

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133571.8 - phase: 0
(495 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
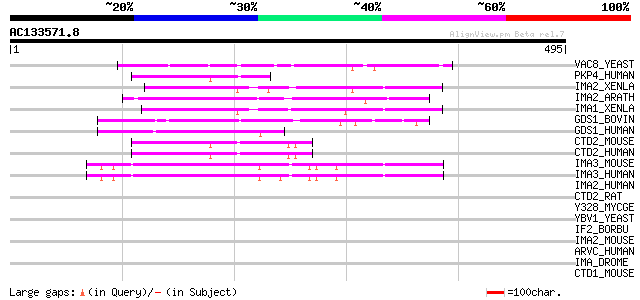
Score E
Sequences producing significant alignments: (bits) Value
VAC8_YEAST (P39968) Vacuolar protein 8 55 5e-07
PKP4_HUMAN (Q99569) Plakophilin 4 (p0071) 50 2e-05
IMA2_XENLA (P52171) Importin alpha-2 subunit (Karyopherin alpha-... 48 5e-05
IMA2_ARATH (O04294) Importin alpha-2 subunit (Karyopherin alpha-... 47 1e-04
IMA1_XENLA (P52170) Importin alpha-1 subunit (Karyopherin alpha-... 47 1e-04
GDS1_BOVIN (Q04173) Rap1 GTPase-GDP dissociation stimulator 1 (S... 47 1e-04
GDS1_HUMAN (P52306) Rap1 GTPase-GDP dissociation stimulator 1 (S... 46 2e-04
CTD2_MOUSE (O35927) Catenin delta-2 (Neural plakophilin-related ... 45 3e-04
CTD2_HUMAN (Q9UQB3) Catenin delta-2 (Delta-catenin) (Neural plak... 45 3e-04
IMA3_MOUSE (O35344) Importin alpha-3 subunit (Karyopherin alpha-... 45 4e-04
IMA3_HUMAN (O00505) Importin alpha-3 subunit (Karyopherin alpha-... 45 4e-04
IMA2_HUMAN (P52292) Importin alpha-2 subunit (Karyopherin alpha-... 44 0.001
CTD2_RAT (O35116) Catenin delta-2 (Fragment) 42 0.003
Y328_MYCGE (Q49419) Hypothetical protein MG328 41 0.008
YBV1_YEAST (P38260) Hypothetical 32.6 kDa protein in VPS15-YMC2 ... 40 0.011
IF2_BORBU (O51741) Translation initiation factor IF-2 40 0.018
IMA2_MOUSE (P52293) Importin alpha-2 subunit (Karyopherin alpha-... 39 0.024
ARVC_HUMAN (O00192) Armadillo repeat protein deleted in velo-car... 38 0.053
IMA_DROME (P52295) Importin alpha subunit (Karyopherin alpha sub... 38 0.069
CTD1_MOUSE (P30999) Catenin delta-1 (p120 catenin) (p120(ctn)) (... 38 0.069
>VAC8_YEAST (P39968) Vacuolar protein 8
Length = 578
Score = 54.7 bits (130), Expect = 5e-07
Identities = 72/303 (23%), Positives = 134/303 (43%), Gaps = 19/303 (6%)
Query: 97 NAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLV 156
N E+K + ++ ++ + G++ + A + +L D + +A GA+ PL
Sbjct: 115 NNENKLLIVEMGGLEPLINQMMGDNVEVQCNAVGCITNLATRDDN-KHKIATSGALIPLT 173
Query: 157 GMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIV 216
+ S+ + Q ++ ALLN+ ++ N+ +V GAV ++ L+ S D V
Sbjct: 174 KLAKSKHIRVQRNATGALLNM-THSEENRKELVNAGAVPVLVSLLSS---TDPDVQYYCT 229
Query: 217 ANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNI 276
++ ++N+ + + P LV L +L +S SS+VK A AL NL+ + +
Sbjct: 230 TALSNIAVDEANRKKLAQTE--PRLVSKLVSLMDS--PSSRVKCQATLALRNLASDTSYQ 285
Query: 277 SFVLETDLVVFLINSIEDMEVSERVLSI--LSNLVSSPEGRKAISAVKDA--ITVLVDVL 332
++ + L+ I+ + + S+ + N+ P + DA + LV +L
Sbjct: 286 LEIVRAGGLPHLVKLIQSDSIPLVLASVACIRNISIHPLNE---GLIVDAGFLKPLVRLL 342
Query: 333 NWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQKRASRILQC 392
++ DS E Q A L +A + +R+ E+G V EL L Q S I C
Sbjct: 343 DYKDSEEIQCHAVSTLRNLAASSEKNRKEFFESGAVEKCKELALDSPVSVQ---SEISAC 399
Query: 393 FRL 395
F +
Sbjct: 400 FAI 402
>PKP4_HUMAN (Q99569) Plakophilin 4 (p0071)
Length = 1211
Score = 49.7 bits (117), Expect = 2e-05
Identities = 37/126 (29%), Positives = 63/126 (49%), Gaps = 4/126 (3%)
Query: 109 EMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQI 168
E+ V+ L + + + AAA ++ L D++ + + LG I LV +LD L Q
Sbjct: 529 ELPEVIHMLEHQFPSVQANAAAYLQHLCFGDNKVKMEVCRLGGIKHLVDLLDHRVLEVQK 588
Query: 169 DSLYALLNL--GIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALD 226
++ AL NL G + D NK A+ +G + +L+L+ +D+ V E + LS+ D
Sbjct: 589 NACGALRNLVFGKSTDENKIAMKNVGGIPALLRLLRKS--IDAEVRELVTGVLWNLSSCD 646
Query: 227 SNKPII 232
+ K I
