

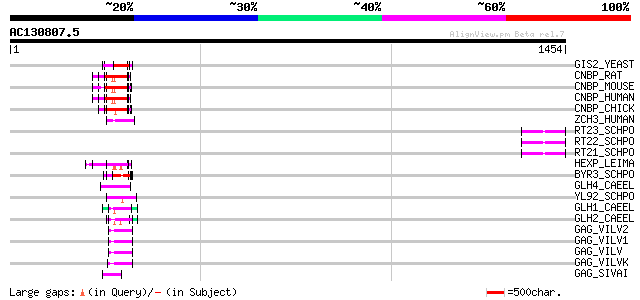
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.5 + phase: 0 /pseudo
(1454 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
GIS2_YEAST (P53849) Zinc-finger protein GIS2 70 5e-11
CNBP_RAT (P62634) Cellular nucleic acid binding protein (CNBP) (... 67 4e-10
CNBP_MOUSE (P53996) Cellular nucleic acid binding protein (CNBP)... 67 4e-10
CNBP_HUMAN (P62633) Cellular nucleic acid binding protein (CNBP)... 67 4e-10
CNBP_CHICK (O42395) Cellular nucleic acid binding protein (CNBP) 67 4e-10
ZCH3_HUMAN (Q9NUD5) Zinc finger CCHC domain containing protein 3 66 6e-10
RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protei... 64 2e-09
RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protei... 64 2e-09
RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protei... 64 2e-09
HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding p... 64 4e-09
BYR3_SCHPO (P36627) Cellular nucleic acid binding protein homolog 62 9e-09
GLH4_CAEEL (O76743) ATP-dependent RNA helicase glh-4 (EC 3.6.1.-... 57 4e-07
YL92_SCHPO (Q9HFF2) Hypothetical protein C683.02c in chromosome I 56 6e-07
GLH1_CAEEL (P34689) ATP-dependent RNA helicase glh-1 (EC 3.6.1.-... 56 6e-07
GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-... 55 1e-06
GAG_VILV2 (P23425) Gag polyprotein [Contains: Core protein p16; ... 52 2e-05
GAG_VILV1 (P23424) Gag polyprotein [Contains: Core protein p16; ... 52 2e-05
GAG_VILV (P03352) Gag polyprotein [Contains: Core protein p16; C... 52 2e-05
GAG_VILVK (P35955) Gag polyprotein [Contains: Core protein p16; ... 50 5e-05
GAG_SIVAI (Q02843) Gag polyprotein [Contains: Core protein p17; ... 49 8e-05
>GIS2_YEAST (P53849) Zinc-finger protein GIS2
Length = 153
Score = 69.7 bits (169), Expect = 5e-11
Identities = 29/69 (42%), Positives = 41/69 (59%), Gaps = 5/69 (7%)
Query: 244 PNRRDASAEIVCFKCGEKGHKSNVCTKDEK----KCFRCGQKGHLLADCKRGDVVCYNCN 299
P + +++ C+KCG H + C K++ KC+ CGQ GH+ DC+ D +CYNCN
Sbjct: 83 PKKTSRFSKVSCYKCGGPNHMAKDCMKEDGISGLKCYTCGQAGHMSRDCQN-DRLCYNCN 141
Query: 300 EEGHISTQC 308
E GHIS C
Sbjct: 142 ETGHISKDC 150
Score = 67.0 bits (162), Expect = 4e-10
Identities = 29/77 (37%), Positives = 43/77 (55%), Gaps = 6/77 (7%)
Query: 248 DASAEIVCFKCGEKGHKSNVCTKDE----KKCFRCGQKGHLLADCKRGDVVCYNCNEEGH 303
D +E +C+ C + GH CT K+C+ CG+ GH+ ++C C+NCN+ GH
Sbjct: 18 DCDSERLCYNCNKPGHVQTDCTMPRTVEFKQCYNCGETGHVRSECTVQR--CFNCNQTGH 75
Query: 304 ISTQCTQPKKDRTGGKV 320
IS +C +PKK KV
Sbjct: 76 ISRECPEPKKTSRFSKV 92
Score = 53.9 bits (128), Expect = 3e-06
Identities = 20/41 (48%), Positives = 28/41 (67%), Gaps = 1/41 (2%)
