

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133139.5 + phase: 0
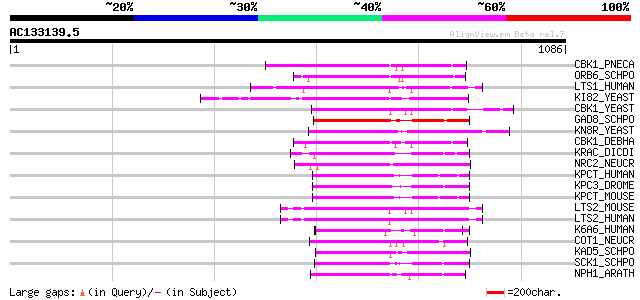
(1086 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CBK1_PNECA (Q6TGC6) Serine/threonine-protein kinase CBK1 (EC 2.7... 247 1e-64
ORB6_SCHPO (O13310) Serine/threonine-protein kinase orb6 (EC 2.7... 244 9e-64
LTS1_HUMAN (O95835) Serine/threonine protein kinase LATS1 (EC 2.... 241 1e-62
KI82_YEAST (P25341) Probable serine/threonine-protein kinase KIN... 240 2e-62
CBK1_YEAST (P53894) Serine/threonine-protein kinase CBK1 (EC 2.7... 238 5e-62
GAD8_SCHPO (Q9P7J8) Serine/threonine-protein kinase gad8 (EC 2.7... 237 1e-61
KN8R_YEAST (P53739) Probable serine/threonine-protein kinase YNR... 235 6e-61
CBK1_DEBHA (Q6BLJ9) Serine/threonine-protein kinase CBK1 (EC 2.7... 233 2e-60
KRAC_DICDI (P54644) RAC-family serine/threonine-protein kinase h... 230 2e-59
NRC2_NEUCR (O42626) Serine/threonine-protein kinase nrc-2 (EC 2.... 228 9e-59
KPCT_HUMAN (Q04759) Protein kinase C, theta type (EC 2.7.1.-) (n... 226 3e-58
KPC3_DROME (P13678) Protein kinase C (EC 2.7.1.-) (PKC) (dPKC98F) 224 7e-58
KPCT_MOUSE (Q02111) Protein kinase C, theta type (EC 2.7.1.-) (n... 223 2e-57
LTS2_MOUSE (Q7TSJ6) Serine/threonine protein kinase LATS2 (EC 2.... 223 3e-57
LTS2_HUMAN (Q9NRM7) Serine/threonine protein kinase LATS2 (EC 2.... 222 4e-57
K6A6_HUMAN (Q9UK32) Ribosomal protein S6 kinase alpha 6 (EC 2.7.... 220 2e-56
COT1_NEUCR (P38679) Serine/threonine-protein kinase cot-1 (EC 2.... 219 2e-56
KAD5_SCHPO (Q09831) Probable serine/threonine-protein kinase C4G... 218 9e-56
SCK1_SCHPO (P50530) Serine/threonine-protein kinase sck1 (EC 2.7... 212 4e-54
NPH1_ARATH (O48963) Nonphototropic hypocotyl protein 1 (EC 2.7.1... 209 2e-53
>CBK1_PNECA (Q6TGC6) Serine/threonine-protein kinase CBK1 (EC
2.7.1.37)
Length = 507
Score = 247 bits (630), Expect = 1e-64
Identities = 151/413 (36%), Positives = 220/413 (52%), Gaps = 21/413 (5%)
Query: 501 NSCDESAFQDIVDCVEDLRCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHDERAE 560
N S D V V D + E A V + K+ E+ + ++ D A+
Sbjct: 29 NKGTSSISYDPVYFVRDASSFTKTTHERAASVKLKLEHLYKVTVEQTVERNQRRMDFEAK 88
Query: 561 SSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRST 620
+ EE + S + + S+ DF +K I +GAFG V L QK T
Sbjct: 89 LAQDRGSEERKKRQLNSLGQKESQFLRLRRTKLSLNDFHTVKVIGKGAFGEVRLVQKIDT 148
Query: 621 GDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNG 680
G ++A+K L K++M +K+ + + AERD+L +P+VV YYSF + LYL+ME+L G
Sbjct: 149 GKIYAMKTLLKSEMFKKDQLAHVKAERDVLAESDSPWVVSLYYSFQDSQYLYLIMEFLPG 208
Query: 681 GDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDF 740
GDL +ML ED+ R YIAE +LA+E +H +HRD+KPDN+LI + GHIKL+DF
Sbjct: 209 GDLMTMLIKYDTFSEDVTRFYIAECILAIEAVHKLGFIHRDIKPDNILIDKTGHIKLSDF 268
Query: 741 GLSKVGLINSTEDL-----------SAPASFTNGFLVD-------DEPKPRHVSKREARQ 782
GLS +G + ++ ++ +S N +VD + K K
Sbjct: 269 GLS-MGFHKTHDNAYYQRLFESKINTSTSSTQNSLMVDTISLTMSSKDKIATWKKNRRIM 327
Query: 783 QQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIIN-R 841
S VGTPDY+APEI G+G DWWS+G I++E L+G PPF +++A + + IIN R
Sbjct: 328 AYSTVGTPDYIAPEIFTQHGYGQECDWWSLGAIMFECLIGWPPFCSENAHETYRKIINWR 387
Query: 842 DIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTL 894
+ + +S EA DL+ KLL + QRLG GA ++K H FF+ VNWDT+
Sbjct: 388 ENLYFPEDLHLSAEAEDLIRKLL-TSADQRLGRYGANDIKLHPFFRGVNWDTI 439
>ORB6_SCHPO (O13310) Serine/threonine-protein kinase orb6 (EC
2.7.1.37)
Length = 469
Score = 244 bits (623), Expect = 9e-64
Identities = 143/356 (40%), Positives = 203/356 (56%), Gaps = 21/356 (5%)
Query: 555 HDERAESSNSMADEESSVDDDTIRSLRAS-----PINGFSKDRTSIEDFEIIKPISRGAF 609
+ R +A E S ++ R LRAS F + R S+EDF IK I +GAF
Sbjct: 46 NQRRINLEQRLATERGS-EERKNRQLRASGEKESQFLRFRRTRLSLEDFSTIKVIGKGAF 104
Query: 610 GRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKE 669
G V L QK TG ++A+K L K +M +++ + + AERD+L+ +P+VV YY+F
Sbjct: 105 GEVRLVQKLDTGKIYAMKSLLKTEMFKRDQLAHVKAERDLLVESDSPWVVSLYYAFQDSL 164
Query: 670 NLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLI 729
LYL+ME+L GGDL +ML N ED+ R Y+AE VLA+ +H +HRD+KPDN+LI
Sbjct: 165 YLYLIMEFLPGGDLMTMLINYDTFSEDVTRFYMAECVLAIADVHRMGYIHRDIKPDNILI 224
Query: 730 GQDGHIKLTDFGLSKVGLINSTEDLSAPASFT-----NGFLVD-------DEPKPRHVSK 777