Sbjct: 647 AVKMTI 652
Score = 31.6 bits (70), Expect = 4.9
Identities = 25/117 (21%), Positives = 58/117 (49%), Gaps = 7/117 (5%)
Query: 155 LVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEA 214
++ ML+ + Q ++ L +L ++ K + ++G + ++ L++ + +
Sbjct: 533 VIHMLEHQFPSVQANAAAYLQHLCFGDNKVKMEVCRLGGIKHLVDLLDHRVLEVQKNACG 592
Query: 215 IVANFLGLSALDSNKPIIGSSGAIPFLVRIL-KNLDNSSKSSSQVKQDALRALYNLS 270
+ N + + D NK + + G IP L+R+L K++D ++V++ L+NLS
Sbjct: 593 ALRNLVFGKSTDENKIAMKNVGGIPALLRLLRKSID------AEVRELVTGVLWNLS 643
>IMA2_XENLA (P52171) Importin alpha-2 subunit (Karyopherin alpha-2
subunit)
Length = 522
Score = 48.1 bits (113), Expect = 5e-05
Identities = 63/275 (22%), Positives = 120/275 (42%), Gaps = 23/275 (8%)
Query: 121 DSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIA 180
+ST + AA + ++ S+ S+ GAI + ++ S LH +++AL N+
Sbjct: 129 NSTLQFEAAWALTNIASGTSDQTKSVVDGGAIPAFISLISSPHLHISEQAVWALGNIAGD 188
Query: 181 NDANKAAIVKIGAVHKMLKLI--ESPCVVDSSVSEAIVANFLGLSALDSNK----PIIGS 234
+ A++ + +L L+ ++P +++ LS L NK P+
Sbjct: 189 GPLYRDALISCNVIPPLLTLVNPQTPLGYLRNITWT-------LSNLCRNKNPYPPMSAV 241
Query: 235 SGAIPFLVRILKNLDNSSKSSSQVKQDALRAL-YNLSINQTNISFVLETDLVVFLINSIE 293
+P L +++ + D + D A+ Y + I V++T LV LI +
Sbjct: 242 LQILPVLTQLMLHED------KDILSDTCWAMSYLTDGSNDRIDVVVKTGLVERLIQLMY 295
Query: 294 DMEVSERVLSI--LSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIM 351
E+S S+ + N+V+ + + + ++VL +L P Q++A++ L +
Sbjct: 296 SPELSILTPSLRTVGNIVTGTDKQTQAAIDAGVLSVLPQLLR-HQKPSIQKEAAWALSNI 354
Query: 352 AHKAYADRQAMIEAGIVSSLLELTLVGTALAQKRA 386
A Q MI G++S L++L G AQK A
Sbjct: 355 AAGPAPQIQQMITCGLLSPLVDLLKKGDFKAQKEA 389
>IMA2_ARATH (O04294) Importin alpha-2 subunit (Karyopherin alpha-2
subunit) (KAP alpha)
Length = 531
Score = 47.0 bits (110), Expect = 1e-04
Identities = 64/280 (22%), Positives = 123/280 (43%), Gaps = 17/280 (6%)
Query: 101 KKKEETLTEMKHVVKDLRGEDSTKRRIAAARV-RSLTKEDSEARGSLAMLGAISP-LVGM 158
+++E+ L E++ K+ R E+ K+R ++ S T + + S L P +V
Sbjct: 27 RRREDNLVEIR---KNKREENLQKKRFTSSMAFGSATGQTEQDLSSANQLKDNLPAMVAG 83
Query: 159 LDSEDLHSQIDSLYALLNL-GIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVA 217
+ SED +SQ+++ L L I + +V+ G V +++K + A
Sbjct: 84 IWSEDSNSQLEATNLLRKLLSIEQNPPINEVVQSGVVPRVVKFLSRDDFPKLQFEAAWAL 143
Query: 218 NFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNI- 276
+ S N +I SGA+P +++L S +S V++ A+ AL N++ +
Sbjct: 144 TNIA-SGTSENTNVIIESGAVPIFIQLL------SSASEDVREQAVWALGNVAGDSPKCR 196
Query: 277 SFVLETDLVVFLINSIEDMEVSERVLSILSNLVSSPEGRK--AISAVKDAITVLVDVLNW 334
VL + L++ + + + L + G+ A + A+ VL ++
Sbjct: 197 DLVLSYGAMTPLLSQFNENTKLSMLRNATWTLSNFCRGKPPPAFEQTQPALPVLERLVQS 256
Query: 335 TDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLEL 374
D E A + L ++ + QA+IEAG+V L++L
Sbjct: 257 MDE-EVLTDACWALSYLSDNSNDKIQAVIEAGVVPRLIQL 295
>IMA1_XENLA (P52170) Importin alpha-1 subunit (Karyopherin alpha-1
subunit)
Length = 521
Score = 47.0 bits (110), Expect = 1e-04
Identities = 61/274 (22%), Positives = 118/274 (42%), Gaps = 15/274 (5%)
Query: 118 RGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNL 177
R ++ST + AA + ++ S+ S+ GAI + ++ S LH +++AL N+
Sbjct: 126 RHDNSTLQFEAAWALTNIASGTSDQTKSVVDGGAIPAFISLISSPHLHISEQAVWALGNI 185
Query: 178 GIANDANKAAIVKIGAVHKMLKLI--ESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSS 235
+ A++ + +L L+ ++P +++ LS L NK
Sbjct: 186 AGDGPLYRDALINCNVIPPLLALVNPQTPLGYLRNITWM-------LSNLCRNKNPYPPM 238
Query: 236 GAIPFLVRILKNLDNSSKSSSQVKQDALRAL-YNLSINQTNISFVLETDLVVFLINSIED 294