Query: 272 EKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPK 312
+K C+ CG+ GHL DC + +CYNCN+ GH+ T CT P+
Sbjct: 3 QKACYVCGKIGHLAEDCD-SERLCYNCNKPGHVQTDCTMPR 42
Score = 42.7 bits (99), Expect = 0.007
Identities = 15/45 (33%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Query: 246 RRDASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKR 290
+ D + + C+ CG+ GH S C D + C+ C + GH+ DC +
Sbjct: 109 KEDGISGLKCYTCGQAGHMSRDCQND-RLCYNCNETGHISKDCPK 152
>CNBP_RAT (P62634) Cellular nucleic acid binding protein (CNBP)
(Zinc finger protein 9)
Length = 177
Score = 67.0 bits (162), Expect = 4e-10
Identities = 29/63 (46%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 248 DASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADC-KRGDVVCYNCNEEGHIST 306
D + E C+ CGE GH CTK KC+RCG+ GH+ +C K +V CY C E GH++
Sbjct: 112 DHADEQKCYSCGEFGHIQKDCTK--VKCYRCGETGHVAINCSKTSEVNCYRCGESGHLAR 169
Query: 307 QCT 309
+CT
Sbjct: 170 ECT 172
Score = 66.6 bits (161), Expect = 5e-10
Identities = 32/97 (32%), Positives = 51/97 (51%), Gaps = 14/97 (14%)
Query: 216 RGKGRQSRPKPYSAPADKGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKDEKKC 275
RG+G +SR + +D+G Q ++ +S +C++CGE GH + C E C
Sbjct: 25 RGRGMRSRGRG-GFTSDRGFQFVS---------SSLPDICYRCGESGHLAKDCDLQEDAC 74
Query: 276 FRCGQKGHLLADCK----RGDVVCYNCNEEGHISTQC 308
+ CG+ GH+ DCK + CYNC + GH++ C
Sbjct: 75 YNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC 111
Score = 63.5 bits (153), Expect = 4e-09
Identities = 29/79 (36%), Positives = 43/79 (53%), Gaps = 6/79 (7%)
Query: 233 KGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTK-DEKKCFRCGQKGHLLADCKRG 291
+G D + P R E C+ CG+ GH + C DE+KC+ CG+ GH+ DC +
Sbjct: 79 RGGHIAKDCKEPKRE---REQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQKDCTK- 134
Query: 292 DVVCYNCNEEGHISTQCTQ 310
V CY C E GH++ C++
Sbjct: 135 -VKCYRCGETGHVAINCSK 152
Score = 61.6 bits (148), Expect = 1e-08
Identities = 30/89 (33%), Positives = 38/89 (41%), Gaps = 28/89 (31%)
Query: 255 CFKCGEKGHKSNVC------------------TKDE----------KKCFRCGQKGHLLA 286
CFKCG GH + C T D C+RCG+ GHL
Sbjct: 6 CFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHLAK 65
Query: 287 DCKRGDVVCYNCNEEGHISTQCTQPKKDR 315
DC + CYNC GHI+ C +PK++R
Sbjct: 66 DCDLQEDACYNCGRGGHIAKDCKEPKRER 94
>CNBP_MOUSE (P53996) Cellular nucleic acid binding protein (CNBP)
(Zinc finger protein 9)
Length = 178
Score = 67.0 bits (162), Expect = 4e-10
Identities = 29/63 (46%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 248 DASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADC-KRGDVVCYNCNEEGHIST 306
