

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133572.2 - phase: 0
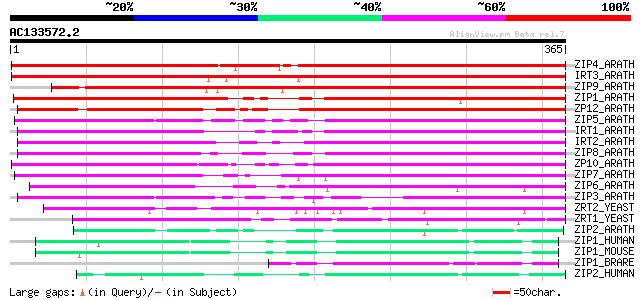
(365 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ZIP4_ARATH (O04089) Zinc transporter 4, chloroplast precursor (Z... 530 e-150
IRT3_ARATH (Q8LE59) Fe(II) transport protein 3, chloroplast prec... 524 e-148
ZIP9_ARATH (O82643) Zinc transporter 9 (ZRT/IRT-like protein 9) 405 e-113
ZIP1_ARATH (O81123) Zinc transporter 1 precursor (ZRT/IRT-like p... 292 8e-79
ZP12_ARATH (Q9FIS2) Probable zinc transporter 12 precursor (ZRT/... 288 2e-77
ZIP5_ARATH (O23039) Zinc transporter 5 precursor (ZRT/IRT-like p... 279 7e-75
IRT1_ARATH (Q38856) Fe(II) transport protein 1 precursor (Iron-r... 266 6e-71
IRT2_ARATH (O81850) Fe(II) transport protein 2 precursor (Iron-r... 262 1e-69
ZIP8_ARATH (Q8S3W4) Probable zinc transporter 8 precursor (ZRT/I... 260 4e-69
ZP10_ARATH (Q8W245) Probable zinc transporter 10 precursor (ZRT/... 256 6e-68
ZIP7_ARATH (Q8W246) Probable zinc transporter 7 precursor (ZRT/I... 237 3e-62
ZIP6_ARATH (O64738) Zinc transporter 6, chloroplast precursor (Z... 214 2e-55
ZIP3_ARATH (Q9SLG3) Zinc transporter 3 precursor (ZRT/IRT-like p... 199 7e-51
ZRT2_YEAST (Q12436) Zinc-regulated transporter 2 (Low-affinity z... 132 1e-30
ZRT1_YEAST (P32804) Zinc-regulated transporter 1 (High-affinity ... 120 5e-27
ZIP2_ARATH (Q9LTH9) Zinc transporter 2 precursor (ZRT/IRT-like p... 74 6e-13
ZIP1_HUMAN (Q9NY26) Zinc transporter ZIP1 (Zinc-iron regulated t... 67 9e-11
ZIP1_MOUSE (Q9QZ03) Zinc transporter ZIP1 (Zinc-iron regulated t... 60 7e-09
ZIP1_BRARE (P59889) Zinc transporter ZIP1 (DrZIP1) 60 1e-08
ZIP2_HUMAN (Q9NP94) Zinc transporter ZIP2 (Eti-1) (6A1) (hZIP2) 58 3e-08
>ZIP4_ARATH (O04089) Zinc transporter 4, chloroplast precursor
(ZRT/IRT-like protein 4)
Length = 374
Score = 530 bits (1364), Expect = e-150
Identities = 272/375 (72%), Positives = 309/375 (81%), Gaps = 17/375 (4%)
Query: 2 TNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAA 61
T C ESDLCRD+ AAF+LKF+A+ASILLAG AG+AIPLIG++RR+L+T+GNLFVAA
Sbjct: 6 TKILCDAGESDLCRDDSAAFLLKFVAIASILLAGAAGVAIPLIGRNRRFLQTEGNLFVAA 65
Query: 62 KAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQ 121
KAFAAGVILATGFVHML+ TEAL++PCLP+FPWSKFPF GFFAM+AAL TLL+DF+GTQ
Sbjct: 66 KAFAAGVILATGFVHMLAGGTEALSNPCLPDFPWSKFPFPGFFAMVAALATLLVDFMGTQ 125
Query: 122 YYERKQGMNRAVDEQARVGTSEEGNV---------GKVFGEEESGGMHIVGMHAHAAHHR 172
YYERKQ N+A E A G+ E V KVFGEE+ GG+HIVG+ AHAAHHR
Sbjct: 126 YYERKQERNQAATEAA-AGSEEIAVVPVVGERVTDNKVFGEEDGGGIHIVGIRAHAAHHR 184
Query: 173 HNHP--HGDACDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLS 230
H+H HG CDG HGH H H ++ E RHVVVSQ+LELGIVSHS+IIGLS
Sbjct: 185 HSHSNSHG-TCDGHA----HGHSHGHMHGNSDVENGARHVVVSQILELGIVSHSIIIGLS 239
Query: 231 LGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIG 290
LGVSQSPC IRPLIAALSFHQFFEGFALGGCISQAQF+ S TIMACFFALTTP+G+GIG
Sbjct: 240 LGVSQSPCTIRPLIAALSFHQFFEGFALGGCISQAQFRNKSATIMACFFALTTPLGIGIG 299
Query: 291 TGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCM 350
T +AS +N +SPGAL+ EGILD+LSAGILVYMALVDLIAADFLSKRMSCN RLQ+VSY M
Sbjct: 300 TAVASSFNSHSPGALVTEGILDSLSAGILVYMALVDLIAADFLSKRMSCNLRLQVVSYVM 359
Query: 351 LFLGAGLMSSLAIWA 365
LFLGAGLMS+LAIWA
Sbjct: 360 LFLGAGLMSALAIWA 374
>IRT3_ARATH (Q8LE59) Fe(II) transport protein 3, chloroplast
precursor (Iron-regulated transporter 3)
Length = 389
Score = 524 bits (1350), Expect = e-148
Identities = 268/384 (69%), Positives = 308/384 (79%), Gaps = 20/384 (5%)
Query: 2 TNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAA 61
+N C +ESDLCRD+ AAF+LKF+A+ASILLAG AG+ IPLIG++RR+L+TDGNLFV A
Sbjct: 6 SNVLCNASESDLCRDDSAAFLLKFVAIASILLAGAAGVTIPLIGRNRRFLQTDGNLFVTA 65
Query: 62 KAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQ 121
