Subtilisin precursor (EC 3.4.21.62) 52 6e-06
SUBD_BACLI (P00781) Subtilisin DY (EC 3.4.21.62) 51 1e-05
SUBT_BACPU (P07518) Subtilisin (EC 3.4.21.62) (Alkaline mesenter... 50 1e-05
>SUBV_BACSU (P29141) Minor extracellular protease vpr precursor (EC
3.4.21.-)
Length = 806
Score = 115 bits (287), Expect = 4e-25
Identities = 136/537 (25%), Positives = 222/537 (41%), Gaps = 88/537 (16%)
Query: 70 YNKQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHT------TRSWEFLGLHGND-- 121
Y + +GF+ L E +L V +V+ + +K T S + + +D
Sbjct: 106 YEQVFSGFSMKLPANEIPKLLAVKDVKAVYPNVTYKTDNMKDKDVTISEDAVSPQMDDSA 165
Query: 122 ----INSAWQKGRFGENTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGSSKKVPCNRKL 177
N AW G G+ +A IDTGV ++ KK K
Sbjct: 166 PYIGANDAWDLGYTGKGIKVAIIDTGV-----------------EYNHPDLKKNFGQYK- 207
Query: 178 IGARFFSDAYERYNGKLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGS 237
G F + Y+ + A D HGTH T N GTIKG +
Sbjct: 208 -GYDFVDNDYDPKETPTGDPRGEATD---HGTHVAGTVAAN------------GTIKGVA 251
Query: 238 PRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEV 297
P A + Y+V L S +V++ +++A+ DG D++++S G +S N+ + T
Sbjct: 252 PDATLLAYRV---LGPGGSGTTENVIAGVERAVQDGADVMNLSLG--NSLNNPDWAT--- 303
Query: 298 SIGAFHALARNILLVASAGNEGPTPGSVVN--VAPWVFTVAASTIDRDFSSTITIGDQII 355
S A++ ++ V S GN GP +V + + +V A+ + + +T G
Sbjct: 304 STALDWAMSEGVVAVTSNGNSGPNGWTVGSPGTSREAISVGATQLPLN-EYAVTFGSY-- 360
Query: 356 RGASLFVDLPPNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIK 415
++ + +N+ + + A +A+ + + GK VA + G I
Sbjct: 361 -SSAKVMGYNKEDDVKALNNKEVELVEAGIGEAK-----DFEGKDLTGK-VAVVKRGSIA 413
Query: 416 SVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDT 475
V + A AGA GM + N +SG + + V G I G
Sbjct: 414 FVDKADNAKKAGAIGMVVYNN--LSGE-------IEANVPGMSVPTIKLSLEDGEKLVSA 464
Query: 476 IESG-TKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFA 534
+++G TK F ++ + +A FSSRGP + +++KPD++APGVNI++
Sbjct: 465 LKAGETKTTFKLTVS---KALGEQVADFSSRGP-VMDTWMIKPDISAPGVNIVSTIPTH- 519
Query: 535 SASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVIT 591
D + + QGTSM+ PH+AG +IK P WS IK+AIM T +T
Sbjct: 520 -------DPDHPYGYGSKQGTSMASPHIAGAVAVIKQAKPKWSVEQIKAAIMNTAVT 569
>P2P_LACLC (P15293) PII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
(LP151)
Length = 1902
Score = 82.0 bits (201), Expect = 4e-15
Identities = 127/541 (23%), Positives = 211/541 (38%), Gaps = 94/541 (17%)
Query: 69 SYNKQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQK 128
SY +NGF+ + + +L + V +V L+K + ++ ++ + W
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKTVTLAKVYYPTDAKANSMA-----NVQAVWSN 203
Query: 129 GRF-GENTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGSSKKVPCNRKLIGARFFSDAY 187
++ GE T+++ ID+G+ P + R K V + + + +
Sbjct: 204 YKYKGEGTVVSVIDSGIDPTHKDM------------RLSDDKDVKLTKSDVEKFTDTAKH 251
Query: 188 ERY-NGKLP-------TSQRTARDFVG--HGTHTLSTAGGNFVPGASIFNIGNG-----T 232
RY N K+P + D V HG H G N G G +
Sbjct: 252 GRYFNSKVPYGFNYADNNDTITDDTVDEQHGMHVAGIIGAN----------GTGDDPAKS 301
Query: 233 IKGGSPRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEI 292
+ G +P A++ KV + +A+ A ++SAI+ + G D++++S G S + E
Sbjct: 302 VVGVAPEAQLLAMKVFTNSDTSATTGSATLVSAIEDSAKIGADVLNMSLGSDSGNQTLE- 360
Query: 293 FTDEVSIGAFHALARNILLVASAGNEGPTPGSVVNV--------------APWVFTVAAS 338
D +A V SAGN G + + V P A +
Sbjct: 361 --DPELAAVQNANESGTAAVISAGNSGTSGSATEGVNKDYYGLQDNEMVGTPGTSRGATT 418
Query: 339 TIDRDFSSTITIGDQIIRGASLFVDLPPN----QSFTLVNSID-AKFSNATTRDARFCRP 393
+ + IT I G L L P S S D KF +
Sbjct: 419 VASAENTDVITQAVTITDGTGL--QLGPETIQLSSNDFTGSFDQKKFYVVKDASGNLSKG 476