+DGHIKL+DFGLS G + S T G +VD + K K
Sbjct: 225 DRDGHIKLSDFGLS-TGFYKQDQSASYMKPRTGNTVKRGQMVDAIWLTMSSKDKMATWKK 283
Query: 778 REARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDN 837
S VGTPDY+APEI L G+G DWWS+G I++E L+G PPF ++++ + +
Sbjct: 284 NRRVMAYSTVGTPDYIAPEIFLQQGYGQDCDWWSLGAIMFECLIGWPPFCSENSHETYRK 343
Query: 838 IIN-RDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWD 892
IIN R+ + +S EA DLM++L+ ++ RLG GA E+ +H FF ++WD
Sbjct: 344 IINWRETLTFPNDIHLSIEARDLMDRLMTDSE-HRLGRGGAIEIMQHPFFTGIDWD 398
>LTS1_HUMAN (O95835) Serine/threonine protein kinase LATS1 (EC
2.7.1.37) (Large tumor suppressor homolog 1) (WARTS
protein kinase) (h-warts)
Length = 1130
Score = 241 bits (614), Expect = 1e-62
Identities = 172/493 (34%), Positives = 254/493 (50%), Gaps = 60/493 (12%)
Query: 471 SVSGRRPISELESYDQ--INKLVEIARAVANANSCDESAFQDIVDCVEDLRCVIQNRKED 528
SV IS+ DQ + K E ++ N +S D+ Q + V +N+K++
Sbjct: 569 SVPPYESISKPSKEDQPSLPKEDESEKSYENVDSGDKEKKQITTSPIT----VRKNKKDE 624
Query: 529 ALI---VDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVD-----DDTIRSL 580
+ ++ + K E+++ + H +R + +E V D +R +
Sbjct: 625 ERRESRIQSYSPQAFKFFMEQHVENVLKSHQQRLHRKKQLENEMMRVGLSQDAQDQMRKM 684
Query: 581 RASPINGFSKDRTSIED---FEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRK 637
+ + + + + D F IK + GAFG V LA+K T L+A K L+K D++ +
Sbjct: 685 LCQKESNYIRLKRAKMDKSMFVKIKTLGIGAFGEVCLARKVDTKALYATKTLRKKDVLLR 744
Query: 638 NAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDM 697
N V + AERDIL N +VVR YYSF K+NLY VM+Y+ GGD+ S+L +G E +
Sbjct: 745 NQVAHVKAERDILAEADNEWVVRLYYSFQDKDNLYFVMDYIPGGDMMSLLIRMGIFPESL 804
Query: 698 ARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGL--------------- 742
AR YIAE+ A+E +H +HRD+KPDN+LI +DGHIKLTDFGL
Sbjct: 805 ARFYIAELTCAVESVHKMGFIHRDIKPDNILIDRDGHIKLTDFGLCTGFRWTHDSKYYQS 864
Query: 743 ---SKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREARQQQ-----SIVGTPDYLA 794
+ ++ + + P+S G D KP + +R ARQ Q S+VGTP+Y+A
Sbjct: 865 GDHPRQDSMDFSNEWGDPSSCRCG----DRLKP--LERRAARQHQRCLAHSLVGTPNYIA 918
Query: 795 PEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPE-EIS 853
PE+LL G+ DWWSVGVIL+E+LVG PPF A + +IN P+ ++S
Sbjct: 919 PEVLLRTGYTQLCDWWSVGVILFEMLVGQPPFLAQTPLETQMKVINWQTSLHIPPQAKLS 978
Query: 854 FEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPY 913
EA DL+ K L P RLG GA E+K H FFK +++ + RQ+ A Y
Sbjct: 979 PEASDLIIK-LCRGPEDRLGKNGADEIKAHPFFKTIDFSSDLRQQ-----------SASY 1026
Query: 914 L-LISHTWDPTNF 925
+ I+H D +NF
Sbjct: 1027 IPKITHPTDTSNF 1039
>KI82_YEAST (P25341) Probable serine/threonine-protein kinase KIN82
(EC 2.7.1.37)
Length = 720
Score = 240 bits (612), Expect = 2e-62
Identities = 172/537 (32%), Positives = 271/537 (50%), Gaps = 47/537 (8%)
Query: 373 AETIEMLLDSLTPTSSLHEEFNELSL--ERNNMSSRCSEDMLDLAPDNTFVADDLNLSRE 430
A T E P S++H E + S+ E N S+ S L P+ + +S
Sbjct: 112 ASTSEKGTHKREPRSTIHTELLQSSIIGEPNVHSTTSST----LIPNEAICSTPNEISGS 167
Query: 431 ISCEAHSLKPDHGAKISSPESLTPRSPLITPRTSQIEMLLSVSGRRPISELESYDQINKL 490
S +A D SS TP SP + S+ LS I+E E ++
Sbjct: 168 SSPDAELFTFDMPTDPSSFH--TPSSPSYIAKDSRN---LSNGSLNDINENEELQNFHRK 222
Query: 491 VE---IARAVAN---ANSCDESAFQDIVDCVEDLRCVIQNRKEDALIVDTFGRRIEKLLQ 544
+ A +AN +NS +S ++ + + I R A + F + L++
Sbjct: 223 ISENGSASPLANLSLSNSPIDSPRKNSETRKDQIPMNITPRLRRAAS-EPFNTAKDGLMR 281
Query: 545 EKYLTLCEQIHDERAESSNSMADEESSVDDDTIRSLRASPI-NGFSKDRTSIEDFEIIKP 603
E Y+ L + S+ D V+ R LR N F + FE I+
Sbjct: 282 EDYIAL---------KQPPSLGD---IVEPRRSRRLRTKSFGNKFQDITVEPQSFEKIRL 329
Query: 604 ISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYY 663
+ +G G+V+L ++R T +FA+KVL K +MI++ ++ +L E++IL + +PF+V Y+
Sbjct: 330 LGQGDVGKVYLMRERDTNQIFALKVLNKHEMIKRKKIKRVLTEQEILATSDHPFIVTLYH 389
Query: 664 SFTCKENLYLVMEYLNGGDLYSML--RNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRD 721
SF K+ LYL MEY GG+ + L R C+ E+ A+ Y +EVV ALEYLH ++RD
Sbjct: 390 SFQTKDYLYLCMEYCMGGEFFRALQTRKSKCIAEEDAKFYASEVVAALEYLHLLGFIYRD 449
Query: 722 LKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREAR 781
LKP+N+L+ Q GH+ L+DF LS + S + +++ + + D
Sbjct: 450 LKPENILLHQSGHVMLSDFDLS-IQATGSKKPTMKDSTYLDTKICSD-----------GF 497
Query: 782 QQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINR 841