A+ ++ +L L + D A+ Y + I V++T +V LI +
Sbjct: 239 SAVLQILPVLTQL--MHHDDKDILSDTCWAMSYLTDGSNDRIDVVVKTGIVDRLIQLMYS 296
Query: 295 MEVS--ERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMA 352
E+S L + N+V+ + + + ++VL +L P Q++A++ + +A
Sbjct: 297 PELSIVTPSLRTVGNIVTGTDKQTQAAIDAGVLSVLPQLLR-HQKPSIQKEAAWAISNIA 355
Query: 353 HKAYADRQAMIEAGIVSSLLELTLVGTALAQKRA 386
Q MI G++S L++L G AQK A
Sbjct: 356 AGPAPQIQQMITCGLLSPLVDLLNKGDFKAQKEA 389
>GDS1_BOVIN (Q04173) Rap1 GTPase-GDP dissociation stimulator 1 (SMG
P21 stimulatory GDP/GTP exchange protein) (SMG GDS
protein) (Exchange factor smgGDS)
Length = 558
Score = 46.6 bits (109), Expect = 1e-04
Identities = 69/308 (22%), Positives = 134/308 (43%), Gaps = 23/308 (7%)
Query: 79 EEKTEKLLDLLNIQVHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKE 138
++ E LD L + + N E+ +K + ++ L + S ++A + + K
Sbjct: 20 DDSLEGCLDCLLQALTQNNMETSEKIQGSGILQVFASLLIPQSSCTAKVANV-IAEIAKN 78
Query: 139 DSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKML 198
+ R G ISPLV +L+S+D + + AL N+ + + +A ++ +G + ++
Sbjct: 79 EF-MRIPCVDAGLISPLVQLLNSKDQEVLLQTGRALGNICYDSHSLQAQLINMGVIPTLV 137
Query: 199 KLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQV 258
KL+ C ++++E + F L+ L+S+K S+ LV++ K K
Sbjct: 138 KLLGIHC-QKAALTEMCLVAFGNLAELESSKEQFASTNIAEELVKLFKKQIEHDK----- 191
Query: 259 KQDALRALYNLSINQTNISFVLETDLVVFLINSIE---DMEVSERVLSILSN-------L 308
++ L L+ N ++E+ LV L+ ++ D + E + + + L
Sbjct: 192 REMVFEVLAPLAENDAIKLQLVESGLVECLLEIVQQKVDSDKEEDIAELKTASDLMVLLL 251
Query: 309 VSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQA--MIEAG 366
+ +K K V VL+W S Q + + L I A+ A D M++ G
Sbjct: 252 LGDESMQKLFEGGKG--NVFQRVLSWIPSNNHQLQLAGALAI-ANFARNDGNCIHMVDNG 308
Query: 367 IVSSLLEL 374
IV L++L
Sbjct: 309 IVEKLMDL 316
>GDS1_HUMAN (P52306) Rap1 GTPase-GDP dissociation stimulator 1 (SMG
P21 stimulatory GDP/GTP exchange protein) (SMG GDS
protein) (Exchange factor smgGDS)
Length = 607
Score = 46.2 bits (108), Expect = 2e-04
Identities = 40/172 (23%), Positives = 80/172 (46%), Gaps = 7/172 (4%)
Query: 79 EEKTEKLLDLLNIQVHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKE 138
E+ E LD L + + N E+ +K + ++ L + S K ++A + + +
Sbjct: 20 EDSLEGCLDCLLQALAQNNTETSEKIQASGILQLFASLLTPQSSCKAKVA--NIIAEVAK 77
Query: 139 DSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKML 198
+ R G ISPLV +L+S+D + + AL N+ + ++A+ + G ++
Sbjct: 78 NEFMRIPCVDAGLISPLVQLLNSKDQEVLLQTGRALGNICYDSHEGRSAVDQAGGAQIVI 137
Query: 199 KLIESPCVVDSSVSEAIVANFLGL-----SALDSNKPIIGSSGAIPFLVRIL 245
+ S C + +E ++ F G+ + DS + + + G IP LV++L
Sbjct: 138 DHLRSLCSITDPANEKLLTVFCGMLMNYSNENDSLQAQLINMGVIPTLVKLL 189
Score = 37.7 bits (86), Expect = 0.069
Identities = 48/213 (22%), Positives = 94/213 (43%), Gaps = 21/213 (9%)
Query: 174 LLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIG 233
L+N ND+ +A ++ +G + ++KL+ C +++++E + F L+ L+S+K
Sbjct: 162 LMNYSNENDSLQAQLINMGVIPTLVKLLGIHC-QNAALTEMCLVAFGNLAELESSKEQFA 220
Query: 234 SSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSIE 293
S+ LV++ K K ++ L L+ N ++E LV L+ ++
Sbjct: 221 STNIAEELVKLFKKQIEHDK-----REMIFEVLAPLAENDAIKLQLVEAGLVECLLEIVQ 275
Query: 294 ---DMEVSERVLSILSN-------LVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEK 343
D + + + + + L+ +K K +V VL+W S Q +
Sbjct: 276 QKVDSDKEDDITELKTGSDLMVLLLLGDESMQKLFEGGKG--SVFQRVLSWIPSNNHQLQ 333
Query: 344 ASYILMIMAHKAYADRQA--MIEAGIVSSLLEL 374
+ L I A+ A D M++ GIV L++L
Sbjct: 334 LAGALAI-ANFARNDANCIHMVDNGIVEKLMDL 365