KAFAAGVILATGFVHML+ TEAL +PCLP+FPWSKFPF GFFAM+AAL TL +DF+GTQ
Sbjct: 66 KAFAAGVILATGFVHMLAGGTEALKNPCLPDFPWSKFPFPGFFAMIAALITLFVDFMGTQ 125
Query: 122 YYERKQGM--NRAVDEQARVGT--------SEEGNVGKVFGEEESGGMHIVGMHAHAAHH 171
YYERKQ + +V+ R + E N GKVFGEE+SGG+HIVG+HAHAAHH
Sbjct: 126 YYERKQEREASESVEPFGREQSPGIVVPMIGEGTNDGKVFGEEDSGGIHIVGIHAHAAHH 185
Query: 172 RHNHPHG-DACDGGGIVK---------EHGHDHSHALIAANEETDVRHVVVSQVLELGIV 221
RH+HP G D+C+G + HGH H H + RH+VVSQVLELGIV
Sbjct: 186 RHSHPPGHDSCEGHSKIDIGHAHAHGHGHGHGHGHVHGGLDAVNGARHIVVSQVLELGIV 245
Query: 222 SHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFAL 281
SHS+IIGLSLGVSQSPC IRPLIAALSFHQFFEGFALGGCISQAQF+ S TIMACFFAL
Sbjct: 246 SHSIIIGLSLGVSQSPCTIRPLIAALSFHQFFEGFALGGCISQAQFRNKSATIMACFFAL 305
Query: 282 TTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNF 341
TTPIG+GIGT +AS +N +S GAL+ EGILD+LSAGILVYMALVDLIAADFLS +M CNF
Sbjct: 306 TTPIGIGIGTAVASSFNSHSVGALVTEGILDSLSAGILVYMALVDLIAADFLSTKMRCNF 365
Query: 342 RLQLVSYCMLFLGAGLMSSLAIWA 365
RLQ+VSY MLFLGAGLMSSLAIWA
Sbjct: 366 RLQIVSYVMLFLGAGLMSSLAIWA 389
>ZIP9_ARATH (O82643) Zinc transporter 9 (ZRT/IRT-like protein 9)
Length = 344
Score = 405 bits (1042), Expect = e-113
Identities = 214/347 (61%), Positives = 259/347 (73%), Gaps = 12/347 (3%)
Query: 28 MASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFAAGVILATGFVHMLSDATEALNS 87
MASIL++G AG++IPL+G L +G L AKAFAAGVILATGFVHMLS ++AL+
Sbjct: 1 MASILISGAAGVSIPLVGT---LLPLNGGLMRGAKAFAAGVILATGFVHMLSGGSKALSD 57
Query: 88 PCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYERKQG--MNRAVDE---QARVGTS 142
PCLPEFPW FPF FFAM+AAL TLL DF+ T YYERKQ MN++V+ Q V +
Sbjct: 58 PCLPEFPWKMFPFPEFFAMVAALLTLLADFMITGYYERKQEKMMNQSVESLGTQVSVMSD 117
Query: 143 EEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHG----DACDGGGIVKEHGHDHSHAL 198
G + +E+ G +HIVGM AHA HHRH+ G +A V HGH HSH
Sbjct: 118 PGLESGFLRDQEDGGALHIVGMRAHAEHHRHSLSMGAEGFEALSKRSGVSGHGHGHSHGH 177
Query: 199 IAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFAL 258
++ VRHVVVSQ+LE+GIVSHS+IIG+SLGVS SPC IRPL+ ALSFHQFFEGFAL
Sbjct: 178 GDVGLDSGVRHVVVSQILEMGIVSHSIIIGISLGVSHSPCTIRPLLLALSFHQFFEGFAL 237
Query: 259 GGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGI 318
GGC+++A+ + +MA FFA+TTPIGV +GT IAS YN YS AL+AEG+LD+LSAGI
Sbjct: 238 GGCVAEARLTPRGSAMMAFFFAITTPIGVAVGTAIASSYNSYSVAALVAEGVLDSLSAGI 297
Query: 319 LVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
LVYMALVDLIAADFLSK+MS +FR+Q+VSYC LFLGAG+MS+LAIWA
Sbjct: 298 LVYMALVDLIAADFLSKKMSVDFRVQVVSYCFLFLGAGMMSALAIWA 344
>ZIP1_ARATH (O81123) Zinc transporter 1 precursor (ZRT/IRT-like
protein 1)
Length = 355
Score = 292 bits (748), Expect = 8e-79
Identities = 170/366 (46%), Positives = 225/366 (61%), Gaps = 42/366 (11%)
Query: 3 NSTCGGAESDLCRDEPA-AFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAA 61
+S C + + +DE A LK ++A +L+AG G+++PLIGK L+ + ++F
Sbjct: 29 SSDCTSHDDPVSQDEAEKATKLKLGSIALLLVAGGVGVSLPLIGKRIPALQPENDIFFMV 88
Query: 62 KAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQ 121
KAFAAGVIL TGFVH+L DA E L+SPCL + KFPF GF AM++A+ TL++D T
Sbjct: 89 KAFAAGVILCTGFVHILPDAFERLSSPCLEDTTAGKFPFAGFVAMLSAMGTLMIDTFATG 148
Query: 122 YYERKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDAC 181
YY+R+ N +Q V EE E +G +HI H HA+H
Sbjct: 149 YYKRQHFSNNHGSKQVNVVVDEE---------EHAGHVHI---HTHASH----------- 185
Query: 182 DGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIR 241
GH H + +R +VSQVLE+GIV HSVIIG+SLG SQS I+
Sbjct: 186 ---------GHTHGSTEL-------IRRRIVSQVLEIGIVVHSVIIGISLGASQSIDTIK 229
Query: 242 PLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIAS--VYNP 299
PL+AALSFHQFFEG LGGCIS A K+ ST +MA FF++T P+G+GIG G++S Y
Sbjct: 230 PLMAALSFHQFFEGLGLGGCISLADMKSKSTVLMATFFSVTAPLGIGIGLGMSSGLGYRK 289
Query: 300 YSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMS 359