+ S VGT +YLAPE++ G GH DWW++G+++YE+L G PF D++ + F NI+ +
Sbjct: 498 RTNSFVGTEEYLAPEVIRGNGHTAAVDWWTLGILIYEMLFGCTPFKGDNSNETFSNILTK 557
Query: 842 DIQWPKHPEEISFEAYDLMNKLLIENPVQRLG-VTGATEVKRHAFFKDVNWDTLARQ 897
D+++P H +E+S DL+ KLL +N +RLG +GA ++KRH FFK V W L Q
Sbjct: 558 DVKFP-HDKEVSKNCKDLIKKLLNKNEAKRLGSKSGAADIKRHPFFKKVQWSFLRNQ 613
>CBK1_YEAST (P53894) Serine/threonine-protein kinase CBK1 (EC
2.7.1.37) (Cell wall biosynthesis kinase)
Length = 756
Score = 238 bits (608), Expect = 5e-62
Identities = 156/432 (36%), Positives = 228/432 (52%), Gaps = 71/432 (16%)
Query: 590 KDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDI 649
+ R S+EDF +K I +GAFG V L QK+ TG ++A+K L K++M +K+ + + AERD+
Sbjct: 344 RTRLSLEDFHTVKVIGKGAFGEVRLVQKKDTGKIYAMKTLLKSEMYKKDQLAHVKAERDV 403
Query: 650 LISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLAL 709
L +P+VV YYSF + LYL+ME+L GGDL +ML ED+ R Y+AE +LA+
Sbjct: 404 LAGSDSPWVVSLYYSFQDAQYLYLIMEFLPGGDLMTMLIRWQLFTEDVTRFYMAECILAI 463
Query: 710 EYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSK--------------VGLINSTEDLS 755
E +H +HRD+KPDN+LI GHIKL+DFGLS + +T +S
Sbjct: 464 ETIHKLGFIHRDIKPDNILIDIRGHIKLSDFGLSTGFHKTHDSNYYKKLLQQDEATNGIS 523
Query: 756 APASFTNGFLVDDEPKPR-----HVSKREARQQQ-------------SIVGTPDYLAPEI 797
P ++ N D K + +S + +QQ S VGTPDY+APEI
Sbjct: 524 KPGTY-NANTTDTANKRQTMVVDSISLTMSNRQQIQTWRKSRRLMAYSTVGTPDYIAPEI 582
Query: 798 LLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIIN--RDIQWPKHPEEISFE 855
L G+G DWWS+G I+YE L+G PPF ++ Q+ + I+N + +Q+P IS+E
Sbjct: 583 FLYQGYGQECDWWSLGAIMYECLIGWPPFCSETPQETYRKIMNFEQTLQFP-DDIHISYE 641
Query: 856 AYDLMNKLLIENPVQRLG-VTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYL 914
A DL+ +LL + QRLG GA E+K H FF+ V+W+T+ +
Sbjct: 642 AEDLIRRLL-THADQRLGRHGGADEIKSHPFFRGVDWNTIRQ------------------ 682
Query: 915 LISHTWDPTNFLQAMFIPS-AEAHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGS 973
++A +IP + DT +F + + NV D A + E +
Sbjct: 683 -----------VEAPYIPKLSSITDTRFFPTDELENVPDSPAMAQAA---KQREQMTKQG 728
Query: 974 GSDSLDEDAMFI 985
GS + ED FI
Sbjct: 729 GSAPVKEDLPFI 740
>GAD8_SCHPO (Q9P7J8) Serine/threonine-protein kinase gad8 (EC
2.7.1.37)
Length = 569
Score = 237 bits (604), Expect = 1e-61
Identities = 118/306 (38%), Positives = 187/306 (60%), Gaps = 33/306 (10%)
Query: 594 SIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISV 653
+I+ FE++K + +G+FG+V +KR T ++A+K +KKA ++ ++ V+ LAER +L V
Sbjct: 226 TIDAFELLKVVGKGSFGKVMQVRKRDTSRIYALKTMKKAHIVSRSEVDHTLAERTVLAQV 285
Query: 654 RNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLH 713
NPF+V +SF LYLV+ ++NGG+L+ L+ GC D A+ YIAE+++ALE LH
Sbjct: 286 NNPFIVPLKFSFQSPGKLYLVLAFVNGGELFHHLQREGCFDTYRAKFYIAELLVALECLH 345
Query: 714 SQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPR 773
++++RDLKP+N+L+ GHI L DFGL K+ + A TN F
Sbjct: 346 EFNVIYRDLKPENILLDYTGHIALCDFGLCKLNM--------AKTDRTNTF--------- 388
Query: 774 HVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQ 833
GTP+YLAPE+LLG G+ DWW++GV+LYE++ G+PPF ++ +
Sbjct: 389 -------------CGTPEYLAPELLLGHGYTKVVDWWTLGVLLYEMITGLPPFYDENINE 435
Query: 834 IFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDT 893
++ I+ +++P + +E +A DL++ LL P +RLG GA E+K H FF D++W
Sbjct: 436 MYRKILQDPLRFPDNIDE---KAKDLLSGLLTRAPEKRLGSGGAQEIKNHPFFDDIDWKK 492
Query: 894 LARQKV 899
L +K+
Sbjct: 493 LCAKKI 498
>KN8R_YEAST (P53739) Probable serine/threonine-protein kinase
YNR047W (EC 2.7.1.37)
Length = 893
Score = 235 bits (599), Expect = 6e-61
Identities = 136/396 (34%), Positives = 215/396 (53%), Gaps = 20/396 (5%)
Query: 586 NGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILA 645
N F + FE I+ + +G G+VFL +++ T ++A+KVL K +MI++N ++ +L
Sbjct: 484 NKFQDIMVGPQSFEKIRLLGQGDVGKVFLVREKKTNRVYALKVLSKDEMIKRNKIKRVLT 543
Query: 646 ERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSML--RNLGCLDEDMARVYIA 703
E++IL + +PF+V Y+SF ++ LYL MEY GG+ + L R C+ ED AR Y +
Sbjct: 544 EQEILATSNHPFIVTLYHSFQSEDYLYLCMEYCMGGEFFRALQTRKTKCICEDDARFYAS 603
Query: 704 EVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNG 763
EV ALEYLH ++RDLKP+N+L+ Q GHI L+DF LS + + A T
Sbjct: 604 EVTAALEYLHLLGFIYRDLKPENILLHQSGHIMLSDFDLSIQAKDSKVPVVKGSAQST-- 661
Query: 764 FLVDDEPKPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGI 823