D + E C+ CGE GH CTK KC+RCG+ GH+ +C K +V CY C E GH++
Sbjct: 113 DHADEQKCYSCGEFGHIQKDCTK--VKCYRCGETGHVAINCSKTSEVNCYRCGESGHLAR 170
Query: 307 QCT 309
+CT
Sbjct: 171 ECT 173
Score = 63.5 bits (153), Expect = 4e-09
Identities = 31/98 (31%), Positives = 54/98 (54%), Gaps = 15/98 (15%)
Query: 216 RGKGRQSRPKPYSAPADKGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVC-TKDEKK 274
RG+G +SR + +D+G Q ++ +S +C++CGE GH + C ++++
Sbjct: 25 RGRGMRSRGRG-GFTSDRGFQFVS---------SSLPDICYRCGESGHLAKDCDLQEDEA 74
Query: 275 CFRCGQKGHLLADCK----RGDVVCYNCNEEGHISTQC 308
C+ CG+ GH+ DCK + CYNC + GH++ C
Sbjct: 75 CYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC 112
Score = 63.5 bits (153), Expect = 4e-09
Identities = 29/79 (36%), Positives = 43/79 (53%), Gaps = 6/79 (7%)
Query: 233 KGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTK-DEKKCFRCGQKGHLLADCKRG 291
+G D + P R E C+ CG+ GH + C DE+KC+ CG+ GH+ DC +
Sbjct: 80 RGGHIAKDCKEPKRE---REQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQKDCTK- 135
Query: 292 DVVCYNCNEEGHISTQCTQ 310
V CY C E GH++ C++
Sbjct: 136 -VKCYRCGETGHVAINCSK 153
Score = 61.6 bits (148), Expect = 1e-08
Identities = 28/75 (37%), Positives = 39/75 (51%), Gaps = 5/75 (6%)
Query: 248 DASAEIVCFKCGEKGHKSNVCTKDEKK----CFRCGQKGHLLADCKRGDVV-CYNCNEEG 302
D + C+ CG GH + C + +++ C+ CG+ GHL DC D CY+C E G
Sbjct: 68 DLQEDEACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEFG 127
Query: 303 HISTQCTQPKKDRTG 317
HI CT+ K R G
Sbjct: 128 HIQKDCTKVKCYRCG 142
Score = 60.1 bits (144), Expect = 4e-08
Identities = 31/90 (34%), Positives = 39/90 (42%), Gaps = 29/90 (32%)
Query: 255 CFKCGEKGHKSNVC------------------TKDE----------KKCFRCGQKGHLLA 286
CFKCG GH + C T D C+RCG+ GHL
Sbjct: 6 CFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHLAK 65
Query: 287 DCK-RGDVVCYNCNEEGHISTQCTQPKKDR 315
DC + D CYNC GHI+ C +PK++R
Sbjct: 66 DCDLQEDEACYNCGRGGHIAKDCKEPKRER 95
>CNBP_HUMAN (P62633) Cellular nucleic acid binding protein (CNBP)
(Zinc finger protein 9)
Length = 177
Score = 67.0 bits (162), Expect = 4e-10
Identities = 29/63 (46%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 248 DASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADC-KRGDVVCYNCNEEGHIST 306
D + E C+ CGE GH CTK KC+RCG+ GH+ +C K +V CY C E GH++
Sbjct: 112 DHADEQKCYSCGEFGHIQKDCTK--VKCYRCGETGHVAINCSKTSEVNCYRCGESGHLAR 169
Query: 307 QCT 309
+CT
Sbjct: 170 ECT 172
Score = 66.6 bits (161), Expect = 5e-10
Identities = 32/97 (32%), Positives = 51/97 (51%), Gaps = 14/97 (14%)
Query: 216 RGKGRQSRPKPYSAPADKGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKDEKKC 275