S A++ EG+L+A SAGIL+YM+LVDL+A DF++ R+ N L L +Y L LGAG MS
Sbjct: 290 ESKEAIMVEGMLNAASAGILIYMSLVDLLATDFMNPRLQSNLWLHLAAYLSLVLGAGSMS 349
Query: 360 SLAIWA 365
LAIWA
Sbjct: 350 LLAIWA 355
>ZP12_ARATH (Q9FIS2) Probable zinc transporter 12 precursor
(ZRT/IRT-like protein 12)
Length = 355
Score = 288 bits (736), Expect = 2e-77
Identities = 163/362 (45%), Positives = 221/362 (61%), Gaps = 40/362 (11%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
CGG++ ++ +A K IA SIL+AG+ G+ +P+ G L+T+ N F+ KAFA
Sbjct: 32 CGGSKGGSAAEKASALKYKIIAFFSILIAGVFGVCLPIFG-----LKTESNFFMYVKAFA 86
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFP-WSKFPFTGFFAMMAALFTLLLDFVGTQYYE 124
AGVILATGFVH+L DATE+L S CL E P W FP TG AM A++ T+L++ + Y
Sbjct: 87 AGVILATGFVHILPDATESLTSSCLGEEPPWGDFPMTGLVAMAASILTMLIESFASGYLN 146
Query: 125 RKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGG 184
R + A+ G + + G GEEE H H HA+
Sbjct: 147 RSR--------LAKEGKTLPVSTG---GEEEHA--HTGSAHTHASQ-------------- 179
Query: 185 GIVKEHGHDHSHALIAANEE-TDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPL 243
GH H LI +++ D+R +V+Q+LELGIV HSVIIG+SLG S S I+PL
Sbjct: 180 ------GHSHGSLLIPQDDDHIDMRKKIVTQILELGIVVHSVIIGISLGASPSVSTIKPL 233
Query: 244 IAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPG 303
IAA++FHQ FEGF LGGCIS+A+F+ +M FFALT PIG+GIG G+A +YN SP
Sbjct: 234 IAAITFHQLFEGFGLGGCISEAKFRVKKIWVMLMFFALTAPIGIGIGIGVAEIYNENSPM 293
Query: 304 ALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAI 363
AL G L+A ++GIL+YMALVDL+A F++++ + ++Q+ L +GAGLMS LAI
Sbjct: 294 ALKVSGFLNATASGILIYMALVDLVAPLFMNQKTQSSMKIQVACSVSLVVGAGLMSLLAI 353
Query: 364 WA 365
WA
Sbjct: 354 WA 355
>ZIP5_ARATH (O23039) Zinc transporter 5 precursor (ZRT/IRT-like
protein 5)
Length = 360
Score = 279 bits (714), Expect = 7e-75
Identities = 160/362 (44%), Positives = 213/362 (58%), Gaps = 30/362 (8%)
Query: 4 STCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKA 63
S C + D ++ A K A+ S+L AG+ G+ PL+GK L+ + F KA
Sbjct: 29 SKCECSHEDDEANKAGAKKYKIAAIPSVLAAGVIGVMFPLLGKFFPSLKPETTFFFVTKA 88
Query: 64 FAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYY 123
FAAGVILATGF+H+L + E L SPCL W +FPFTGF AM+AA+ TL +D T Y+
Sbjct: 89 FAAGVILATGFMHVLPEGYEKLTSPCLKGEAW-EFPFTGFIAMVAAILTLSVDSFATSYF 147
Query: 124 ERKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDG 183
+ R+G EE + G GG +G+H HA H H HG
Sbjct: 148 HKAH-----FKTSKRIGDGEEQDAG-----GGGGGGDELGLHVHA----HGHTHGIV--- 190
Query: 184 GGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPL 243
G+ H R VV+QVLE+GI+ HSV+IG+SLG SQSP + L
Sbjct: 191 -GVESGESQVQLH-----------RTRVVAQVLEVGIIVHSVVIGISLGASQSPDTAKAL 238
Query: 244 IAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPG 303
AAL FHQ FEG LGGCI+Q F S TIM+ FF++TTP+G+ +G I+S Y+ SP
Sbjct: 239 FAALMFHQCFEGLGLGGCIAQGNFNCMSITIMSIFFSVTTPVGIAVGMAISSSYDDSSPT 298
Query: 304 ALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAI 363
ALI +G+L+A SAGIL+YM+LVD +AADF+ +M N RLQ++++ L +GAG+MS LA
Sbjct: 299 ALIVQGVLNAASAGILIYMSLVDFLAADFMHPKMQSNTRLQIMAHISLLVGAGVMSLLAK 358
Query: 364 WA 365
WA
Sbjct: 359 WA 360
>IRT1_ARATH (Q38856) Fe(II) transport protein 1 precursor
(Iron-regulated transporter 1)
Length = 339
Score = 266 bits (680), Expect = 6e-71
Identities = 149/360 (41%), Positives = 206/360 (56%), Gaps = 46/360 (12%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
CG ++ C ++ A LK IA+ IL+A M G+ PL ++ +L+ DGN+F K FA
Sbjct: 26 CGSESANPCVNKAKALPLKVIAIFVILIASMIGVGAPLFSRNVSFLQPDGNIFTIIKCFA 85
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
+G+IL TGF+H+L D+ E L+S CL E PW KFPF+GF AM++ L TL +D + T Y
Sbjct: 86 SGIILGTGFMHVLPDSFEMLSSICLEENPWHKFPFSGFLAMLSGLITLAIDSMATSLYTS 145
Query: 126 KQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGG 185
K VG+ H H H HG A D
Sbjct: 146 KNA---------------------------------VGIMPHG----HGHGHGPANDVTL 168