LVD + + + S VGT +Y+APE++ G GH DWW++G+++YE+L G
Sbjct: 662 -LVDTK------ICSDGFRTNSFVGTEEYIAPEVIRGNGHTAAVDWWTLGILIYEMLFGF 714
Query: 824 PPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGV-TGATEVKR 882
PF D+ + F NI+ ++ +P + EIS DL+ KLL +N +RLG GA +VK+
Sbjct: 715 TPFKGDNTNETFTNILKNEVSFPNN-NEISRTCKDLIKKLLTKNESKRLGCKMGAADVKK 773
Query: 883 HAFFKDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPSAEAHDTSYF 942
H FFK V W L Q+ ++ + D + L S+ T+ + H
Sbjct: 774 HPFFKKVQWSLLRNQEPPLIPVLSEDGYDFAKLSSNKKRQTS-------QDSHKHLDEQE 826
Query: 943 MSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSL 978
+ + VE D+ + F+D + S ++S+
Sbjct: 827 KNMFEERVEYDDEVSEDDPFHDFNSMSLMEQDNNSM 862
>CBK1_DEBHA (Q6BLJ9) Serine/threonine-protein kinase CBK1 (EC
2.7.1.37)
Length = 716
Score = 233 bits (594), Expect = 2e-60
Identities = 146/387 (37%), Positives = 213/387 (54%), Gaps = 53/387 (13%)
Query: 555 HDERAESSNSMADEESSVDDDT----IRSL--RASPINGFSKDRTSIEDFEIIKPISRGA 608
+ R + + + +EES ++ +++L + S + + S+EDF +K I +GA
Sbjct: 266 NQRRLDLEHKLLNEESGSSEERKNRQLQNLGKKESQFLRLRRTKLSLEDFNTVKVIGKGA 325
Query: 609 FGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCK 668
FG V L QKR TG ++A+K L K++M +K+ + + AERD+L +P+VV YYSF
Sbjct: 326 FGEVRLVQKRDTGKIYAMKTLLKSEMYKKDQLAHVKAERDVLAGSDSPWVVSLYYSFQDA 385
Query: 669 ENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLL 728
+ LYL+ME+L GGDL +ML ED+ R Y+AE VLA+E +H +HRD+KPDN+L
Sbjct: 386 QYLYLIMEFLPGGDLMTMLIRWQIFTEDITRFYMAECVLAIEAIHKLGFIHRDIKPDNIL 445
Query: 729 IGQDGHIKLTDFGLSKVGLINSTED----------------------LSAPASFTNG--- 763
I GHIKL+DFGLS + T D L AP+ TN
Sbjct: 446 IDIRGHIKLSDFGLSTG--FHKTHDSNYYKKLLEKENPHHTNPQNGNLQAPSMATNNRNS 503
Query: 764 FLVDDEPKPRHVSKREARQQQ-----------SIVGTPDYLAPEILLGMGHGTTADWWSV 812
+VD H++ +Q Q S VGTPDY+APEI + G+G DWWS+
Sbjct: 504 MMVD----AIHLTMSNRQQMQTWRKSRRLMAYSTVGTPDYIAPEIFVHQGYGQECDWWSL 559
Query: 813 GVILYELLVGIPPFNADHAQQIFDNIIN--RDIQWPKHPEEISFEAYDLMNKLLIENPVQ 870
G I++E L+G PPF ++ + + I+N +Q P +S E+ DL+ KLL N
Sbjct: 560 GAIMFECLIGWPPFCSETPHETYRKILNWQETLQIP-DDIHLSPESEDLIRKLL-TNAEN 617
Query: 871 RLG-VTGATEVKRHAFFKDVNWDTLAR 896
RLG GA E+K H FF+ V+WDT+ +
Sbjct: 618 RLGRYNGADELKLHPFFRGVDWDTIRK 644
>KRAC_DICDI (P54644) RAC-family serine/threonine-protein kinase
homolog (EC 2.7.1.37)
Length = 444
Score = 230 bits (586), Expect = 2e-59
Identities = 123/354 (34%), Positives = 198/354 (55%), Gaps = 50/354 (14%)
Query: 550 LCEQIHDERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTS----IEDFEIIKPIS 605
LC + +ERA+ + +E + +NG + + S + DFE++ +
Sbjct: 80 LCSETEEERAKWIEILINERELL------------LNGGKQPKKSEKVGVADFELLNLVG 127
Query: 606 RGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSF 665
+G+FG+V +K+ TG+++A+KVL K ++ N VE L+ER+IL + +PF+V YSF
Sbjct: 128 KGSFGKVIQVRKKDTGEVYAMKVLSKKHIVEHNEVEHTLSERNILQKINHPFLVNLNYSF 187
Query: 666 TCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPD 725
++ LY +++Y+NGG+L+ L+ ED R Y AE+VLALE+LH +++RDLKP+
Sbjct: 188 QTEDKLYFILDYVNGGELFYHLQKDKKFTEDRVRYYGAEIVLALEHLHLSGVIYRDLKPE 247
Query: 726 NLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREARQQQS 785
NLL+ +GHI +TDFGL K GL+ T+ +
Sbjct: 248 NLLLTNEGHICMTDFGLCKEGLLTPTDKTG-----------------------------T 278
Query: 786 IVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQW 845
GTP+YLAPE+L G G+G DWWS G +LYE+L G+PPF Q+++ I+ + +
Sbjct: 279 FCGTPEYLAPEVLQGNGYGKQVDWWSFGSLLYEMLTGLPPFYNQDVQEMYRKIMMEKLSF 338
Query: 846 PKHPEEISFEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKV 899
P IS +A L+ +LL +P +RL +KRH FF+ ++W+ L ++ +
Sbjct: 339 PHF---ISPDARSLLEQLLERDPEKRL--ADPNLIKRHPFFRSIDWEQLFQKNI 387
>NRC2_NEUCR (O42626) Serine/threonine-protein kinase nrc-2 (EC
2.7.1.37) (Nonrepressible conidiation protein 2)
Length = 623
Score = 228 bits (580), Expect = 9e-59
Identities = 133/378 (35%), Positives = 206/378 (54%), Gaps = 37/378 (9%)
Query: 557 ERAESSNSMADEESSVDDDTIRSLRASPING------------------FSKDRTSIEDF 598
E ++ SN++ E ++D+ SL A +G +S + + +
Sbjct: 176 EESKDSNTVGFAEQKPNNDSSTSLAAPDADGLGALPPPIRQSPLAFRRTYSSNSIKVRNV 235
Query: 599 EI-------IKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILI 651
E+ IK I +G G+V+L +++ +G L+A+KVL K +MI++N ++ LAE++IL