Query: 394 RTLD-PSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLS 452
+ D + KGKI A + G++ + + A +AGA G+ + N + T ++ + +
Sbjct: 477 KVADYTADAKGKI-AIVKRGELTFADKQKYAQAAGAAGLIIVNNDGTA--TPVTSMALTT 533
Query: 453 TVGGNGQAAITAPPRLGVTATDTIES-GTKIRFSQAITLIGRK--PAPVMASFSSRGPNQ 509
T G +++T + A +S G KI A+TL+ + M+ F+S GP
Sbjct: 534 TFPTFGLSSVTGQKLVDWVAAHPDDSLGVKI----ALTLVPNQKYTEDKMSDFTSYGP-- 587
Query: 510 VQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLI 569
V KPD+TAPG NI + T N G + M GTSM+ P +AG+ L+
Sbjct: 588 VSNLSFKPDITAPGGNIWS------------TQNNNG--YTNMSGTSMASPFIAGSQALL 633
Query: 570 K 570
K
Sbjct: 634 K 634
>P2P_LACPA (Q02470) PII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
(LP151)
Length = 1902
Score = 80.1 bits (196), Expect = 2e-14
Identities = 130/544 (23%), Positives = 211/544 (37%), Gaps = 100/544 (18%)
Query: 69 SYNKQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQK 128
SY +NGF+ + + +L + V +V L+K + ++ ++ + W
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKTVTLAKVYYPTDAKANSMA-----NVQAVWSN 203
Query: 129 GRF-GENTIIANIDTGVWP---ESRSFSDRGIGPIPAKWRGGSSKKVPCNRKLIGARFFS 184
++ GE T+++ IDTG+ P + R D+ + K +K K R+F+
Sbjct: 204 YKYKGEGTVVSVIDTGIDPTHKDMRLSDDKDV-----KLTKYDVEKFTDTAK--HGRYFT 256
Query: 185 DAYERYNGKLPTSQRTARDFVG--HGTHTLSTAGGNFVPGASIFNIGNG-----TIKGGS 237
+ D V HG H G N G G ++ G +
Sbjct: 257 SKVPYGFNYADNNDTITDDTVDEQHGMHVAGIIGAN----------GTGDDPTKSVVGVA 306
Query: 238 PRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEV 297
P A++ KV + +A+ A ++SAI+ + G D++++S G S + E D
Sbjct: 307 PEAQLLAMKVFTNSDTSATTGSATLVSAIEDSAKIGADVLNMSLGSDSGNQTLE---DPE 363
Query: 298 SIGAFHALARNILLVASAGNEGPTPGSVVNVAPWVF-----------------TVAASTI 340
+A V SAGN G + + V + T AS
Sbjct: 364 IAAVQNANESGTAAVISAGNSGTSGSATQGVNKDYYGLQDNEMVGTPGTSRGATTVASAE 423
Query: 341 DRDFSS---TITIGDQIIRGASLFVDLPPN--------QSFTLVNSIDAKFSNATTRDAR 389
+ D S TIT G + G + L N + F +V S D
Sbjct: 424 NTDVISQAVTITDGKDLQLGPET-IQLSSNDFTGSFDQKKFYVVKDASGDLSKGAAADYT 482
Query: 390 FCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPH 449
+ KGKI A + G++ + + A +AGA G+ + N + T L+
Sbjct: 483 ---------ADAKGKI-AIVKRGELNFADKQKYAQAAGAAGLIIVNNDGTA--TPLTSIR 530
Query: 450 VLSTVGGNGQAAITAPPRLG-VTATDTIESGTKIRFSQAITLIGRKP--APVMASFSSRG 506
+ +T G ++ T + VTA G KI A+TL+ + M+ F+S G
Sbjct: 531 LTTTFPTFGLSSKTGQKLVDWVTAHPDDSLGVKI----ALTLLPNQKYTEDKMSDFTSYG 586
Query: 507 PNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTA 566
P V KPD+TAPG NI + T N G+ M GTSM+ P +AG+
Sbjct: 587 P--VSNLSFKPDITAPGGNIWS------------TQNNNGY--TNMSGTSMASPFIAGSQ 630
Query: 567 GLIK 570
L+K
Sbjct: 631 ALLK 634
>P1P_LACLC (P16271) PI-type proteinase precursor (EC 3.4.21.-)
(Wall-associated serine proteinase)
Length = 1902
Score = 79.3 bits (194), Expect = 3e-14
Identities = 127/541 (23%), Positives = 210/541 (38%), Gaps = 94/541 (17%)
Query: 69 SYNKQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQK 128
SY +NGF+ + + +L + V +V L+K + ++ ++ + W
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKTVTLAKVYYPTDAKANSMA-----NVQAVWSN 203
Query: 129 GRF-GENTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGSSKKVPCNRKLIGARFFSDAY 187
++ GE T+++ ID+G+ P + R K V + + + +
Sbjct: 204 YKYKGEGTVVSVIDSGIDPTHKDM------------RLSDDKDVKLTKSDVEKFTDTAKH 251
Query: 188 ERY-NGKLP-------TSQRTARDFVG--HGTHTLSTAGGNFVPGASIFNIGNG-----T 232
RY N K+P + D V HG H G N G G +
Sbjct: 252 GRYFNSKVPYGFNYADNNDTITDDTVDEQHGMHVAGIIGAN----------GTGDDPAKS 301
Query: 233 IKGGSPRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEI 292
+ G +P A++ KV + +A+ + ++SAI+ + G D++++S G S + E