Query: 186 IVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIA 245
+KE D S+A + +R+ V++ VLELGI+ HSV+IGLSLG + C I+ LIA
Sbjct: 169 PIKED--DSSNAQL-------LRYRVIAMVLELGIIVHSVVIGLSLGATSDTCTIKGLIA 219
Query: 246 ALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGAL 305
AL FHQ FEG LGGCI QA++ +MA FFA+TTP G+ +G +++VY SP AL
Sbjct: 220 ALCFHQMFEGMGLGGCILQAEYTNMKKFVMAFFFAVTTPFGIALGIALSTVYQDNSPKAL 279
Query: 306 IAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
I G+L+A SAG+L+YMALVDL+AA+F+ ++ + ++Q LG G MS +A WA
Sbjct: 280 ITVGLLNACSAGLLIYMALVDLLAAEFMGPKLQGSIKMQFKCLIAALLGCGGMSIIAKWA 339
>IRT2_ARATH (O81850) Fe(II) transport protein 2 precursor
(Iron-regulated transporter 2)
Length = 350
Score = 262 bits (669), Expect = 1e-69
Identities = 140/360 (38%), Positives = 202/360 (55%), Gaps = 38/360 (10%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C + C ++ A LK +A+ +IL + G+ PL ++ +LR DGN F+ K F+
Sbjct: 29 CDSGFDNPCINKAKALPLKIVAIVAILTTSLIGVTSPLFSRYISFLRPDGNGFMIVKCFS 88
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
+G+IL TGF+H+L D+ E L+S CL + PW KFPF GF AMM+ L TL +D + T Y
Sbjct: 89 SGIILGTGFMHVLPDSFEMLSSKCLSDNPWHKFPFAGFVAMMSGLVTLAIDSITTSLYTG 148
Query: 126 KQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGG 185
K + DE+ +G ++ +H+VG HNH HG
Sbjct: 149 KNSVGPVPDEE--------------YGIDQEKAIHMVG---------HNHSHG------- 178
Query: 186 IVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIA 245
H L ++ +R+ V++ VLE+GI+ HSV+IGLSLG + C I+ LI
Sbjct: 179 --------HGVVLATKDDGQLLRYQVIAMVLEVGILFHSVVIGLSLGATNDSCTIKGLII 230
Query: 246 ALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGAL 305
AL FH FEG LGGCI QA F +MA FF TTP G+ +G ++S+Y SP AL
Sbjct: 231 ALCFHHLFEGIGLGGCILQADFTNVKKFLMAFFFTGTTPCGIFLGIALSSIYRDNSPTAL 290
Query: 306 IAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
I G+L+A SAG+L+YMALVDL+A +F+ + + +LQ+ + LG +MS +A+WA
Sbjct: 291 ITIGLLNACSAGMLIYMALVDLLATEFMGSMLQGSIKLQIKCFTAALLGCAVMSVVAVWA 350
>ZIP8_ARATH (Q8S3W4) Probable zinc transporter 8 precursor
(ZRT/IRT-like protein 8)
Length = 347
Score = 260 bits (664), Expect = 4e-69
Identities = 150/360 (41%), Positives = 204/360 (56%), Gaps = 45/360 (12%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C +D C D+ A LK +A+ +IL+ M G+A PL ++ +L DG +F+ K FA
Sbjct: 33 CETDSTDSCIDKTKALPLKIVAIVAILVTSMIGVAAPLFSRYVTFLHPDGKIFMIIKCFA 92
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
+G+IL TGF+H+L D+ E L+SPCL + PW KFPFTGF AM++ L TL +D + T Y +
Sbjct: 93 SGIILGTGFMHVLPDSFEMLSSPCLEDNPWHKFPFTGFVAMLSGLVTLAIDSIATSLYTK 152
Query: 126 KQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGG 185
K AV + + EE + M I H
Sbjct: 153 K-----AVADDS---------------EERTTPMIIQIDHLPLT---------------- 176
Query: 186 IVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIA 245
KE S L+ R+ V++ VLELGI+ HSV+IGLSLG + C I+ LIA
Sbjct: 177 -TKERSSTCSKQLL--------RYRVIATVLELGIIVHSVVIGLSLGATNDTCTIKGLIA 227
Query: 246 ALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGAL 305
AL FHQ FEG LGGCI QA++ +MA FFA+TTP G+ +G ++SVY SP AL
Sbjct: 228 ALCFHQMFEGMGLGGCILQAEYTNVKKFVMAFFFAVTTPSGIALGIALSSVYKDNSPTAL 287
Query: 306 IAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
I G+L+A SAG+L+YMALVDL+AA+F+ + + +LQL + LG G MS LA WA
Sbjct: 288 ITVGLLNACSAGLLIYMALVDLLAAEFMGSMLQRSVKLQLNCFGAALLGCGGMSVLAKWA 347
>ZP10_ARATH (Q8W245) Probable zinc transporter 10 precursor
(ZRT/IRT-like protein 10)
Length = 364
Score = 256 bits (654), Expect = 6e-68
Identities = 145/364 (39%), Positives = 214/364 (57%), Gaps = 29/364 (7%)
Query: 2 TNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAA 61
+N C + C D+ A LK +++ SIL+ + G+ +P + + + + F+
Sbjct: 30 SNKDCQSKSNYSCIDKNKALDLKLLSIFSILITSLIGVCLPFFARSIPAFQPEKSHFLIV 89
Query: 62 KAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQ 121