Sbjct: 236 EVGPQSFDKIKLIGKGDVGKVYLVKEKKSGRLYAMKVLSKKEMIKRNKIKRALAEQEILA 295
Query: 652 SVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSML--RNLGCLDEDMARVYIAEVVLAL 709
+ +PF+V Y+SF ++ LYL MEY +GG+ + L R C+ ED AR Y AEV AL
Sbjct: 296 TSNHPFIVTLYHSFQSEDYLYLCMEYCSGGEFFRALQTRPGKCIPEDDARFYAAEVTAAL 355
Query: 710 EYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASF--TNGFLVD 767
EYLH ++RDLKP+N+L+ Q GHI L+DF LSK ++ P NG
Sbjct: 356 EYLHLMGFIYRDLKPENILLHQSGHIMLSDFDLSK-----QSDPGGKPTMIIGKNGTSTS 410
Query: 768 DEPKPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFN 827
P S + S VGT +Y+APE++ G GH + DWW++G+++YE+L G PF
Sbjct: 411 SLPTIDTKSCIANFRTNSFVGTEEYIAPEVIKGSGHTSAVDWWTLGILIYEMLYGTTPFK 470
Query: 828 ADHAQQIFDNIINRDIQWPKH--PEEISFEAYDLMNKLLIENPVQRLGV-TGATEVKRHA 884
+ F NI+ DI +P H +IS L+ KLLI++ +RLG GA+++K H
Sbjct: 471 GKNRNATFANILREDIPFPDHAGAPQISNLCKSLIRKLLIKDENRRLGARAGASDIKTHP 530
Query: 885 FFKDVNWDTLARQKVVIL 902
FF+ W + K I+
Sbjct: 531 FFRTTQWALIRHMKPPIV 548
>KPCT_HUMAN (Q04759) Protein kinase C, theta type (EC 2.7.1.-)
(nPKC-theta)
Length = 706
Score = 226 bits (575), Expect = 3e-58
Identities = 119/309 (38%), Positives = 185/309 (59%), Gaps = 36/309 (11%)
Query: 592 RTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDIL- 650
+ IEDF + K + +G+FG+VFLA+ + T FAIK LKK ++ + VE + E+ +L
Sbjct: 374 KLKIEDFILHKMLGKGSFGKVFLAEFKKTNQFFAIKALKKDVVLMDDDVECTMVEKRVLS 433
Query: 651 ISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALE 710
++ +PF+ + +F KENL+ VMEYLNGGDL +++ D A Y AE++L L+
Sbjct: 434 LAWEHPFLTHMFCTFQTKENLFFVMEYLNGGDLMYHIQSCHKFDLSRATFYAAEIILGLQ 493
Query: 711 YLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEP 770
+LHS+ IV+RDLK DN+L+ +DGHIK+ DFG+ K ++ + TN F
Sbjct: 494 FLHSKGIVYRDLKLDNILLDKDGHIKIADFGMCKENMLGDAK--------TNTF------ 539
Query: 771 KPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADH 830
GTPDY+APEILLG + + DWWS GV+LYE+L+G PF+
Sbjct: 540 ----------------CGTPDYIAPEILLGQKYNHSVDWWSFGVLLYEMLIGQSPFHGQD 583
Query: 831 AQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVN 890
+++F +I + P +P + EA DL+ KL + P +RLGV G ++++H F+++N
Sbjct: 584 EEELFHSI---RMDNPFYPRWLEKEAKDLLVKLFVREPEKRLGVRG--DIRQHPLFREIN 638
Query: 891 WDTLARQKV 899
W+ L R+++
Sbjct: 639 WEELERKEI 647
>KPC3_DROME (P13678) Protein kinase C (EC 2.7.1.-) (PKC) (dPKC98F)
Length = 634
Score = 224 bits (572), Expect = 7e-58
Identities = 115/310 (37%), Positives = 185/310 (59%), Gaps = 35/310 (11%)
Query: 592 RTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDIL- 650
+ S+ DF IK + +G+FG+V LA+K+ T +++AIKVLKK +I+ + V+ + E+ IL
Sbjct: 297 KCSLLDFNFIKVLGKGSFGKVMLAEKKGTDEIYAIKVLKKDAIIQDDDVDCTMTEKRILA 356
Query: 651 ISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALE 710
++ +PF+ + F + L+ VMEY+NGGDL ++ + A Y AEV LAL+
Sbjct: 357 LAANHPFLTALHSCFQTPDRLFFVMEYVNGGDLMFQIQKARRFEASRAAFYAAEVTLALQ 416
Query: 711 YLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEP 770
+LH+ +++RDLK DN+L+ Q+GH KL DFG+ K G++N G L
Sbjct: 417 FLHTHGVIYRDLKLDNILLDQEGHCKLADFGMCKEGIMN-------------GMLTT--- 460
Query: 771 KPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADH 830
+ GTPDY+APEIL +G + DWW++GV++YE++ G PPF AD+
Sbjct: 461 --------------TFCGTPDYIAPEILKEQEYGASVDWWALGVLMYEMMAGQPPFEADN 506
Query: 831 AQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTG-ATEVKRHAFFKDV 889
++FD+I++ D+ +P +S EA ++ L +NP QRLG TG E+++H FF +
Sbjct: 507 EDELFDSIMHDDVLYPVW---LSREAVSILKGFLTKNPEQRLGCTGDENEIRKHPFFAKL 563
Query: 890 NWDTLARQKV 899
+W L ++ +
Sbjct: 564 DWKELEKRNI 573
>KPCT_MOUSE (Q02111) Protein kinase C, theta type (EC 2.7.1.-)
(nPKC-theta)
Length = 707
Score = 223 bits (569), Expect = 2e-57
Identities = 118/309 (38%), Positives = 185/309 (59%), Gaps = 36/309 (11%)
Query: 592 RTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDIL- 650
+ I+DF + K + +G+FG+VFLA+ + T FAIK LKK ++ + VE + E+ +L
Sbjct: 374 KLKIDDFILHKMLGKGSFGKVFLAEFKRTNQFFAIKALKKDVVLMDDDVECTMVEKRVLS 433
Query: 651 ISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALE 710
++ +PF+ + +F KENL+ VMEYLNGGDL +++ D A Y AEV+L L+
Sbjct: 434 LAWEHPFLTHMFCTFQTKENLFFVMEYLNGGDLMYHIQSCHKFDLSRATFYAAEVILGLQ 493
Query: 711 YLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEP 770