Sbjct: 302 VVGVAPEAQLLAMKVFTNSDTSATTGSSTLVSAIEDSAKIGADVLNMSLGSDSGNQTLE- 360
Query: 293 FTDEVSIGAFHALARNILLVASAGNEGPTPGSVVNV--------------APWVFTVAAS 338
D +A V SAGN G + + V P A +
Sbjct: 361 --DPELAAVQNANESGTAAVISAGNSGTSGSATEGVNKDYYGLQDNEMVGTPGTSRGATT 418
Query: 339 TIDRDFSSTITIGDQIIRGASLFVDLPPN----QSFTLVNSIDAK-FSNATTRDARFCRP 393
+ + IT I G L L P S S D K F +
Sbjct: 419 VASAENTDVITQAVTITDGTGL--QLGPGTIQLSSNDFTGSFDQKKFYVVKDASGNLSKG 476
Query: 394 RTLD-PSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLS 452
D + KGKI A + G++ + + A +AGA G+ + N + T ++ + +
Sbjct: 477 ALADYTADAKGKI-AIVKRGELSFDDKQKYAQAAGAAGLIIVNNDGTA--TPVTSMALTT 533
Query: 453 TVGGNGQAAITAPPRLG-VTATDTIESGTKIRFSQAITLIGRKP--APVMASFSSRGPNQ 509
T G +++T + VTA G KI A+TL+ + M+ F+S GP
Sbjct: 534 TFPTFGLSSVTGQKLVDWVTAHPDDSLGVKI----ALTLVPNQKYTEDKMSDFTSYGP-- 587
Query: 510 VQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLI 569
V KPD+TAPG NI + T N G+ M GTSM+ P +AG+ L+
Sbjct: 588 VSNLSFKPDITAPGGNIWS------------TQNNNGY--TNMSGTSMASPFIAGSQALL 633
Query: 570 K 570
K
Sbjct: 634 K 634
>P3P_LACLC (P15292) PIII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
Length = 1902
Score = 77.0 bits (188), Expect = 1e-13
Identities = 126/539 (23%), Positives = 207/539 (38%), Gaps = 90/539 (16%)
Query: 69 SYNKQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQK 128
SY +NGF+ + + +L + V +V L+K + ++ ++ + W
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKTVTLAKVYYPTDAKANSMA-----NVQAVWSN 203
Query: 129 GRF-GENTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGSSKKVPCNRKLIGARFFSDAY 187
++ GE T+++ ID+G+ P + R K V + + + +
Sbjct: 204 YKYKGEGTVVSVIDSGIDPTHKDM------------RLSDDKDVKLTKSDVEKFTDTVKH 251
Query: 188 ERY-NGKLP-------TSQRTARDFVG--HGTHTLSTAGGNFVPGASIFNIGNG-----T 232
RY N K+P + D V HG H G N G G +
Sbjct: 252 GRYFNSKVPYGFNYADNNDTITDDKVDEQHGMHVAGIIGAN----------GTGDDPAKS 301
Query: 233 IKGGSPRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEI 292
+ G +P A++ KV + +A A V+SAI+ + G D++++S G S + +
Sbjct: 302 VVGVAPEAQLLAMKVFSNSDTSAKTGSATVVSAIEDSAKIGADVLNMSLG---SNSGNQT 358
Query: 293 FTDEVSIGAFHALARNILLVASAGNEGPTPGSVVNV--------------APWVFTVAAS 338
D +A V SAGN G + + V +P A +
Sbjct: 359 LEDPELAAVQNANESGTAAVISAGNSGTSGSATEGVNKDYYGLQDNEMVGSPGTSRGATT 418
Query: 339 TIDRDFSSTITIGDQIIRGASLFVDLPPN----QSFTLVNSID-AKFSNATTRDARFCRP 393
+ + IT I G L L P S S D KF +
Sbjct: 419 VASAENTDVITQAVTITDGTGL--QLGPETIQLSSHDFTGSFDQKKFYIVKDASGNLSKG 476
Query: 394 RTLD-PSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLS 452
D + KGKI A + G+ + + A +AGA G+ + N + T ++ + +
Sbjct: 477 ALADYTADAKGKI-AIVKRGEFSFDDKQKYAQAAGAAGLIIVNTDGTA--TPMTSIALTT 533
Query: 453 TVGGNGQAAITAPPRLG-VTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQ 511
T G +++T + VTA G KI + A+ + M+ F+S GP V
Sbjct: 534 TFPTFGLSSVTGQKLVDWVTAHPDDSLGVKI--TLAMLPNQKYTEDKMSDFTSYGP--VS 589
Query: 512 PYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIK 570
KPD+TAPG NI + T N G + M GTSM+ P +AG+ L+K
Sbjct: 590 NLSFKPDITAPGGNIWS------------TQNNNG--YTNMSGTSMASPFIAGSQALLK 634
>SUBS_BACLE (P29600) Subtilisin Savinase (EC 3.4.21.62) (Alkaline
protease)
Length = 269
Score = 59.3 bits (142), Expect = 3e-08
Identities = 47/185 (25%), Positives = 77/185 (41%), Gaps = 29/185 (15%)
Query: 404 KIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAIT 463
K++ + G + S+A+G E AG GM + N S + + +++ G +
Sbjct: 92 KVLGASGSGSVSSIAQGLEW--AGNNGMHVANLSLGSPSPSATLEQAVNSATSRGVLVVA 149
Query: 464 APPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPG 523
A G + R++ A+ + ASFS G D+ APG
Sbjct: 150 ASGNSGAGSISY-----PARYANAMAVGATDQNNNRASFSQYGAGL--------DIVAPG 196