K+FA+G+IL+TGF+H+L D+ E L+SPCL + PW KFPF GF AMM+A+FTL++D + T
Sbjct: 90 KSFASGIILSTGFMHVLPDSFEMLSSPCLNDNPWHKFPFAGFVAMMSAVFTLMVDSITTS 149
Query: 122 YYERKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDAC 181
+ K G + A V T ++ +G V +H H H H PH
Sbjct: 150 VF-TKSGRKDLRADVASVETPDQ-EIGH------------VQVHGHV--HSHTLPH---- 189
Query: 182 DGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIR 241
HG + + +R+ +++ VLELGIV S++IGLS+G + + C I+
Sbjct: 190 ------NLHGENDKE---LGSYLQLLRYRILAIVLELGIVVQSIVIGLSVGDTNNTCTIK 240
Query: 242 PLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYS 301
L+AAL FHQ FEG LGGCI QA++ +MA FFA+TTP GV +G ++ Y S
Sbjct: 241 GLVAALCFHQMFEGMGLGGCILQAEYGWVKKAVMAFFFAVTTPFGVVLGMALSKTYKENS 300
Query: 302 PGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSL 361
P +LI G+L+A SAG+L+YMALVDL+AADF+ ++M + +LQL SY + LGAG MS +
Sbjct: 301 PESLITVGLLNASSAGLLIYMALVDLLAADFMGQKMQRSIKLQLKSYAAVLLGAGGMSVM 360
Query: 362 AIWA 365
A WA
Sbjct: 361 AKWA 364
>ZIP7_ARATH (Q8W246) Probable zinc transporter 7 precursor
(ZRT/IRT-like protein 7)
Length = 365
Score = 237 bits (605), Expect = 3e-62
Identities = 142/368 (38%), Positives = 203/368 (54%), Gaps = 43/368 (11%)
Query: 4 STCGGAESDL-CRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAK 62
S C DL C + A LK IA+ SIL+A M G+++PL + L D + V K
Sbjct: 35 SECKAESGDLSCHNNKEAQKLKIIAIPSILVASMIGVSLPLFSRSIPALGPDREMSVIVK 94
Query: 63 AFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQY 122
A+GVILATGF+H+L D+ + L S CLPE PW KFPF F M++AL L+++
Sbjct: 95 TLASGVILATGFMHVLPDSFDDLTSKCLPEDPWQKFPFATFITMISALLVLMIESFAMCA 154
Query: 123 YERKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACD 182
Y R+ + EG V + E+G ++ D
Sbjct: 155 YARRT-------------SKREGEVVPL----ENGS--------------------NSVD 177
Query: 183 GGGIVK--EHGHDHSHALIAANEETD---VRHVVVSQVLELGIVSHSVIIGLSLGVSQSP 237
++ E+G + NE+ +R+ V++Q+LELGIV HSV+IGL++G S +
Sbjct: 178 TQNDIQTLENGSSYVEKQEKVNEDKTSELLRNKVIAQILELGIVVHSVVIGLAMGASDNK 237
Query: 238 CAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVY 297
C ++ LIAAL FHQ FEG LGG I QAQFK+ + M FF++TTP G+ +G I +Y
Sbjct: 238 CTVQSLIAALCFHQLFEGMGLGGSILQAQFKSKTNWTMVFFFSVTTPFGIVLGMAIQKIY 297
Query: 298 NPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGL 357
+ SP ALI G+L+A SAG+L+YMALV+L+A +F ++ N +L ++ Y F GA
Sbjct: 298 DETSPTALIVVGVLNACSAGLLIYMALVNLLAHEFFGPKIQGNIKLHVLGYVATFTGAAG 357
Query: 358 MSSLAIWA 365
MS +A WA
Sbjct: 358 MSLMAKWA 365
>ZIP6_ARATH (O64738) Zinc transporter 6, chloroplast precursor
(ZRT/IRT-like protein 6)
Length = 341
Score = 214 bits (546), Expect = 2e-55
Identities = 141/364 (38%), Positives = 195/364 (52%), Gaps = 51/364 (14%)
Query: 14 CRDEPAAFVLKFIAMASILLAGMAGIAIP-LIGKHRRYLRTDGNLFVAAKAFAAGVILAT 72
CRD A LK +A+ +I L + G+ P L+ K+ + K FAAGVIL+T
Sbjct: 17 CRDGEEASHLKIVAVFAIFLTSVFGVWGPVLLAKYFHGKPLYDKAILVIKCFAAGVILST 76
Query: 73 GFVHMLSDATEAL-NSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYERKQGMNR 131
VH+L +A E+L + PW FPF G M+ A+ LL+D +++
Sbjct: 77 SLVHVLPEAFESLADCQVSSRHPWKDFPFAGLVTMIGAITALLVDLTASEH--------- 127
Query: 132 AVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGGIVKEHG 191
+G G GGM + P G A GG+ + G
Sbjct: 128 ---------------MGHGGGGGGDGGMEYM-------------PVGKAV--GGLEMKEG 157
Query: 192 HDHSHALIAANEETDV---RHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALS 248
+ I N E ++ + +VSQVLE+GI+ HSVIIG+++G+SQ+ C IRPLIAALS
Sbjct: 158 KCGADLEIQENSEEEIVKMKQRLVSQVLEIGIIFHSVIIGVTMGMSQNKCTIRPLIAALS 217
Query: 249 FHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGI--ASVYNPYSPGALI 306
FHQ FEG LGGCI+QA FKA + M FA+TTP+G+ +G I A+ Y+ +P ALI
Sbjct: 218 FHQIFEGLGLGGCIAQAGFKAGTVVYMCLMFAVTTPLGIVLGMVIFAATGYDDQNPNALI 277
Query: 307 AEGILDALSAGILVYMALVDLIAADFLSKRM-----SCNFRLQLVSYCMLFLGAGLMSSL 361
EG+L + S+GIL+YMALVDLIA DF +M RL+ + + L LG+ MS L