+LHS+ IV+RDLK DN+L+ +DGHIK+ DFG+ K ++ + TN F
Sbjct: 494 FLHSKGIVYRDLKLDNILLDRDGHIKIADFGMCKENMLGDAK--------TNTF------ 539
Query: 771 KPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADH 830
GTPDY+APEILLG + + DWWS GV++YE+L+G PF+
Sbjct: 540 ----------------CGTPDYIAPEILLGQKYNHSVDWWSFGVLVYEMLIGQSPFHGQD 583
Query: 831 AQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVN 890
+++F +I + P +P + EA DL+ KL + P +RLGV G ++++H F+++N
Sbjct: 584 EEELFHSI---RMDNPFYPRWLEREAKDLLVKLFVREPEKRLGVRG--DIRQHPLFREIN 638
Query: 891 WDTLARQKV 899
W+ L R+++
Sbjct: 639 WEELERKEI 647
>LTS2_MOUSE (Q7TSJ6) Serine/threonine protein kinase LATS2 (EC
2.7.1.37) (Large tumor suppressor homolog 2)
(Serine/threonine kinase kpm) (Kinase phosphorylated
during mitosis protein)
Length = 1042
Score = 223 bits (567), Expect = 3e-57
Identities = 152/415 (36%), Positives = 218/415 (51%), Gaps = 45/415 (10%)
Query: 531 IVDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDDDTIRSLRASPINGFSK 590
++ T+ +++ + LQ + E A++ A++E I + S N +
Sbjct: 571 VIKTYQQKVSRRLQ---------LEQEMAKAGLCEAEQEQM---RKILYQKESNYNRLKR 618
Query: 591 DRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDIL 650
+ F IK + GAFG V LA K T L+A+K L+K D++ +N V + AERDIL
Sbjct: 619 AKMDKSMFVKIKTLGIGAFGEVCLACKLDTHALYAMKTLRKKDVLNRNQVAHVKAERDIL 678
Query: 651 ISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALE 710
N +VV+ YYSF K++LY VM+Y+ GGD+ S+L + E +AR YIAE+ LA+E
Sbjct: 679 AEADNEWVVKLYYSFQDKDSLYFVMDYIPGGDMMSLLIRMEVFPEHLARFYIAELTLAIE 738
Query: 711 YLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGL------SKVGLINSTEDLSAPASFTNGF 764
+H +HRD+KPDN+LI DGHIKLTDFGL + + S G
Sbjct: 739 SVHKMGFIHRDIKPDNILIDLDGHIKLTDFGLCTGFRWTHNSKYYQKGNHMRQDSMEPGD 798
Query: 765 LVDDEPKPR------HVSKREARQQQ-----SIVGTPDYLAPEILLGMGHGTTADWWSVG 813
L DD R + +R +Q Q S+VGTP+Y+APE+LL G+ DWWSVG
Sbjct: 799 LWDDVSNCRCGDRLKTLEQRAQKQHQRCLAHSLVGTPNYIAPEVLLRKGYTQLCDWWSVG 858
Query: 814 VILYELLVGIPPFNADHAQQIFDNIINRD--IQWPKHPEEISFEAYDLMNKLLIENPVQR 871
VIL+E+LVG PPF A + +IN + + P +S EA DL+ KL R
Sbjct: 859 VILFEMLVGQPPFLAPTPTETQLKVINWESTLHIPTQ-VRLSAEARDLITKLCCAADC-R 916
Query: 872 LGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYL-LISHTWDPTNF 925
LG GA ++K H FF +++ R++ APY+ ISH D +NF
Sbjct: 917 LGRDGADDLKAHPFFNTIDFSRDIRKQP-----------APYVPTISHPMDTSNF 960
>LTS2_HUMAN (Q9NRM7) Serine/threonine protein kinase LATS2 (EC
2.7.1.37) (Large tumor suppressor homolog 2)
(Serine/threonine kinase kpm) (Kinase phosphorylated
during mitosis protein) (Warts-like kinase)
Length = 1088
Score = 222 bits (566), Expect = 4e-57
Identities = 152/415 (36%), Positives = 220/415 (52%), Gaps = 45/415 (10%)
Query: 531 IVDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDDDTIRSLRASPINGFSK 590
++ T+ +++ + LQ + E A++ A++E I + S N +
Sbjct: 613 VIKTYQQKVNRRLQ---------LEQEMAKAGLCEAEQEQM---RKILYQKESNYNRLKR 660
Query: 591 DRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDIL 650
+ F IK + GAFG V LA K T L+A+K L+K D++ +N V + AERDIL
Sbjct: 661 AKMDKSMFVKIKTLGIGAFGEVCLACKVDTHALYAMKTLRKKDVLNRNQVVHVKAERDIL 720
Query: 651 ISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALE 710
N +VV+ YYSF K++LY VM+Y+ GGD+ S+L + E +AR YIAE+ LA+E
Sbjct: 721 AEADNEWVVKLYYSFQDKDSLYFVMDYIPGGDMMSLLIRMEVFPEHLARFYIAELTLAIE 780
Query: 711 YLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGL---------SKVGLINS--TEDLSAPAS 759
+H +HRD+KPDN+LI DGHIKLTDFGL SK S +D P+
Sbjct: 781 SVHKMGFIHRDIKPDNILIDLDGHIKLTDFGLCTGFRWTHNSKYYQKGSHVRQDSMEPSD 840
Query: 760 F---TNGFLVDDEPKPRHVSKREARQQ---QSIVGTPDYLAPEILLGMGHGTTADWWSVG 813
+ D K R+ Q+ S+VGTP+Y+APE+LL G+ DWWSVG
Sbjct: 841 LWDDVSNCRCGDRLKTLEQRARKQHQRCLAHSLVGTPNYIAPEVLLRKGYTQLCDWWSVG 900
Query: 814 VILYELLVGIPPFNADHAQQIFDNIIN--RDIQWPKHPEEISFEAYDLMNKLLIENPVQR 871
VIL+E+LVG PPF A + +IN + P ++S EA DL+ KL + R
Sbjct: 901 VILFEMLVGQPPFLAPTPTETQLKVINWENTLHIPAQ-VKLSPEARDLITKLCC-SADHR 958
Query: 872 LGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYL-LISHTWDPTNF 925
LG GA ++K H FF +++ + R++ APY+ ISH D +NF
Sbjct: 959 LGRNGADDLKAHPFFSAIDFSSDIRKQP-----------APYVPTISHPMDTSNF 1002
>K6A6_HUMAN (Q9UK32) Ribosomal protein S6 kinase alpha 6 (EC
2.7.1.37) (S6K-alpha 6) (90 kDa ribosomal protein S6