Query: 524 VNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKS 583
VN+ + Y G + + GTSM+ PHVAG A L+K +P+WS I++
Sbjct: 197 VNVQSTYP--------------GSTYASLNGTSMATPHVAGAAALVKQKNPSWSNVQIRN 242
Query: 584 AIMTT 588
+ T
Sbjct: 243 HLKNT 247
>SUBB_BACLE (P29599) Subtilisin BL (EC 3.4.21.62) (Alkaline
protease)
Length = 269
Score = 59.3 bits (142), Expect = 3e-08
Identities = 48/185 (25%), Positives = 77/185 (40%), Gaps = 29/185 (15%)
Query: 404 KIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAIT 463
K++ G I S+A+G E AG GM + N S + + +++ G +
Sbjct: 92 KVLGADGRGAISSIAQGLEW--AGNNGMHVANLSLGSPSPSATLEQAVNSATSRGVLVVA 149
Query: 464 APPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPG 523
A G ++ R++ A+ + ASFS G D+ APG
Sbjct: 150 ASGNSGASSISY-----PARYANAMAVGATDQNNNRASFSQYGAGL--------DIVAPG 196
Query: 524 VNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKS 583
VN+ + Y G + + GTSM+ PHVAG A L+K +P+WS I++
Sbjct: 197 VNVQSTYP--------------GSTYASLNGTSMATPHVAGAAALVKQKNPSWSNVQIRN 242
Query: 584 AIMTT 588
+ T
Sbjct: 243 HLKNT 247
>PRTM_BACSK (Q99405) M-protease (EC 3.4.21.-)
Length = 269
Score = 59.3 bits (142), Expect = 3e-08
Identities = 47/185 (25%), Positives = 77/185 (41%), Gaps = 29/185 (15%)
Query: 404 KIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAIT 463
K++ + G + S+A+G E AG GM + N S + + +++ G +
Sbjct: 92 KVLGASGSGSVSSIAQGLEW--AGNNGMHVANLSLGSPSPSATLEQAVNSATSRGVLVVA 149
Query: 464 APPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPG 523
A G + R++ A+ + ASFS G D+ APG
Sbjct: 150 ASGNSGAGSISY-----PARYANAMAVGATDQNNNRASFSQYGAGL--------DIVAPG 196
Query: 524 VNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKS 583
VN+ + Y G + + GTSM+ PHVAG A L+K +P+WS I++
Sbjct: 197 VNVQSTYP--------------GSTYASLNGTSMATPHVAGVAALVKQKNPSWSNVQIRN 242
Query: 584 AIMTT 588
+ T
Sbjct: 243 HLKNT 247
>ELYA_BACCS (P41362) Alkaline protease precursor (EC 3.4.21.-)
Length = 380
Score = 59.3 bits (142), Expect = 3e-08
Identities = 47/185 (25%), Positives = 77/185 (41%), Gaps = 29/185 (15%)
Query: 404 KIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAIT 463
K++ + G + S+A+G E AG GM + N S + + +++ G +
Sbjct: 203 KVLGASGSGSVSSIAQGLEW--AGNNGMHVANLSLGSPSPSATLEQAVNSATSRGVLVVA 260
Query: 464 APPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPG 523
A G + R++ A+ + ASFS G D+ APG
Sbjct: 261 ASGNSGAGSISY-----PARYANAMAVGATDQNNNRASFSQYGAGL--------DIVAPG 307
Query: 524 VNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKS 583
VN+ + Y G + + GTSM+ PHVAG A L+K +P+WS I++
Sbjct: 308 VNVQSTYP--------------GSTYASLNGTSMATPHVAGAAALVKQKNPSWSNVQIRN 353
Query: 584 AIMTT 588
+ T
Sbjct: 354 HLKNT 358
>ELYA_BACAO (P27693) Alkaline protease precursor (EC 3.4.21.-)
Length = 380
Score = 59.3 bits (142), Expect = 3e-08
Identities = 47/185 (25%), Positives = 77/185 (41%), Gaps = 29/185 (15%)
Query: 404 KIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAIT 463
K++ + G + S+A+G E AG GM + N S + + +++ G +
Sbjct: 203 KVLGASGSGSVSSIAQGLEW--AGNNGMHVANLSLGSPSPSATLEQAVNSATSRGVLVVA 260
Query: 464 APPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPG 523
A G + R++ A+ + ASFS G D+ APG
Sbjct: 261 ASGNSGAGSISY-----PARYANAMAVGATDQNNNRASFSQYGAGL--------DIVAPG 307
Query: 524 VNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKS 583
VN+ + Y G + + GTSM+ PHVAG A L+K +P+WS I++
Sbjct: 308 VNVQSTYP--------------GSTYASLNGTSMATPHVAGAAALVKQKNPSWSNVQIRN 353
Query: 584 AIMTT 588
+ T
Sbjct: 354 HLKNT 358
>SUBT_BACAM (P00782) Subtilisin BPN' precursor (EC 3.4.21.62)
(Subtilisin Novo) (Subtilisin DFE) (Alkaline protease)
Length = 382