Sbjct: 278 MEGLLGSFSSGILIYMALVDLIALDFFHNKMLTTCGESGSRLKKLCFVALVLGSASMSLL 337
Query: 362 AIWA 365
A+WA
Sbjct: 338 ALWA 341
>ZIP3_ARATH (Q9SLG3) Zinc transporter 3 precursor (ZRT/IRT-like
protein 3)
Length = 339
Score = 199 bits (507), Expect = 7e-51
Identities = 137/364 (37%), Positives = 195/364 (52%), Gaps = 64/364 (17%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C + D ++ A K A+ ++L+AG+ G+ PL+GK LR + F KAFA
Sbjct: 36 CECSHEDDHENKAGARKYKIAAIPTVLIAGIIGVLFPLLGKVFPSLRPETCFFFVTKAFA 95
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLD-FVGTQYYE 124
AGVILATGF+H+L +A E LNSPCL W +FPFTGF AM+AA+ TL +D F + +Y+
Sbjct: 96 AGVILATGFMHVLPEAYEMLNSPCLTSEAW-EFPFTGFIAMIAAILTLSVDTFATSSFYK 154
Query: 125 RKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGG 184
++ V + G + E +V +S + I+ A + G
Sbjct: 155 SHCKASKRVSD----GETGESSV-------DSEKVQILRTRVIA----------QVLELG 193
Query: 185 GIVKEHGHDHSHAL---IAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIR 241
IV HS + + A++ D ++ L + ++ H GL L
Sbjct: 194 IIV------HSVVIGISLGASQSPD-----AAKALFIALMFHQCFEGLGL---------- 232
Query: 242 PLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYS 301
GGCI+Q +FK S TIM+ FFA+TTPIG+ +G GIA+ Y+ S
Sbjct: 233 -----------------GGCIAQGKFKCLSVTIMSTFFAITTPIGIVVGMGIANSYDESS 275
Query: 302 PGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSL 361
P ALI +G+L+A SAGIL+YM+LVDL+AADF +M N LQ++++ L LGAGLMS L
Sbjct: 276 PTALIVQGVLNAASAGILIYMSLVDLLAADFTHPKMQSNTGLQIMAHIALLLGAGLMSLL 335
Query: 362 AIWA 365
A WA
Sbjct: 336 AKWA 339
>ZRT2_YEAST (Q12436) Zinc-regulated transporter 2 (Low-affinity zinc
transport protein ZRT2)
Length = 422
Score = 132 bits (333), Expect = 1e-30
Identities = 119/414 (28%), Positives = 176/414 (41%), Gaps = 88/414 (21%)
Query: 23 LKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFAAGVILATGFVHMLSDAT 82
L+ +A+ IL++ G+ P++ ++R F AK F +GVI+AT FVH+L A
Sbjct: 26 LRILAVFIILISSGLGVYFPILSSRYSFIRLPNWCFFIAKFFGSGVIVATAFVHLLQPAA 85
Query: 83 EALNSPCL----PEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYERKQGMNRAVDEQAR 138
EAL CL E+PW+ F + +LF L + T Y+ V +
Sbjct: 86 EALGDECLGGTFAEYPWA------FGICLMSLFLLFFTEIITHYF---------VAKTLG 130
Query: 139 VGTSEEGNVGKVFGEEESGGMHI-------VGMHAHAAHHRHNHPHGDACDGGGIV---- 187
+ G V + + S G I V + AA+ HN IV
Sbjct: 131 HDHGDHGEVTSIDVDAPSSGFVIRNMDSDPVSFNNEAAYSIHNDKTPYTTRNEEIVATPI 190
Query: 188 --KEHGHD---------------------HSHALIAA-----------NEETDVRHVV-- 211
KE G + SHA A N+ DV +
Sbjct: 191 KEKEPGSNVTNYDLEPGKTESLANELVPTSSHATNLASVPGKDHYSHENDHQDVSQLATR 250
Query: 212 ---------VSQVL-----ELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFA 257
++Q+L E GI+ HSV +GLSL V+ L L+FHQ FEG
Sbjct: 251 IEEEDKEQYLNQILAVFILEFGIIFHSVFVGLSLSVAGE--EFETLFIVLTFHQMFEGLG 308
Query: 258 LGGCISQAQFKASS---TTIMACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILDAL 314
LG +++ + S +M F LT+PI V +G G+ + P S ALIA G+ D++
Sbjct: 309 LGTRVAETNWPESKKYMPWLMGLAFTLTSPIAVAVGIGVRHSWIPGSRRALIANGVFDSI 368
Query: 315 SAGILVYMALVDLIAADFLSKRM---SCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
S+GIL+Y LV+L+A +FL + L +Y ++ GA LM+ L WA
Sbjct: 369 SSGILIYTGLVELMAHEFLYSNQFKGPDGLKKMLSAYLIMCCGAALMALLGKWA 422
>ZRT1_YEAST (P32804) Zinc-regulated transporter 1 (High-affinity
zinc transport protein ZRT1)
Length = 376
Score = 120 bits (301), Expect = 5e-27
Identities = 97/332 (29%), Positives = 154/332 (46%), Gaps = 31/332 (9%)
Query: 42 PLIGKHRRYLRTDGNLFVAAKAFAAGVILATGFVHMLSDATEALN-SPCLPEFP-WSKFP 99
PLI + LR +++ AK F +GVI+AT F+H++ A A+ + C+ + W +
Sbjct: 68 PLISTKVKRLRIPLYVYLFAKYFGSGVIVATAFIHLMDPAYGAIGGTTCVGQTGNWGLYS 127
Query: 100 FTGFFAMMAALFTLLLDFVGTQYYERKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGM 159
+ + + FT L D + + ERK G++ T + G
Sbjct: 128 WCPAIMLTSLTFTFLTDLFSSVWVERKYGLSHDHTHDEIKDTVVRNTAAVSSENDNENGT 187