kinase 6) (p90-RSK 6) (Ribosomal S6 kinase 4) (RSK-4)
(pp90RSK4)
Length = 745
Score = 220 bits (560), Expect = 2e-56
Identities = 117/305 (38%), Positives = 180/305 (58%), Gaps = 37/305 (12%)
Query: 598 FEIIKPISRGAFGRVFLAQKRS---TGDLFAIKVLKKADMIRKNAVEGILAERDILISVR 654
FE++K + +G+FG+VFL +K++ G L+A+KVLKKA + ++ V + ERDIL+ V
Sbjct: 73 FELLKVLGQGSFGKVFLVRKKTGPDAGQLYAMKVLKKASLKVRDRVRTKM-ERDILVEVN 131
Query: 655 NPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHS 714
+PF+V+ +Y+F + LYL++++L GGD+++ L E+ + Y+AE+ LAL++LH
Sbjct: 132 HPFIVKLHYAFQTEGKLYLILDFLRGGDVFTRLSKEVLFTEEDVKFYLAELALALDHLHQ 191
Query: 715 QSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRH 774
IV+RDLKP+N+L+ + GHIKLTDFGLSK VD E K
Sbjct: 192 LGIVYRDLKPENILLDEIGHIKLTDFGLSKES-------------------VDQEKK--- 229
Query: 775 VSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQI 834
S GT +Y+APE++ GH +ADWWS GV+++E+L G PF +
Sbjct: 230 --------AYSFCGTVEYMAPEVVNRRGHSQSADWWSYGVLMFEMLTGTLPFQGKDRNET 281
Query: 835 FDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTL 894
+ I+ + P+ +S EA L+ L NP RLG G E+KRH FF +++WD L
Sbjct: 282 MNMILKAKLGMPQF---LSAEAQSLLRMLFKRNPANRLGSEGVEEIKRHLFFANIDWDKL 338
Query: 895 ARQKV 899
+++V
Sbjct: 339 YKREV 343
Score = 82.8 bits (203), Expect = 5e-15
Identities = 72/302 (23%), Positives = 130/302 (42%), Gaps = 55/302 (18%)
Query: 596 EDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRN 655
E +E+ + I G++ +T FA+K++ K+ +E ++ ++
Sbjct: 424 EVYELKEDIGVGSYSVCKRCIHATTNMEFAVKIIDKSKRDPSEEIEILMRYG------QH 477
Query: 656 PFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQ 715
P ++ F +YLV + + GG+L + C E A + + ++YLH Q
Sbjct: 478 PNIITLKDVFDDGRYVYLVTDLMKGGELLDRILKQKCFSEREASDILYVISKTVDYLHCQ 537
Query: 716 SIVHRDLKPDNLLIGQDG----HIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPK 771
+VHRDLKP N+L + I++ DFG +K
Sbjct: 538 GVVHRDLKPSNILYMDESASADSIRICDFGFAK--------------------------- 570
Query: 772 PRHVSKREARQQQSIVGTP----DYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPF- 826
+ R + ++ TP +++APE+L+ G+ D WS+GV+ Y +L G PF
Sbjct: 571 -------QLRGENGLLLTPCYTANFVAPEVLMQQGYDAACDIWSLGVLFYTMLAGYTPFA 623
Query: 827 --NADHAQQIFDNIINRDIQWP-KHPEEISFEAYDLMNKLLIENPVQRLGVTGATEVKRH 883
D ++I I N + + IS A DL++ +L +P QR A ++ +H
Sbjct: 624 NGPNDTPEEILLRIGNGKFSLSGGNWDNISDGAKDLLSHMLHMDPHQRY---TAEQILKH 680
Query: 884 AF 885
++
Sbjct: 681 SW 682
>COT1_NEUCR (P38679) Serine/threonine-protein kinase cot-1 (EC
2.7.1.37) (Colonial temperature-sensitive 1)
Length = 598
Score = 219 bits (559), Expect = 2e-56
Identities = 131/331 (39%), Positives = 194/331 (58%), Gaps = 28/331 (8%)
Query: 588 FSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAER 647
F + + E+++ IK I +GAFG V L QK++ G ++A+K L K +M +K+ + + AER
Sbjct: 204 FLRTKDKPENYQTIKIIGKGAFGEVKLVQKKADGKVYAMKSLIKTEMFKKDQLAHVRAER 263
Query: 648 DILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVL 707
DIL +P+VV+ Y +F LY++ME+L GGDL +ML ED+ R YIAE+VL
Sbjct: 264 DILAESDSPWVVKLYTTFQDANFLYMLMEFLPGGDLMTMLIKYEIFSEDITRFYIAEIVL 323
Query: 708 ALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLS----KVGLINSTEDL-----SAPA 758
A++ +H +HRD+KPDN+L+ + GH+KLTDFGLS K+ N L + P
Sbjct: 324 AIDAVHKLGFIHRDIKPDNILLDRGGHVKLTDFGLSTGFHKLHDNNYYTQLLQGKSNKPR 383
Query: 759 SFTNGFLVDD-----EPKPRHVSKREARQQQ--SIVGTPDYLAPEILLGMGHGTTADWWS 811
N +D + + R +R+ S VGTPDY+APEI G G+ DWWS
Sbjct: 384 DNRNSVAIDQINLTVSNRAQINDWRRSRRLMAYSTVGTPDYIAPEIFTGHGYSFDCDWWS 443
Query: 812 VGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKH----PEEISF--EAYDLMNKLLI 865
+G I++E LVG PPF A+ + + I+N W +H P++I+ +A +L+ L+
Sbjct: 444 LGTIMFECLVGWPPFCAEDSHDTYRKIVN----W-RHSLYFPDDITLGVDAENLIRSLIC 498
Query: 866 ENPVQRLGVTGATEVKRHAFFKDVNWDTLAR 896
N RLG GA E+K HAFF+ V +D+L R
Sbjct: 499 -NTENRLGRGGAHEIKSHAFFRGVEFDSLRR 528
>KAD5_SCHPO (Q09831) Probable serine/threonine-protein kinase
C4G8.05 (EC 2.7.1.37)
Length = 566
Score = 218 bits (554), Expect = 9e-56
Identities = 121/314 (38%), Positives = 185/314 (58%), Gaps = 16/314 (5%)
Query: 598 FEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPF 657
FE + + +G GRV+L +++ +G +A+KVL K +MI++N + AE+ IL + +PF
Sbjct: 195 FEKVFLLGKGDVGRVYLVREKKSGKFYAMKVLSKQEMIKRNKSKRAFAEQHILATSNHPF 254