Score = 58.2 bits (139), Expect = 6e-08
Identities = 75/305 (24%), Positives = 125/305 (40%), Gaps = 64/305 (20%)
Query: 72 KQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQKGRF 131
K ++ +A L E+ +L K+P V ++ ++H H G+ + +G
Sbjct: 72 KYVDAASATLNEKAVKELKKDPSVA--YVEEDHVAHAYAQSVPYGVSQIKAPALHSQGYT 129
Query: 132 GENTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGSSKKVPCNRKLIGARFFSDAYERYN 191
G N +A ID+G+ S D K GG+S
Sbjct: 130 GSNVKVAVIDSGI---DSSHPD-------LKVAGGASM---------------------- 157
Query: 192 GKLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSL 251
+P+ +D HGTH T N G + G +P A + KV +
Sbjct: 158 --VPSETNPFQDNNSHGTHVAGTVAA--------LNNSIGVL-GVAPSASLYAVKVLGA- 205
Query: 252 TDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILL 311
D + + +++ I+ AI + +D+I++S GGPS + + + D+ A+A +++
Sbjct: 206 -DGSGQYSW-IINGIEWAIANNMDVINMSLGGPSGSAALKAAVDK-------AVASGVVV 256
Query: 312 VASAGNEGPTPGSVVNVA-----PWVFTVAASTIDRDFSSTITIG---DQIIRGASLFVD 363
VA+AGNEG T GS V P V V A +S ++G D + G S+
Sbjct: 257 VAAAGNEG-TSGSSSTVGYPGKYPSVIAVGAVDSSNQRASFSSVGPELDVMAPGVSIQST 315
Query: 364 LPPNQ 368
LP N+
Sbjct: 316 LPGNK 320
Score = 53.1 bits (126), Expect = 2e-06
Identities = 35/89 (39%), Positives = 45/89 (50%), Gaps = 22/89 (24%)
Query: 500 ASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSC 559
ASFSS GP DV APGV+I + L N+ G GTSM+
Sbjct: 294 ASFSSVGPEL--------DVMAPGVSI----------QSTLPGNKYG----AYNGTSMAS 331
Query: 560 PHVAGTAGLIKTLHPNWSPAAIKSAIMTT 588
PHVAG A LI + HPNW+ ++S++ T
Sbjct: 332 PHVAGAAALILSKHPNWTNTQVRSSLENT 360
>SUBT_BACLI (P00780) Subtilisin Carlsberg precursor (EC 3.4.21.62)
Length = 379
Score = 57.0 bits (136), Expect = 1e-07
Identities = 78/344 (22%), Positives = 135/344 (38%), Gaps = 65/344 (18%)
Query: 74 INGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQKGRFGE 133
IN A L++E ++ +P V ++ ++H H G+ + +G G
Sbjct: 72 INAAKAKLDKEALKEVKNDPDVA--YVEEDHVAHALAQTVPYGIPLIKADKVQAQGFKGA 129
Query: 134 NTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGSSKKVPCNRKLIGARFFSDAYERYNGK 193
N +A +DTG+ + G GA F A E YN
Sbjct: 130 NVKVAVLDTGIQASHPDLNVVG-----------------------GASFV--AGEAYN-- 162
Query: 194 LPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGT-IKGGSPRARVATYKVCWSLT 252
D GHGTH T + N T + G +P + KV L
Sbjct: 163 --------TDGNGHGTHVAGTVAA----------LDNTTGVLGVAPSVSLYAVKV---LN 201
Query: 253 DAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLV 312
+ S + ++S I+ A +G+D+I++S GGPS + + + D +A AR +++V
Sbjct: 202 SSGSGTYSGIVSGIEWATTNGMDVINMSLGGPSGSTAMKQAVD-------NAYARGVVVV 254
Query: 313 ASAGNEGPT--PGSVVNVAPWVFTVAASTIDRD-----FSSTITIGDQIIRGASLFVDLP 365
A+AGN G + ++ A + +A +D + FSS + + GA ++ P
Sbjct: 255 AAAGNSGSSGNTNTIGYPAKYDSVIAVGAVDSNSNRASFSSVGAELEVMAPGAGVYSTYP 314
Query: 366 PNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVACA 409
+ TL + A A + L S+V+ ++ + A
Sbjct: 315 TSTYATLNGTSMASPHVAGAAALILSKHPNLSASQVRNRLSSTA 358
Score = 47.8 bits (112), Expect = 9e-05
Identities = 47/191 (24%), Positives = 77/191 (39%), Gaps = 28/191 (14%)
Query: 404 KIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAIT 463
K++ + G + G E + + + SG+T + + + G +
Sbjct: 198 KVLNSSGSGTYSGIVSGIEWATTNGMDVINMSLGGPSGSTAMKQ--AVDNAYARGVVVVA 255
Query: 464 APPRLGVTA-TDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAP 522
A G + T+TI G ++ I + ASFSS G +V AP
Sbjct: 256 AAGNSGSSGNTNTI--GYPAKYDSVIAVGAVDSNSNRASFSSVGAEL--------EVMAP 305
Query: 523 GVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIK 582
G + + Y A+ + GTSM+ PHVAG A LI + HPN S + ++
Sbjct: 306 GAGVYSTYPTSTYAT--------------LNGTSMASPHVAGAAALILSKHPNLSASQVR 351