Query: 160 HIVGMHAHAAHHRHNHPHGDACDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELG 219
A+ +H N G+ D + + + + ++ LE G
Sbjct: 188 ------ANGSHDTKN----------GVEYYEDSDATSMDVVQSFQAQFYAFLI---LEFG 228
Query: 220 IVSHSVIIGLSLG-VSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTT---IM 275
++ HSV+IGL+LG V ++ P++ FHQ FEG +G +S +F S +
Sbjct: 229 VIFHSVMIGLNLGSVGDEFSSLYPVLV---FHQSFEGLGIGARLSAIEFPRSKRWWPWAL 285
Query: 276 ACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFL-- 333
+ LTTPI V IG G+ + Y S AL+ G+LDA+SAGIL+Y LV+L+A DF+
Sbjct: 286 CVAYGLTTPICVAIGLGVRTRYVSGSYTALVISGVLDAISAGILLYTGLVELLARDFIFN 345
Query: 334 SKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
+R L C LF GAG+M+ + WA
Sbjct: 346 PQRTKDLRELSFNVICTLF-GAGIMALIGKWA 376
>ZIP2_ARATH (Q9LTH9) Zinc transporter 2 precursor (ZRT/IRT-like
protein 2)
Length = 353
Score = 73.9 bits (180), Expect = 6e-13
Identities = 81/325 (24%), Positives = 131/325 (39%), Gaps = 52/325 (16%)
Query: 43 LIGKHRRYLRTDGNLFVAAKAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTG 102
L G + R + + + F+ G+ LAT +H LSDA E E+P++
Sbjct: 77 LAGVSPYFYRWNESFLLLGTQFSGGIFLATALIHFLSDANETFRGLKHKEYPYA------ 130
Query: 103 FFAMMAALFTL-LLDFVGTQYYERKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHI 161
F + AA + L +L V + N A VG S E + V +E G I
Sbjct: 131 -FMLAAAGYCLTMLADVAVAFVAAGSNNNHV---GASVGESREDDDVAV---KEEGRREI 183
Query: 162 VGMHAHAAHHRHNHPHGDACDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIV 221
+ G D S ALI + D + +L +
Sbjct: 184 ---------------------------KSGVDVSQALIRTSGFGD------TALLIFALC 210
Query: 222 SHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASS--TTIMACFF 279
HS+ G+++G+S + + +S H+ F A+G + + K T + + F
Sbjct: 211 FHSIFEGIAIGLSDTKSDAWRNLWTISLHKVFAAVAMGIALLKLIPKRPFFLTVVYSFAF 270
Query: 280 ALTTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSC 339
+++PIGVGIG GI + G I L+ G+ VY+A+ LI+ + R C
Sbjct: 271 GISSPIGVGIGIGINA--TSQGAGGDWTYAISMGLACGVFVYVAVNHLISKGY-KPREEC 327
Query: 340 NFRLQLVSYCMLFLGAGLMSSLAIW 364
F + + +FLG L+S + IW
Sbjct: 328 YFDKPIYKFIAVFLGVALLSVVMIW 352
>ZIP1_HUMAN (Q9NY26) Zinc transporter ZIP1 (Zinc-iron regulated
transporter-like) (CGI-08/CGI-71) (hZIP1)
Length = 324
Score = 66.6 bits (161), Expect = 9e-11
Identities = 69/349 (19%), Positives = 132/349 (37%), Gaps = 58/349 (16%)
Query: 18 PAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNL-----FVAAKAFAAGVILAT 72
P +K A+ +L+ + +P+ R +G+ FA GV LAT
Sbjct: 24 PVGLEVKLGALVLLLVLTLLCSLVPICVLRRPGANHEGSASRQKALSLVSCFAGGVFLAT 83
Query: 73 GFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYERKQGMNRA 132
+ +L D A++ +FP F M L+++ + T Y+ + G +
Sbjct: 84 CLLDLLPDYLAAIDEALAALHVTLQFPLQEFILAMGFFLVLVMEQI-TLAYKEQSGPSPL 142
Query: 133 VDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGGIVKEHGH 192
+ +A +GT G H H DG G+ + G
Sbjct: 143 EETRALLGTVNGG-----------------------PQHWH--------DGPGVPQASGA 171
Query: 193 DHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFHQF 252
+ + + A VL + HSV GL++G+ + L AL H+
Sbjct: 172 PATPSALRAC------------VLVFSLALHSVFEGLAVGLQRDRARAMELCLALLLHKG 219
Query: 253 FEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILD 312
+L + Q+ +A F+ TP+G+G+G +A P +A+ +L+
Sbjct: 220 ILAVSLSLRLLQSHLRAQVVAGCGILFSCMTPLGIGLGAALAESAGPLHQ---LAQSVLE 276
Query: 313 ALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSL 361
++AG +Y+ ++++ + S +++ +L G L++ L
Sbjct: 277 GMAAGTFLYITFLEILPQELASSEQ------RILKVILLLAGFALLTGL 319
>ZIP1_MOUSE (Q9QZ03) Zinc transporter ZIP1 (Zinc-iron regulated
transporter-like)
Length = 324
Score = 60.5 bits (145), Expect = 7e-09
Identities = 67/349 (19%), Positives = 128/349 (36%), Gaps = 58/349 (16%)
Query: 18 PAAFVLKFIAMASILLAGMAGIAIPLI-----GKHRRYLRTDGNLFVAAKAFAAGVILAT 72
P +K A+ +LL + +P+ G + + FA GV LAT