>CTD2_MOUSE (O35927) Catenin delta-2 (Neural plakophilin-related
ARM-repeat protein) (NPRAP) (Neurojungin)
Length = 1247
Score = 45.4 bits (106), Expect = 3e-04
Identities = 48/175 (27%), Positives = 81/175 (45%), Gaps = 15/175 (8%)
Query: 109 EMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQI 168
E+ V++ L+ + + + AAA ++ L D++ + + G I LV +LD
Sbjct: 548 ELPEVIQMLQHQFPSVQSNAAAYLQHLCFGDNKIKAEIRRQGGIQLLVDLLDHRMTEVHR 607
Query: 169 DSLYALLNL--GIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALD 226
+ AL NL G AND NK A+ G + +++L+ D + E + LS+ D
Sbjct: 608 SACGALRNLVYGKANDDNKIALKNCGGIPALVRLLRK--TTDLEIRELVTGVLWNLSSCD 665
Query: 227 SNK-PIIGSSGAIPFLVRILKN-------LDNSSK---SSSQVKQDALRALYNLS 270
+ K PII + A+ I+ + L + K SSQV ++A L N+S
Sbjct: 666 ALKMPIIQDALAVLTNAVIIPHSGWENSPLQDDRKIQLHSSQVLRNATGCLRNVS 720
>CTD2_HUMAN (Q9UQB3) Catenin delta-2 (Delta-catenin) (Neural
plakophilin-related ARM-repeat protein) (NPRAP)
(Neurojungin) (GT24)
Length = 1225
Score = 45.4 bits (106), Expect = 3e-04
Identities = 48/175 (27%), Positives = 81/175 (45%), Gaps = 15/175 (8%)
Query: 109 EMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQI 168
E+ V++ L+ + + + AAA ++ L D++ + + G I LV +LD
Sbjct: 551 ELPEVIQMLQHQFPSVQSNAAAYLQHLCFGDNKIKAEIRRQGGIQLLVDLLDHRMTEVHR 610
Query: 169 DSLYALLNL--GIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALD 226
+ AL NL G AND NK A+ G + +++L+ D + E + LS+ D
Sbjct: 611 SACGALRNLVYGKANDDNKIALKNCGGIPALVRLLRK--TTDLEIRELVTGVLWNLSSCD 668
Query: 227 SNK-PIIGSSGAIPFLVRILKN-------LDNSSK---SSSQVKQDALRALYNLS 270
+ K PII + A+ I+ + L + K SSQV ++A L N+S
Sbjct: 669 ALKMPIIQDALAVLTNAVIIPHSGWENSPLQDDRKIQLHSSQVLRNATGCLRNVS 723
>IMA3_MOUSE (O35344) Importin alpha-3 subunit (Karyopherin alpha-3
subunit) (Importin alpha Q2)
Length = 521
Score = 45.1 bits (105), Expect = 4e-04
Identities = 74/370 (20%), Positives = 150/370 (40%), Gaps = 55/370 (14%)
Query: 69 TVKELRGEVQEE-----KTEKLLDLLNI----QVHETNAESKKKEETLTEMKHVVKDLRG 119
T++ R EV E + E LL N+ + +++ ++ K + +T ++ ++++
Sbjct: 24 TMRRHRNEVTVELRKNKRDEHLLKKRNVPQEESLEDSDVDADFKAQNVT-LEAILQNATS 82
Query: 120 EDSTKRRIAAARVRSLTKEDSEAR-GSLAMLGAISPLVGMLDSEDLHS-QIDSLYALLNL 177
++ + A R L D L G + LV L+ +D S Q ++ +AL N+
Sbjct: 83 DNPVVQLSAVQAARKLLSSDRNPPIDDLIKSGILPILVKCLERDDNPSLQFEAAWALTNI 142
Query: 178 GIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLG---------------- 221
A A+V+ AV L+L+ SP + + N +G
Sbjct: 143 ASGTSAQTQAVVQSNAVPLFLRLLHSPHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVK 202
Query: 222 --LSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRAL----YNLSIN--- 272
LS ++ + PI +V + +N D + Q+ L AL Y+ IN
Sbjct: 203 PLLSFINPSIPITFLRNVTWVIVNLCRNKD--PPPPMETVQEILPALCVLIYHTDINILV 260
Query: 273 -------------QTNISFVLETDLVVFLIN--SIEDMEVSERVLSILSNLVSSPEGRKA 317
I V+++ +V FL+ S ++++V L + N+V+ + +
Sbjct: 261 DTVWALSYLTDGGNEQIQMVIDSGVVPFLVPLLSHQEVKVQTAALRAVGNIVTGTDEQTQ 320
Query: 318 ISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLV 377
+ D ++ ++L+ + ++A + L + QA+I+AG++ ++
Sbjct: 321 VVLNCDVLSHFPNLLS-HPKEKINKEAVWFLSNITAGNQQQVQAVIDAGLIPMIIHQLAK 379
Query: 378 GTALAQKRAS 387
G QK A+
Sbjct: 380 GDFGTQKEAA 389
>IMA3_HUMAN (O00505) Importin alpha-3 subunit (Karyopherin alpha-3
subunit) (SRP1-gamma)
Length = 521
Score = 45.1 bits (105), Expect = 4e-04
Identities = 76/370 (20%), Positives = 154/370 (41%), Gaps = 55/370 (14%)
Query: 69 TVKELRGEVQEE-----KTEKLLDLLNI----QVHETNAESKKKEETLTEMKHVVKDLRG 119
T++ R EV E + E LL N+ + +++ ++ K + +T ++ ++++