Query: 583 SAIMTTVITYL 593
+ + +T TYL
Sbjct: 352 NRLSSTA-TYL 361
>ISP1_BACSU (P11018) Major intracellular serine protease precursor
(EC 3.4.21.-) (ISP-1)
Length = 319
Score = 56.6 bits (135), Expect = 2e-07
Identities = 53/194 (27%), Positives = 79/194 (40%), Gaps = 47/194 (24%)
Query: 126 WQKGRFGENTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGSSKKVPCNRKLIGARFFSD 185
W KG G+N +A +DTG + ++IG + F+D
Sbjct: 35 WAKGVKGKNIKVAVLDTGC----------------------DTSHPDLKNQIIGGKNFTD 72
Query: 186 AYERYNGKLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATY 245
GK + D+ GHGTH V G N NG I G +P A +
Sbjct: 73 DD---GGK----EDAISDYNGHGTH---------VAGTIAANDSNGGIAGVAPEASLLIV 116
Query: 246 KVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHAL 305
KV + +++ I+ A++ VDIIS+S GGPS E+ +A+
Sbjct: 117 KVLGGENGSGQYEW--IINGINYAVEQKVDIISMSLGGPSD-------VPELKEAVKNAV 167
Query: 306 ARNILLVASAGNEG 319
+L+V +AGNEG
Sbjct: 168 KNGVLVVCAAGNEG 181
Score = 32.0 bits (71), Expect = 4.8
Identities = 19/54 (35%), Positives = 28/54 (51%), Gaps = 14/54 (25%)
Query: 518 DVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKT 571
D+ APG NIL+ N++ + + GTSM+ PHV+G LIK+
Sbjct: 222 DLVAPGENILSTLP-----------NKK---YGKLTGTSMAAPHVSGALALIKS 261
>SCA2_STRPY (P58099) C5a peptidase precursor (EC 3.4.21.-) (SCP)
Length = 1181
Score = 53.9 bits (128), Expect = 1e-06
Identities = 102/406 (25%), Positives = 165/406 (40%), Gaps = 92/406 (22%)
Query: 207 HGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAASCFGADVLSAI 266
HGTH GN A ++G P A++ +V + + + + + AI
Sbjct: 193 HGTHVSGILSGN----APSETKEPYRLEGAMPEAQLLLMRV--EIVNGLADYARNYAQAI 246
Query: 267 DQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNEGPTPGS-- 324
A++ G +I++S G + + DE +A ++ + +V SAGN+ G
Sbjct: 247 IDAVNLGAKVINMSFGNAALAYAN--LPDETKKAFDYAKSKGVSIVTSAGNDSSFGGKTR 304
Query: 325 ----------VVNV---APWVFTVAASTIDRDFSSTITIGDQIIRGASLFVDLPPNQSFT 371
VV A TVA+ + D+ + T T+ + + V L N+ F
Sbjct: 305 LPLADHPDYGVVGTPAAADSTLTVASYSPDKQLTETATVKTADQQDKEMPV-LSTNR-FE 362
Query: 372 LVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGM 431
+ D ++N ++ F VKGKI R G I + A AGA G+
Sbjct: 363 PNKAYDYAYANRGMKEDDF--------KDVKGKIALIER-GDIDFKDKIANAKKAGAVGV 413
Query: 432 FL-ENQPKVSGNTLLSEPHV----LSTVGG-----NGQAAIT--APPRLGVTATDTIESG 479
+ +NQ K L + + +S G N Q IT A P++ TA SG
Sbjct: 414 LIYDNQDKGFPIELPNVDQMPAAFISRKDGLLLKENPQKTITFNATPKVLPTA-----SG 468
Query: 480 TKI-RFSQ-AITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASAS 537
TK+ RFS +T G +KPD+ APG +IL++
Sbjct: 469 TKLSRFSSWGLTADGN---------------------IKPDIAAPGQDILSS-------- 499
Query: 538 NLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLI----KTLHPNWSPA 579
+ +N+ + + GTSMS P VAG GL+ +T +P+ +P+
Sbjct: 500 --VANNK----YAKLSGTSMSAPLVAGIMGLLQKQYETQYPDMTPS 539
>SCA1_STRPY (P15926) C5a peptidase precursor (EC 3.4.21.-) (SCP)
Length = 1167
Score = 53.5 bits (127), Expect = 2e-06
Identities = 94/393 (23%), Positives = 158/393 (39%), Gaps = 66/393 (16%)
Query: 207 HGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAASCFGADVLSAI 266
HGTH GN A ++G P A++ +V + + + + + AI
Sbjct: 193 HGTHVSGILSGN----APSETKEPYRLEGAMPEAQLLLMRV--EIVNGLADYARNYAQAI 246
Query: 267 DQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNEGPTPGS-- 324
A++ G +I++S G + + DE +A ++ + +V SAGN+ G
Sbjct: 247 RDAVNLGAKVINMSFGNAALAYAN--LPDETKKAFDYAKSKGVSIVTSAGNDSSFGGKTR 304
Query: 325 ----------VVNV---APWVFTVAASTIDRDFSSTITIGDQIIRGASLFVDLPPNQSFT 371
VV A TVA+ + D+ + T + + + V L N+ F
Sbjct: 305 LPLADHPDYGVVGTPAAADSTLTVASYSPDKQLTETAMVKTDDQQDKEMPV-LSTNR-FE 362