RG+G +SR + +D+G Q ++ +S +C++CGE GH + C E C
Sbjct: 25 RGRGMRSRGRG-GFTSDRGFQFVS---------SSLPDICYRCGESGHLAKDCDLQEDAC 74
Query: 276 FRCGQKGHLLADCK----RGDVVCYNCNEEGHISTQC 308
+ CG+ GH+ DCK + CYNC + GH++ C
Sbjct: 75 YNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC 111
Score = 63.5 bits (153), Expect = 4e-09
Identities = 29/79 (36%), Positives = 43/79 (53%), Gaps = 6/79 (7%)
Query: 233 KGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTK-DEKKCFRCGQKGHLLADCKRG 291
+G D + P R E C+ CG+ GH + C DE+KC+ CG+ GH+ DC +
Sbjct: 79 RGGHIAKDCKEPKRE---REQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQKDCTK- 134
Query: 292 DVVCYNCNEEGHISTQCTQ 310
V CY C E GH++ C++
Sbjct: 135 -VKCYRCGETGHVAINCSK 152
Score = 61.6 bits (148), Expect = 1e-08
Identities = 30/89 (33%), Positives = 38/89 (41%), Gaps = 28/89 (31%)
Query: 255 CFKCGEKGHKSNVC------------------TKDE----------KKCFRCGQKGHLLA 286
CFKCG GH + C T D C+RCG+ GHL
Sbjct: 6 CFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHLAK 65
Query: 287 DCKRGDVVCYNCNEEGHISTQCTQPKKDR 315
DC + CYNC GHI+ C +PK++R
Sbjct: 66 DCDLQEDACYNCGRGGHIAKDCKEPKRER 94
>CNBP_CHICK (O42395) Cellular nucleic acid binding protein (CNBP)
Length = 172
Score = 67.0 bits (162), Expect = 4e-10
Identities = 29/63 (46%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 248 DASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADC-KRGDVVCYNCNEEGHIST 306
D + E C+ CGE GH CTK KC+RCG+ GH+ +C K +V CY C E GH++
Sbjct: 107 DHADEQKCYSCGEFGHIQKDCTK--VKCYRCGETGHVAINCSKTSEVNCYRCGESGHLAR 164
Query: 307 QCT 309
+CT
Sbjct: 165 ECT 167
Score = 63.5 bits (153), Expect = 4e-09
Identities = 29/79 (36%), Positives = 43/79 (53%), Gaps = 6/79 (7%)
Query: 233 KGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTK-DEKKCFRCGQKGHLLADCKRG 291
+G D + P R E C+ CG+ GH + C DE+KC+ CG+ GH+ DC +
Sbjct: 74 RGGHIAKDCKEPKRE---REQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQKDCTK- 129
Query: 292 DVVCYNCNEEGHISTQCTQ 310
V CY C E GH++ C++
Sbjct: 130 -VKCYRCGETGHVAINCSK 147
Score = 62.4 bits (150), Expect = 9e-09
Identities = 23/60 (38%), Positives = 37/60 (61%), Gaps = 5/60 (8%)
Query: 254 VCFKCGEKGHKSNVCT-KDEKKCFRCGQKGHLLADCK----RGDVVCYNCNEEGHISTQC 308
+C++CGE GH + C +++K C+ CG+ GH+ DCK + CYNC + GH++ C
Sbjct: 47 ICYRCGESGHLAKDCDLQEDKACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC 106
Score = 61.6 bits (148), Expect = 1e-08
Identities = 28/75 (37%), Positives = 39/75 (51%), Gaps = 5/75 (6%)
Query: 248 DASAEIVCFKCGEKGHKSNVCTKDEKK----CFRCGQKGHLLADCKRGDVV-CYNCNEEG 302
D + C+ CG GH + C + +++ C+ CG+ GHL DC D CY+C E G