Sbjct: 24 TMRRHRNEVTVELRKNKRDEHLLKKRNVPQEESLEDSDVDADFKAQNVT-LEAILQNATS 82
Query: 120 EDSTKRRIAAARVRSLTKEDSEAR-GSLAMLGAISPLVGMLDSEDLHS-QIDSLYALLNL 177
++ + A R L D L G + LV L+ +D S Q ++ +AL N+
Sbjct: 83 DNPVVQLSAVQAARKLLSSDRNPPIDDLIKSGILPILVKCLERDDNPSLQFEAAWALTNI 142
Query: 178 GIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLG--------LSALDSNK 229
A A+V+ AV L+L+ SP + + N +G + +L K
Sbjct: 143 ASGTSAQTQAVVQSNAVPLFLRLLRSPHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVK 202
Query: 230 PIIG-SSGAIPF---------LVRILKNLDNSSKSSSQVKQDALRAL----YNLSIN--- 272
P++ S +IP +V + +N D + Q+ L AL Y+ IN
Sbjct: 203 PLLSFISPSIPITFLRNVTWVIVNLCRNKD--PPPPMETVQEILPALCVLIYHTDINILV 260
Query: 273 -------------QTNISFVLETDLVVFLIN--SIEDMEVSERVLSILSNLVSSPEGRKA 317
I V+++ +V FL+ S ++++V L + N+V+ + +
Sbjct: 261 DTVWALSYLTDGGNEQIQMVIDSGVVPFLVPLLSHQEVKVQTAALRAVGNIVTGTDEQTQ 320
Query: 318 ISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLV 377
+ D ++ ++L+ + ++A + L + QA+I+AG++ ++
Sbjct: 321 VVLNCDVLSHFPNLLS-HPKEKINKEAVWFLSNITAGNQQQVQAVIDAGLIPMIIHQLAK 379
Query: 378 GTALAQKRAS 387
G QK A+
Sbjct: 380 GDFGTQKEAA 389
>IMA2_HUMAN (P52292) Importin alpha-2 subunit (Karyopherin alpha-2
subunit) (SRP1-alpha) (RAG cohort protein 1)
Length = 529
Score = 43.9 bits (102), Expect = 0.001
Identities = 61/267 (22%), Positives = 113/267 (41%), Gaps = 20/267 (7%)
Query: 128 AAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAA 187
A + S T E ++A + GAI + +L S H +++AL N+ +
Sbjct: 143 ALTNIASGTSEQTKA---VVDGGAIPAFISLLASPHAHISEQAVWALGNIAGDGSVFRDL 199
Query: 188 IVKIGAVHKMLKLIESPCVVDSSVSEAIVANFL-GLSALDSNK----PIIGSSGAIPFLV 242
++K GAV +L L+ P + SS++ + N LS L NK PI +P LV
Sbjct: 200 VIKYGAVDPLLALLAVPDM--SSLACGYLRNLTWTLSNLCRNKNPAPPIDAVEQILPTLV 257
Query: 243 RILKNLDNSSKSSSQVKQDALRALYNLSIN-QTNISFVLETDLVVFLINSI--EDMEVSE 299
R+L + D +V D A+ L+ I V++T +V L+ + ++ +
Sbjct: 258 RLLHHDD------PEVLADTCWAISYLTDGPNERIGMVVKTGVVPQLVKLLGASELPIVT 311
Query: 300 RVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADR 359
L + N+V+ + + + A+ V +L Q++A++ + +
Sbjct: 312 PALRAIGNIVTGTDEQTQVVIDAGALAVFPSLLT-NPKTNIQKEATWTMSNITAGRQDQI 370
Query: 360 QAMIEAGIVSSLLELTLVGTALAQKRA 386
Q ++ G+V L+ + QK A
Sbjct: 371 QQVVNHGLVPFLVSVLSKADFKTQKEA 397
Score = 37.7 bits (86), Expect = 0.069
Identities = 78/383 (20%), Positives = 152/383 (39%), Gaps = 24/383 (6%)
Query: 48 ASRHHRHYHREEEISSVASTATVKELRGEVQE-EKTEKLLDLLNIQVHETNAESKKKEET 106
A+R HR ++ ++ + + E+ E+++ +K +++L N+ +A S +E
Sbjct: 11 AARLHRFKNKGKDSTEMRRRRI--EVNVELRKAKKDDQMLKRRNVSSFPDDATSPLQENR 68
Query: 107 LTE------MKHVVKDLRGEDSTKRRIAAARVRSL-TKEDSEARGSLAMLGAISPLVGML 159
+ + +VK + + + A R L ++E ++ G I V L
Sbjct: 69 NNQGTVNWSVDDIVKGINSSNVENQLQATQAARKLLSREKQPPIDNIIRAGLIPKFVSFL 128
Query: 160 DSEDLHS-QIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVAN 218
D Q +S +AL N+ A+V GA+ + L+ SP S + + N
Sbjct: 129 GRTDCSPIQFESAWALTNIASGTSEQTKAVVDGGAIPAFISLLASPHAHISEQAVWALGN 188
Query: 219 FLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQT---- 274
G ++ + ++ GA+ L+ +L D SS + ++ + L NL N+
Sbjct: 189 IAGDGSV--FRDLVIKYGAVDPLLALLAVPDMSSLACGYLR-NLTWTLSNLCRNKNPAPP 245
Query: 275 --NISFVLETDLVVFLINSIEDMEVSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVL 332
+ +L T +V L++ +D EV +S L P R + + LV +L
Sbjct: 246 IDAVEQILPT--LVRLLHH-DDPEVLADTCWAISYLTDGPNERIGMVVKTGVVPQLVKLL 302
Query: 333 NWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQKRASRILQC 392
++ P I I+ Q +I+AG ++ L QK A+ +
Sbjct: 303 GASELPIVTPALRAIGNIVT-GTDEQTQVVIDAGALAVFPSLLTNPKTNIQKEATWTMSN 361