Query: 372 LVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGM 431
+ D ++N ++ F VKGKI R G I + A AGA G+
Sbjct: 363 PNKAYDYAYANRGMKEDDF--------KDVKGKIALIER-GDIDFKDKVANAKKAGAVGV 413
Query: 432 FL-ENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITL 490
+ +NQ K P L V A I+ G+ D + I F+ +
Sbjct: 414 LIYDNQDK-------GFPIELPNVDQMPAAFISRKD--GLLLKDNPQK--TITFNATPKV 462
Query: 491 IGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFN 550
+ ++ FSS G +KPD+ APG +IL++ + +N+ +
Sbjct: 463 LPTASGTKLSRFSSWG--LTADGNIKPDIAAPGQDILSS----------VANNK----YA 506
Query: 551 VMQGTSMSCPHVAGTAGLI----KTLHPNWSPA 579
+ GTSMS P VAG GL+ +T +P+ +P+
Sbjct: 507 KLSGTSMSAPLVAGIMGLLQKQYETQYPDMTPS 539
>MS1P_HUMAN (Q14703) Membrane-bound transcription factor site-1
protease precursor (EC 3.4.21.-) (Site-1 protease)
(Subtilisin/kexin-isozyme-1) (SKI-1)
Length = 1052
Score = 52.4 bits (124), Expect = 3e-06
Identities = 56/216 (25%), Positives = 90/216 (40%), Gaps = 56/216 (25%)
Query: 126 WQKGRFGENTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGSSKKVPCNRKLIGARFFSD 185
WQ G G N +A DTG+ S+K P F +
Sbjct: 203 WQMGYTGANVRVAVFDTGL-----------------------SEKHP---------HFKN 230
Query: 186 AYERYNGKLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATY 245
ER N T++RT D +GHGT FV G +G +P A + +
Sbjct: 231 VKERTNW---TNERTLDDGLGHGT---------FVAGVIA---SMRECQGFAPDAELHIF 275
Query: 246 KVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHAL 305
+V T+ + + L A + AI +D++++S GGP + F D+V +
Sbjct: 276 RV---FTNNQVSYTSWFLDAFNYAILKKIDVLNLSIGGPDFMDHP--FVDKV----WELT 326
Query: 306 ARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTID 341
A N+++V++ GN+GP G++ N A + + ID
Sbjct: 327 ANNVIMVSAIGNDGPLYGTLNNPADQMDVIGVGGID 362
>MS1P_CRIGR (Q9Z2A8) Membrane-bound transcription factor site-1
protease precursor (EC 3.4.21.-) (Site-1 protease)
(Subtilisin/kexin-isozyme-1) (SKI-1) (Sterol-regulated
luminal protease)
Length = 1052
Score = 52.4 bits (124), Expect = 3e-06
Identities = 56/216 (25%), Positives = 90/216 (40%), Gaps = 56/216 (25%)
Query: 126 WQKGRFGENTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGSSKKVPCNRKLIGARFFSD 185
WQ G G N +A DTG+ S+K P F +
Sbjct: 203 WQMGYTGANVRVAVFDTGL-----------------------SEKHP---------HFKN 230
Query: 186 AYERYNGKLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATY 245
ER N T++RT D +GHGT FV G +G +P A + +
Sbjct: 231 VKERTNW---TNERTLDDGLGHGT---------FVAGVIA---SMRECQGFAPDAELHIF 275
Query: 246 KVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHAL 305
+V T+ + + L A + AI +D++++S GGP + F D+V +
Sbjct: 276 RV---FTNNQVSYTSWFLDAFNYAILKKIDVLNLSIGGPDFMDHP--FVDKV----WELT 326
Query: 306 ARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTID 341
A N+++V++ GN+GP G++ N A + + ID
Sbjct: 327 ANNVIMVSAIGNDGPLYGTLNNPADQMDVIGVGGID 362
>SUBT_BACS9 (P28842) Subtilisin precursor (EC 3.4.21.62)
Length = 420
Score = 51.6 bits (122), Expect = 6e-06
Identities = 49/169 (28%), Positives = 78/169 (45%), Gaps = 24/169 (14%)
Query: 197 SQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAAS 256
+ + D GHGTH AG G + GNG + G +P A + YKV L D S
Sbjct: 172 TNNSCTDRQGHGTHV---AGSALADGGT----GNG-VYGVAPDADLWAYKV---LGDDGS 220
Query: 257 CFGADVLSAIDQAIDDGVD-----IISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILL 311
+ D+ +AI A D +I++S G S+ + T+ V+ ++ + +L+
Sbjct: 221 GYADDIAAAIRHAGDQATALNTKVVINMSLG---SSGESSLITNAVN----YSYNKGVLI 273
Query: 312 VASAGNEGPTPGSVVNVAPWVFTVAASTIDRDF-SSTITIGDQIIRGAS 359
+A+AGN GP GS+ V VA + ++ + T + D RG S
Sbjct: 274 IAAAGNSGPYQGSIGYPGALVNAVAVAALENKVENGTYRVADFSSRGYS 322
>SUBD_BACLI (P00781) Subtilisin DY (EC 3.4.21.62)