Sbjct: 62 DLQEDKACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEFG 121
Query: 303 HISTQCTQPKKDRTG 317
HI CT+ K R G
Sbjct: 122 HIQKDCTKVKCYRCG 136
Score = 61.2 bits (147), Expect = 2e-08
Identities = 29/84 (34%), Positives = 38/84 (44%), Gaps = 23/84 (27%)
Query: 255 CFKCGEKGHKSNVCTKDEKK----------------------CFRCGQKGHLLADCK-RG 291
CFKCG GH + C + C+RCG+ GHL DC +
Sbjct: 6 CFKCGRTGHWARECPTGIGRGRGMRSRGRAGFQFMSSSLPDICYRCGESGHLAKDCDLQE 65
Query: 292 DVVCYNCNEEGHISTQCTQPKKDR 315
D CYNC GHI+ C +PK++R
Sbjct: 66 DKACYNCGRGGHIAKDCKEPKRER 89
>ZCH3_HUMAN (Q9NUD5) Zinc finger CCHC domain containing protein 3
Length = 404
Score = 66.2 bits (160), Expect = 6e-10
Identities = 28/71 (39%), Positives = 42/71 (58%), Gaps = 3/71 (4%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKKD 314
CFKCG + H S CT+D +CFRCG++GHL C++G +VC C + GH QC + +
Sbjct: 336 CFKCGSRTHMSGSCTQD--RCFRCGEEGHLSPYCRKG-IVCNLCGKRGHAFAQCPKAVHN 392
Query: 315 RTGGKVFALTG 325
++ + G
Sbjct: 393 SVAAQLTGVAG 403
Score = 41.2 bits (95), Expect = 0.021
Identities = 15/36 (41%), Positives = 21/36 (57%), Gaps = 2/36 (5%)
Query: 273 KKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQC 308
K CF+CG + H+ C + C+ C EEGH+S C
Sbjct: 334 KTCFKCGSRTHMSGSCTQDR--CFRCGEEGHLSPYC 367
>RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protein
type 3
Length = 1333
Score = 64.3 bits (155), Expect = 2e-09
Identities = 36/113 (31%), Positives = 60/113 (52%), Gaps = 2/113 (1%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQ+ERT Q++E LLR TW H+ L++ +YNN HS+ M PFE ++ R
Sbjct: 1080 QTDGQTERTNQTVEKLLRCVCSTHPNTWVDHISLVQQSYNNAIHSATQMTPFEIVH--RY 1137
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
L E E Q+T + + ++E + ++ + K Y D + +++E
Sbjct: 1138 SPALSPLELPSFSDKTDENSQETIQVFQTVKEHLNTNNIKMKKYFDMKIQEIE 1190
>RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protein
type 2
Length = 1333
Score = 64.3 bits (155), Expect = 2e-09
Identities = 36/113 (31%), Positives = 60/113 (52%), Gaps = 2/113 (1%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQ+ERT Q++E LLR TW H+ L++ +YNN HS+ M PFE ++ R
Sbjct: 1080 QTDGQTERTNQTVEKLLRCVCSTHPNTWVDHISLVQQSYNNAIHSATQMTPFEIVH--RY 1137
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
L E E Q+T + + ++E + ++ + K Y D + +++E
Sbjct: 1138 SPALSPLELPSFSDKTDENSQETIQVFQTVKEHLNTNNIKMKKYFDMKIQEIE 1190
>RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protein
type 1
Length = 1333
Score = 64.3 bits (155), Expect = 2e-09
Identities = 36/113 (31%), Positives = 60/113 (52%), Gaps = 2/113 (1%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQ+ERT Q++E LLR TW H+ L++ +YNN HS+ M PFE ++ R