Query: 393 FRLDKGKQVSRSCDGGNLGLTVS 415
+ Q+ + + G + VS
Sbjct: 362 ITAGRQDQIQQVVNHGLVPFLVS 384
>CTD2_RAT (O35116) Catenin delta-2 (Fragment)
Length = 264
Score = 42.4 bits (98), Expect = 0.003
Identities = 45/156 (28%), Positives = 71/156 (44%), Gaps = 15/156 (9%)
Query: 128 AAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNL--GIANDANK 185
AAA ++ L D++ + + G I LV +LD + AL NL G AND NK
Sbjct: 8 AAAYLQHLCFGDNKIKAEIRRQGGIQLLVDLLDHRMTEVHRSACGALRNLVYGKANDDNK 67
Query: 186 AAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNK-PIIGSSGAIPFLVRI 244
A+ G + +++L+ D + E + LS+ D+ K PII + A+ I
Sbjct: 68 IALKNCGGIPALVRLLRK--TTDLEIRELVTGVLWNLSSCDALKMPIIQDALAVLTNAVI 125
Query: 245 LKN-------LDNSSK---SSSQVKQDALRALYNLS 270
+ + L + K SSQV ++A L N+S
Sbjct: 126 IPHSGWENSPLQDDRKIQLHSSQVLRNATGCLRNVS 161
>Y328_MYCGE (Q49419) Hypothetical protein MG328
Length = 756
Score = 40.8 bits (94), Expect = 0.008
Identities = 65/279 (23%), Positives = 125/279 (44%), Gaps = 47/279 (16%)
Query: 73 LRGEVQEEKTEKLLDLLNIQVHETNAESKKKEETLTEMKH-VVKDLRGE-DSTKRRIAAA 130
L+ + +K ++L D N ++ NA+ KK+ + L E + + DL E D KRR++
Sbjct: 224 LKEQHPTKKVDELDDYNNKELLLENADLKKQIDDLKENNNDQIFDLEQEIDDLKRRLSEE 283
Query: 131 RVRSLTKED------SEARGSLAMLG----AISPLVGMLDSEDLHSQIDSLYALLNLGIA 180
+ + L + E R L AI+PL S++++ ++++L +
Sbjct: 284 KSKHLHTKKLQDDLLQENRDLYEQLQNKPVAINPL-----SDEVNEELENLKQ--EKALL 336
Query: 181 NDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPF 240
+D A K V + L L+ V+++ ++E + N L L+A ++N+ + F
Sbjct: 337 SDQLDALKNKSSNVQQQLALLP---VLNNQINE--LQNQL-LTAREANQRLDLVEQENDF 390
Query: 241 LVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSIEDMEVSER 300
L LK L +++ + K D L Y L ++ F + ++V ++
Sbjct: 391 LKNELKKLHDNTSNDENEKYDDLLNQYELLFDENETKF--------------DKLQVKQQ 436
Query: 301 VLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPE 339
L++ + +K ISA+K VL+D + WT S +
Sbjct: 437 ALNL--------DYQKTISALKHENDVLLDEIEWTRSKD 467
>YBV1_YEAST (P38260) Hypothetical 32.6 kDa protein in VPS15-YMC2
intergenic region
Length = 290
Score = 40.4 bits (93), Expect = 0.011
Identities = 50/207 (24%), Positives = 93/207 (44%), Gaps = 43/207 (20%)
Query: 136 TKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVH 195
TK++ +L+++G + + LDS++ + D+ L + IA+D K V+ A +
Sbjct: 98 TKDEELRAAALSIIG--TAVQNNLDSQNNFMKYDNGLRSL-IEIASDKTKPLDVRTKAFY 154
Query: 196 KMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSS 255
+ LI + +SE F L+ LD P++ DN++K
Sbjct: 155 ALSNLIRN----HKDISE----KFFKLNGLDCIAPVLS---------------DNTAKPK 191
Query: 256 SQVKQDALRALY--NLSINQTNISFVLETDLVVFLINSIED---MEVSERVLSILSNLVS 310
+++ AL Y ++ I++ IS + + ++ I + D + + +RVLS LS+L+S
Sbjct: 192 LKMRAIALLTAYLSSVKIDENIISVLRKDGVIESTIECLSDESNLNIIDRVLSFLSHLIS 251
Query: 311 S------------PEGRKAISAVKDAI 325
S EG K I +KD +
Sbjct: 252 SGIKFNEQELHKLNEGYKHIEPLKDRL 278
>IF2_BORBU (O51741) Translation initiation factor IF-2
Length = 882
Score = 39.7 bits (91), Expect = 0.018
Identities = 63/325 (19%), Positives = 137/325 (41%), Gaps = 41/325 (12%)
Query: 20 AKNTTAGNFRFSASSFRRIIFDALSCGGASRHHRHYHREEEISSVASTATVKELRGEVQE 79
AKN + + ++ SFRR+I + +S + A + KEL ++ E
Sbjct: 207 AKNNSQNKYTTTSMSFRRLIKTKVPAIVSS------------TPAADSENSKELNRKLGE 254
Query: 80 EKTEKLLDLLNIQVHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKED 139
+K ++ + + +S K+++ TE K + + + + K+R + + +
Sbjct: 255 KKKQQ----------QESQKSYKRKKAETESKTIEQKVFEQLQKKKR---ENLANPIPKS 301
Query: 140 SEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLK 199