Length = 274
Score = 50.8 bits (120), Expect = 1e-05
Identities = 53/212 (25%), Positives = 93/212 (43%), Gaps = 28/212 (13%)
Query: 206 GHGTHTLSTAGGNFVPGASIFNIGNGT-IKGGSPRARVATYKVCWSLTDAASCFGADVLS 264
GHGTH T + N T + G +P + KV L + S + ++S
Sbjct: 62 GHGTHVAGTVAA----------LDNTTGVLGVAPNVSLYAIKV---LNSSGSGTYSAIVS 108
Query: 265 AIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNEGP--TP 322
I+ A +G+D+I++S GGPS + + + D+ A A I++VA+AGN G +
Sbjct: 109 GIEWATQNGLDVINMSLGGPSGSTALKQAVDK-------AYASGIVVVAAAGNSGSSGSQ 161
Query: 323 GSVVNVAPWVFTVAASTIDRD-----FSSTITIGDQIIRGASLFVDLPPNQSFTLVNSID 377
++ A + +A +D + FSS + + G S++ P N +L +
Sbjct: 162 NTIGYPAKYDSVIAVGAVDSNKNRASFSSVGAELEVMAPGVSVYSTYPSNTYTSLNGTSM 221
Query: 378 AKFSNATTRDARFCRPRTLDPSKVKGKIVACA 409
A A + TL S+V+ ++ + A
Sbjct: 222 ASPHVAGAAALILSKYPTLSASQVRNRLSSTA 253
Score = 42.7 bits (99), Expect = 0.003
Identities = 43/185 (23%), Positives = 75/185 (40%), Gaps = 25/185 (13%)
Query: 404 KIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAIT 463
K++ + G ++ G E + + + SG+T L + + +G +
Sbjct: 93 KVLNSSGSGTYSAIVSGIEWATQNGLDVINMSLGGPSGSTALKQ--AVDKAYASGIVVVA 150
Query: 464 APPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPG 523
A G + + G ++ I + ASFSS G +V APG
Sbjct: 151 AAGNSGSSGSQNT-IGYPAKYDSVIAVGAVDSNKNRASFSSVGAEL--------EVMAPG 201
Query: 524 VNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKS 583
V++ + Y SN T + GTSM+ PHVAG A LI + +P S + +++
Sbjct: 202 VSVYSTYP-----SNTYTS---------LNGTSMASPHVAGAAALILSKYPTLSASQVRN 247
Query: 584 AIMTT 588
+ +T
Sbjct: 248 RLSST 252
>SUBT_BACPU (P07518) Subtilisin (EC 3.4.21.62) (Alkaline
mesentericopeptidase)
Length = 275
Score = 50.4 bits (119), Expect = 1e-05
Identities = 39/122 (31%), Positives = 54/122 (43%), Gaps = 25/122 (20%)
Query: 472 ATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYS 531
+T T+ G ++ I + A ASFSS G DV APGV+I +
Sbjct: 161 STSTV--GYPAKYPSTIAVGAVNSANQRASFSSAGSEL--------DVMAPGVSIQSTLP 210
Query: 532 LFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVIT 591
G + GTSM+ PHVAG A LI + HP W+ A ++ + +T T
Sbjct: 211 --------------GGTYGAYNGTSMATPHVAGAAALILSKHPTWTNAQVRDRLESTA-T 255
Query: 592 YL 593
YL
Sbjct: 256 YL 257
Score = 46.6 bits (109), Expect = 2e-04
Identities = 47/180 (26%), Positives = 82/180 (45%), Gaps = 28/180 (15%)
Query: 194 LPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTD 253
+P+ +D HGTH T N G + G +P + + KV L
Sbjct: 51 VPSETNPYQDGSSHGTHVAGTIAA--------LNNSIGVL-GVAPSSALYAVKV---LDS 98
Query: 254 AASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVA 313
S + +++ I+ AI + +D+I++S GGP+ + + + D+ A++ I++ A
Sbjct: 99 TGSGQYSWIINGIEWAISNNMDVINMSLGGPTGSTALKTVVDK-------AVSSGIVVAA 151
Query: 314 SAGNEGPTPGSVVNV---APWVFTVAASTID-----RDFSSTITIGDQIIRGASLFVDLP 365
+AGNEG + GS V A + T+A ++ FSS + D + G S+ LP
Sbjct: 152 AAGNEG-SSGSTSTVGYPAKYPSTIAVGAVNSANQRASFSSAGSELDVMAPGVSIQSTLP 210
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.134 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 71,262,691
Number of Sequences: 164201
Number of extensions: 3040841
Number of successful extensions: 7524
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 48
Number of HSP's that attempted gapping in prelim test: 7351
Number of HSP's gapped (non-prelim): 152
length of query: 609
length of database: 59,974,054
effective HSP length: 116
effective length of query: 493
effective length of database: 40,926,738
effective search space: 20176881834
effective search space used: 20176881834
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC133779.10