Query: 658 VVRFYYSFTCKENLYLVMEYLNGGDLYSML--RNLGCLDEDMARVYIAEVVLALEYLHSQ 715
+V Y+SF E LYL MEY GG+ + L R CL E+ A+ YIAEV ALEYLH
Sbjct: 255 IVTLYHSFQSDEYLYLCMEYCMGGEFFRALQRRPGRCLSENEAKFYIAEVTAALEYLHLM 314
Query: 716 SIVHRDLKPDNLLIGQDGHIKLTDFGLSK----VGLINSTEDLSAPASFTNGFLVDDEPK 771
++RDLKP+N+L+ + GHI L+DF LSK G + +AP S N + +D +
Sbjct: 315 GFIYRDLKPENILLHESGHIMLSDFDLSKQSNSAGAPTVIQARNAP-SAQNAYALDTK-- 371
Query: 772 PRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHA 831
S + S VGT +Y+APE++ G GH + DWW++G++ YE+L PF +
Sbjct: 372 ----SCIADFRTNSFVGTEEYIAPEVIKGCGHTSAVDWWTLGILFYEMLYATTPFKGKNR 427
Query: 832 QQIFDNIINRDIQWPKHPE--EISFEAYDLMNKLLIENPVQRLG-VTGATEVKRHAFFKD 888
F NI+++D+ +P++ + IS +L+ KLL+++ RLG GA +VK H FFK+
Sbjct: 428 NMTFSNILHKDVIFPEYADAPSISSLCKNLIRKLLVKDENDRLGSQAGAADVKLHPFFKN 487
Query: 889 VNWDTLARQKVVIL 902
V W L + I+
Sbjct: 488 VQWALLRHTEPPII 501
>SCK1_SCHPO (P50530) Serine/threonine-protein kinase sck1 (EC
2.7.1.37)
Length = 696
Score = 212 bits (540), Expect = 4e-54
Identities = 124/309 (40%), Positives = 181/309 (58%), Gaps = 37/309 (11%)
Query: 596 EDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISV-- 653
EDF ++ I +G FG+V+L +K T ++A+K + K ++RK V L ER+IL+
Sbjct: 300 EDFTALRLIGKGTFGQVYLVRKNDTNRIYAMKKISKKLIVRKKEVTHTLGERNILVRTSL 359
Query: 654 -RNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYL 712
+PF+V +SF +LYL+ +Y++GG+L+ L++ G E A+ YIAE+VLALE+L
Sbjct: 360 DESPFIVGLKFSFQTASDLYLITDYMSGGELFWHLQHEGRFPEQRAKFYIAELVLALEHL 419
Query: 713 HSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKP 772
H I++RDLKP+N+L+ DGHI L DFGLSK +LSA A+ TN F
Sbjct: 420 HKHDIIYRDLKPENILLDADGHIALCDFGLSKA-------NLSANAT-TNTF-------- 463
Query: 773 RHVSKREARQQQSIVGTPDYLAPEILL-GMGHGTTADWWSVGVILYELLVGIPPFNADHA 831
GT +YLAPE+LL G+ D+WS+GV+++E+ G PF A
Sbjct: 464 --------------CGTTEYLAPEVLLEDKGYTKQVDFWSLGVLVFEMCCGWSPFYAPDV 509
Query: 832 QQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLG-VTGATEVKRHAFFKDVN 890
QQ++ NI +++PK +S E + LL NP RLG V TE+K H FF D+N
Sbjct: 510 QQMYRNIAFGKVRFPK--GVLSSEGRSFVRGLLNRNPNHRLGAVADTTELKEHPFFADIN 567
Query: 891 WDTLARQKV 899
WD L+++KV
Sbjct: 568 WDLLSKKKV 576
>NPH1_ARATH (O48963) Nonphototropic hypocotyl protein 1 (EC
2.7.1.37) (Phototropin)
Length = 996
Score = 209 bits (533), Expect = 2e-53
Identities = 118/314 (37%), Positives = 182/314 (57%), Gaps = 21/314 (6%)
Query: 589 SKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERD 648
S + ++ F+ +KP+ G G V L + T LFA+K + KA M+ +N V AER+
Sbjct: 654 SGEPIGLKHFKPVKPLGSGDTGSVHLVELVGTDQLFAMKAMDKAVMLNRNKVHRARAERE 713
Query: 649 ILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSML--RNLGCLDEDMARVYIAEVV 706
IL + +PF+ Y SF K ++ L+ +Y GG+L+ +L + L ED R Y A+VV
Sbjct: 714 ILDLLDHPFLPALYASFQTKTHICLITDYYPGGELFMLLDRQPRKVLKEDAVRFYAAQVV 773
Query: 707 LALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLV 766
+ALEYLH Q I++RDLKP+N+LI +G I L+DF LS L + L P+ +
Sbjct: 774 VALEYLHCQGIIYRDLKPENVLIQGNGDISLSDFDLSC--LTSCKPQLLIPS-------I 824
Query: 767 DDEPKPRHVSKREA--------RQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYE 818
D++ K + ++ R S VGT +Y+APEI+ G GH + DWW++G+++YE
Sbjct: 825 DEKKKKKQQKSQQTPIFMAEPMRASNSFVGTEEYIAPEIISGAGHTSAVDWWALGILMYE 884
Query: 819 LLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLG-VTGA 877
+L G PF Q+ F N++ +D+++P S + L+ +LL +P +RLG GA
Sbjct: 885 MLYGYTPFRGKTRQKTFTNVLQKDLKFPA-SIPASLQVKQLIFRLLQRDPKKRLGCFEGA 943
Query: 878 TEVKRHAFFKDVNW 891
EVK+H+FFK +NW
Sbjct: 944 NEVKQHSFFKGINW 957
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 130,719,029
Number of Sequences: 164201
Number of extensions: 5759248
Number of successful extensions: 21616
Number of sequences better than 10.0: 1716
Number of HSP's better than 10.0 without gapping: 1536
Number of HSP's successfully gapped in prelim test: 180
Number of HSP's that attempted gapping in prelim test: 16173
Number of HSP's gapped (non-prelim): 3190
length of query: 1086
length of database: 59,974,054
effective HSP length: 121
effective length of query: 965
effective length of database: 40,105,733
effective search space: 38702032345
effective search space used: 38702032345
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC133139.5