+ GS+ + + L S DL +++ +L ++ + D++ A I+ V +
Sbjct: 302 IDIMGSITVSDLARKM--NLKSSDLIAKLMALGVMVTINEKIDSDTATIL----VEEYGS 355
Query: 200 LIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPF----LVRILKNLDNSSKSS 255
+ + D +V E V + S P+I G + L+ +L+N+D + S
Sbjct: 356 KVNVVSIYDETVIEEEVED---QSKRVEKPPVITIMGHVDHGKTKLLSVLQNIDINQTES 412
Query: 256 SQVKQDALRALYNLSINQTNISFVLETDLVVFLINSIEDMEVSERVLSILSNLVS-SPEG 314
+ Q Y + N I+F+ F + +V++ V+ ++S + P+
Sbjct: 413 GGITQHI--GAYTIVYNDREITFLDTPGHEAFTMMRSRGAQVTDIVVLVVSAIDGVMPQT 470
Query: 315 RKAISAVKDAITVLVDVLNWTDSPE 339
+AI+ K+A ++ +N D P+
Sbjct: 471 IEAINHAKEANVPIIVAINKIDLPD 495
>IMA2_MOUSE (P52293) Importin alpha-2 subunit (Karyopherin alpha-2
subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin)
(Pore targeting complex 58 kDa subunit) (PTAC58)
(Importin alpha P1)
Length = 529
Score = 39.3 bits (90), Expect = 0.024
Identities = 61/273 (22%), Positives = 110/273 (39%), Gaps = 20/273 (7%)
Query: 80 EKTEKLLDLLNIQVHETNAESKKKEETLTE------MKHVVKDLRGEDSTKRRIAAARVR 133
+K E++L N+ +A S +E + ++ +VK + + + A R
Sbjct: 42 KKDEQMLKRRNVSSFPDDATSPLQENRNNQGTVNWSVEDIVKGINSNNLESQLQATQAAR 101
Query: 134 SL-TKEDSEARGSLAMLGAISPLVGMLDSEDLHS-QIDSLYALLNLGIANDANKAAIVKI 191
L ++E ++ G I V L D Q +S +AL N+ A+V
Sbjct: 102 KLLSREKQPPIDNIIRAGLIPKFVSFLGKTDCSPIQFESAWALTNIASGTSEQTKAVVDG 161
Query: 192 GAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNS 251
GA+ + L+ SP S + + N G + + + ++ GAI L+ +L D S
Sbjct: 162 GAIPAFISLLASPHAHISEQAVWALGNIAGDGS--AFRDLVIKHGAIDPLLALLAVPDLS 219
Query: 252 SKSSSQVKQDALRALYNLSINQT------NISFVLETDLVVFLINSIEDMEVSERVLSIL 305
+ + ++ + L NL N+ + +L T +V L++ D EV +
Sbjct: 220 TLACGYLR-NLTWTLSNLCRNKNPAPPLDAVEQILPT--LVRLLHH-NDPEVLADSCWAI 275
Query: 306 SNLVSSPEGRKAISAVKDAITVLVDVLNWTDSP 338
S L P R + K + LV +L T+ P
Sbjct: 276 SYLTDGPNERIEMVVKKGVVPQLVKLLGATELP 308
Score = 37.7 bits (86), Expect = 0.069
Identities = 58/268 (21%), Positives = 114/268 (41%), Gaps = 20/268 (7%)
Query: 128 AAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAA 187
A + S T E ++A + GAI + +L S H +++AL N+ A +
Sbjct: 143 ALTNIASGTSEQTKA---VVDGGAIPAFISLLASPHAHISEQAVWALGNIAGDGSAFRDL 199
Query: 188 IVKIGAVHKMLKLIESPCVVDSSVSEAIVANFL-GLSALDSNK----PIIGSSGAIPFLV 242
++K GA+ +L L+ P + S+++ + N LS L NK P+ +P LV
Sbjct: 200 VIKHGAIDPLLALLAVPDL--STLACGYLRNLTWTLSNLCRNKNPAPPLDAVEQILPTLV 257
Query: 243 RILKNLDNSSKSSSQVKQDALRALYNLSIN-QTNISFVLETDLVVFLINSI--EDMEVSE 299
R+L + D +V D+ A+ L+ I V++ +V L+ + ++ +
Sbjct: 258 RLLHHND------PEVLADSCWAISYLTDGPNERIEMVVKKGVVPQLVKLLGATELPIVT 311
Query: 300 RVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADR 359
L + N+V+ + + A+ V +L Q++A++ + +
Sbjct: 312 PALRAIGNIVTGTDEQTQKVIDAGALAVFPSLLT-NPKTNIQKEATWTMSNITAGRQDQI 370
Query: 360 QAMIEAGIVSSLLELTLVGTALAQKRAS 387
Q ++ G+V L+ + QK A+
Sbjct: 371 QQVVNHGLVPFLVGVLSKADFKTQKEAA 398
>ARVC_HUMAN (O00192) Armadillo repeat protein deleted in
velo-cardio-facial syndrome
Length = 962
Score = 38.1 bits (87), Expect = 0.053
Identities = 35/129 (27%), Positives = 57/129 (44%), Gaps = 3/129 (2%)
Query: 109 EMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQI 168
E+ V+ LR + AAA ++ L E+ + + L + LV +LD +
Sbjct: 359 ELPEVLAMLRHPVDPVKANAAAYLQHLCFENEGVKRRVRQLRGLPLLVALLDHPRAEVRR 418
Query: 169 DSLYALLNLGIANDA-NKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDS 227
+ AL NL D NKAAI G V +++L+ + D+ V E + LS+ +
Sbjct: 419 RACGALRNLSYGRDTDNKAAIRDCGGVPALVRLLRA--ARDNEVRELVTGTLWNLSSYEP 476
Query: 228 NKPIIGSSG 236