Sbjct: 24 PVGLEVKLGALVLLLLLTLICSLVPVCVLRRSGANHEASASGQKALSLVSCFAGGVFLAT 83
Query: 73 GFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYERKQGMNRA 132
+ +L D A++ +FP F M L+++ + T Y+ +
Sbjct: 84 CLLDLLPDYLAAIDEALEALHVTLQFPLQEFILAMGFFLVLVMEQI-TLAYKEQSSPPHP 142
Query: 133 VDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGGIVKEHGH 192
+ +A +GT G H H DG GI + G
Sbjct: 143 EETRALLGTVNGG-----------------------PQHWH--------DGPGIPQAGGT 171
Query: 193 DHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFHQF 252
+ + + A VL + HSV GL++G+ + L AL H+
Sbjct: 172 PAAPSALRAC------------VLVFSLALHSVFEGLAVGLQRDRARAMELCLALLLHKG 219
Query: 253 FEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILD 312
+L + Q+ + F+ TP+G+G+G +A P +A+ +L+
Sbjct: 220 ILAVSLSLRLLQSHLRVQVVAGCGILFSCMTPLGIGLGAALAESAGPLHQ---LAQSVLE 276
Query: 313 ALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSL 361
++AG +Y+ ++++ + + +++ +L G L++ L
Sbjct: 277 GMAAGTFLYITFLEILPQELATSEQ------RILKVILLLAGFALLTGL 319
>ZIP1_BRARE (P59889) Zinc transporter ZIP1 (DrZIP1)
Length = 302
Score = 59.7 bits (143), Expect = 1e-08
Identities = 47/191 (24%), Positives = 84/191 (43%), Gaps = 22/191 (11%)
Query: 171 HRHNHPHGDACDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLS 230
H H HP + +G G H H HA + S +L L + HSV GL+
Sbjct: 130 HAHGHPSVNDLEGSG---HHVHVDFHAHSSFR----------SFMLFLSLSLHSVFEGLA 176
Query: 231 LGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIG 290
+G+ + + + A+ H+ F+L + Q+ K + FA+ +P+G+GI
Sbjct: 177 IGLQTTNAKVLEICIAILVHKSIIVFSLSVKLVQSAVKPLWVVLYVTVFAIMSPLGIGI- 235
Query: 291 TGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCM 350
GI + G LI + +L+ L+AG +Y+ ++++ + S L+
Sbjct: 236 -GIVVIETERQAGGLI-QAVLEGLAAGTFIYITFLEILPHELNSSER------PLLKVLF 287
Query: 351 LFLGAGLMSSL 361
L G +M++L
Sbjct: 288 LLCGFSIMAAL 298
>ZIP2_HUMAN (Q9NP94) Zinc transporter ZIP2 (Eti-1) (6A1) (hZIP2)
Length = 309
Score = 58.2 bits (139), Expect = 3e-08
Identities = 68/338 (20%), Positives = 120/338 (35%), Gaps = 84/338 (24%)
Query: 45 GKHRRYLRTDGNLFVAAKAFAAGVILATGFVHMLSDATEAL-----------------NS 87
G HRR LR G +AGV L GF+HM ++A E + NS
Sbjct: 39 GHHRRVLRLLG-------CISAGVFLGAGFMHMTAEALEEIESQIQKFMVQNRSASERNS 91
Query: 88 PCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYERKQGMNRAVDEQARVGTSEEGNV 147
+ ++P+ + L+ + Q G
Sbjct: 92 SGDADSAHMEYPYGELIISLGFFLVFFLESLALQCCPGAAG------------------- 132
Query: 148 GKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGGIVKEHGHDHSHALIAANEETDV 207
G +EE GG HI +H+H GH S +
Sbjct: 133 GSTVQDEEWGGAHIFELHSH-----------------------GHLPSPS---------- 159
Query: 208 RHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQF 267
+ + + VL L + HSV GL++G+ + A L A+ H+ F +G +
Sbjct: 160 KGPLRALVLLLSLSFHSVFEGLAVGLQPTVAATVQLCLAVLAHKGLVVFGVGMRLVHLGT 219
Query: 268 KASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDL 327
+ AL +P+G+ +G + G +A+ +L+ ++AG +Y+ +++
Sbjct: 220 SSRWAVFSILLLALMSPLGLAVGLAVTG--GDSEGGRGLAQAVLEGVAAGTFLYVTFLEI 277
Query: 328 IAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
+ + S L + + G M+ +A+WA
Sbjct: 278 LPRELASP------EAPLAKWSCVAAGFAFMAFIALWA 309
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.139 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,716,273
Number of Sequences: 164201
Number of extensions: 1741514
Number of successful extensions: 7244
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 45
Number of HSP's that attempted gapping in prelim test: 6781
Number of HSP's gapped (non-prelim): 275
length of query: 365
length of database: 59,974,054
effective HSP length: 112
effective length of query: 253
effective length of database: 41,583,542
effective search space: 10520636126
effective search space used: 10520636126
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 66 (30.0 bits)
Medicago: description of AC133572.2