K +I G
Sbjct: 477 LKMVIIDHG 485
>IMA_DROME (P52295) Importin alpha subunit (Karyopherin alpha
subunit) (Pendulin)
Length = 522
Score = 37.7 bits (86), Expect = 0.069
Identities = 73/356 (20%), Positives = 138/356 (38%), Gaps = 41/356 (11%)
Query: 72 ELRGEVQEEKTEKLLDLLNIQVHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAAR 131
ELR +E++ K ++ + + E + + +V + ED ++ +
Sbjct: 33 ELRKSKKEDQMFKRRNINDEDLTSPLKELNGQSPVQLSVDEIVAAMNSEDQERQFLGMQS 92
Query: 132 VRSLTKEDSEARGSLAMLGAISPLV--GMLDSEDLHSQIDSLYALLNLGIANDANKAAIV 189
R + + L + I P+ + ++ + Q ++ +AL N+ ++
Sbjct: 93 ARKMLSRERNPPIDLMIGHGIVPICIRFLQNTNNSMLQFEAAWALTNIASGTSDQTRCVI 152
Query: 190 KIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSA-----------LDSNKPIIGSSGAI 238
+ AV + L++S + + + + N G A +D P+I + +
Sbjct: 153 EHNAVPHFVALLQSKSMNLAEQAVWALGNIAGDGAAARDIVIHHNVIDGILPLINNETPL 212
Query: 239 PFL---VRILKNL---DNSSKSSSQVKQ------------------DALRAL-YNLSINQ 273
FL V ++ NL N S QVK+ DA AL Y +
Sbjct: 213 SFLRNIVWLMSNLCRNKNPSPPFDQVKRLLPVLSQLLLSQDIQVLADACWALSYVTDDDN 272
Query: 274 TNISFVLETDLVVFLINSIEDMEVSERVLSILS--NLVSSPEGRKAISAVKDAITVLVDV 331
T I V+++D V L+ ++ E S V ++ S N+V+ + + + + L +
Sbjct: 273 TKIQAVVDSDAVPRLVKLLQMDEPSIIVPALRSVGNIVTGTDQQTDVVIASGGLPRLGLL 332
Query: 332 LNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQKRAS 387
L S +E A + I A QA+I+AGI L + G AQK A+
Sbjct: 333 LQHNKSNIVKEAAWTVSNITAGNQ-KQIQAVIQAGIFQQLRTVLEKGDFKAQKEAA 387
Score = 31.6 bits (70), Expect = 4.9
Identities = 36/186 (19%), Positives = 87/186 (46%), Gaps = 17/186 (9%)
Query: 158 MLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLI--ESPCVVDSSVSEAI 215
+L S+D+ D+ +AL + ++ A+V AV +++KL+ + P ++ ++
Sbjct: 248 LLLSQDIQVLADACWALSYVTDDDNTKIQAVVDSDAVPRLVKLLQMDEPSIIVPALRS-- 305
Query: 216 VANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSI-NQT 274
V N ++ D ++ +SG +P L +L++ + S + ++A + N++ NQ
Sbjct: 306 VGNI--VTGTDQQTDVVIASGGLPRLGLLLQH------NKSNIVKEAAWTVSNITAGNQK 357
Query: 275 NISFVLETDLVVFLINSIE--DMEVSERVLSILSNLVSSPEGRKAISAVK--DAITVLVD 330
I V++ + L +E D + + ++N +S + + ++ + +D
Sbjct: 358 QIQAVIQAGIFQQLRTVLEKGDFKAQKEAAWAVTNTTTSGTPEQIVDLIEKYKILKPFID 417
Query: 331 VLNWTD 336
+L+ D
Sbjct: 418 LLDTKD 423
>CTD1_MOUSE (P30999) Catenin delta-1 (p120 catenin) (p120(ctn))
(Cadherin-associated Src substrate) (CAS) (p120(cas))
Length = 911
Score = 37.7 bits (86), Expect = 0.069
Identities = 30/103 (29%), Positives = 47/103 (45%), Gaps = 3/103 (2%)
Query: 128 AAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDA-NKA 186
AAA ++ L + + + +A L I LVG+LD + + AL N+ D NK
Sbjct: 386 AAAYLQHLCYRNDKVKTDVAKLKGIPILVGLLDHPKKEVHLGACGALKNISFGRDQDNKI 445
Query: 187 AIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNK 229
AI V +++L+ D ++E I LS+ DS K
Sbjct: 446 AIKNCDGVPALVRLLRK--ARDMDLTEVITGTLWNLSSHDSIK 486
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.129 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,387,751
Number of Sequences: 164201
Number of extensions: 1842542
Number of successful extensions: 7935
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 74
Number of HSP's that attempted gapping in prelim test: 7864
Number of HSP's gapped (non-prelim): 128
length of query: 495
length of database: 59,974,054
effective HSP length: 114
effective length of query: 381
effective length of database: 41,255,140
effective search space: 15718208340
effective search space used: 15718208340
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC133571.8