Sbjct: 1080 QTDGQTERTNQTVEKLLRCVCSTHPNTWVDHISLVQQSYNNAIHSATQMTPFEIVH--RY 1137
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
L E E Q+T + + ++E + ++ + K Y D + +++E
Sbjct: 1138 SPALSPLELPSFSDKTDENSQETIQVFQTVKEHLNTNNIKMKKYFDMKIQEIE 1190
>HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding
protein)
Length = 271
Score = 63.5 bits (153), Expect = 4e-09
Identities = 28/78 (35%), Positives = 37/78 (46%), Gaps = 14/78 (17%)
Query: 255 CFKCGEKGHKSNVCTKDEKK-------CFRCGQKGHLLADCKRG-------DVVCYNCNE 300
CF+CGE+GH S C + + CFRCG+ GH+ DC CY C +
Sbjct: 45 CFRCGEEGHMSRECPNEARSGAAGAMTCFRCGEAGHMSRDCPNSAKPGAAKGFECYKCGQ 104
Query: 301 EGHISTQCTQPKKDRTGG 318
EGH+S C + GG
Sbjct: 105 EGHLSRDCPSSQGGSRGG 122
Score = 63.2 bits (152), Expect = 5e-09
Identities = 36/116 (31%), Positives = 49/116 (42%), Gaps = 18/116 (15%)
Query: 218 KGRQSRPKPYSAPADKGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKDE----- 272
+G SR P S +G R + S + C+KCG+ GH S C +
Sbjct: 105 EGHLSRDCPSSQGGSRGGYGQKRGRSGAQGGYSGDRTCYKCGDAGHISRDCPNGQGGYSG 164
Query: 273 ---KKCFRCGQKGHLLADCKRG--------DVVCYNCNEEGHISTQCTQPKKDRTG 317
+ C++CG GH+ DC G D CY C E GH+S +C P TG
Sbjct: 165 AGDRTCYKCGDAGHISRDCPNGQGGYSGAGDRKCYKCGESGHMSREC--PSAGSTG 218
Score = 62.0 bits (149), Expect = 1e-08
Identities = 44/144 (30%), Positives = 59/144 (40%), Gaps = 39/144 (27%)
Query: 198 CRICEEDTKAHY-KVMSERRGKGRQSRPKPYSAPADKGKQRLNDERRPN--RRDASAEIV 254
CR C ++ HY + E KG + + G++ PN R A+ +
Sbjct: 18 CRNCGKE--GHYARECPEADSKGDERSTTCFRC----GEEGHMSRECPNEARSGAAGAMT 71
Query: 255 CFKCGEKGHKSNVCTKDEK-------KCFRCGQKGHLLADC-------------KR---- 290
CF+CGE GH S C K +C++CGQ+GHL DC KR
Sbjct: 72 CFRCGEAGHMSRDCPNSAKPGAAKGFECYKCGQEGHLSRDCPSSQGGSRGGYGQKRGRSG 131
Query: 291 ------GDVVCYNCNEEGHISTQC 308
GD CY C + GHIS C
Sbjct: 132 AQGGYSGDRTCYKCGDAGHISRDC 155
Score = 60.5 bits (145), Expect = 3e-08
Identities = 25/70 (35%), Positives = 37/70 (52%), Gaps = 14/70 (20%)
Query: 255 CFKCGEKGHKSNVCTKDE--------KKCFRCGQKGHLLADCKR------GDVVCYNCNE 300
C+KCG+ GH S C + +KC++CG+ GH+ +C GD CY C +
Sbjct: 170 CYKCGDAGHISRDCPNGQGGYSGAGDRKCYKCGESGHMSRECPSAGSTGSGDRACYKCGK 229
Query: 301 EGHISTQCTQ 310
GHIS +C +
Sbjct: 230 PGHISRECPE 239
Score = 59.7 bits (143), Expect = 6e-08
Identities = 26/71 (36%), Positives = 34/71 (47%), Gaps = 17/71 (23%)
Query: 255 CFKCGEKGHKSNVCTK------DEKKCFRCGQKGHLLADCKR-----------GDVVCYN 297
C+KCGE GH S C ++ C++CG+ GH+ +C GD CY
Sbjct: 198 CYKCGESGHMSRECPSAGSTGSGDRACYKCGKPGHISRECPEAGGSYGGSRGGGDRTCYK 257
Query: 298 CNEEGHISTQC 308
C E GHIS C
Sbjct: 258 CGEAGHISRDC 268
Score = 43.9 bits (102), Expect = 0.003
Identities = 19/56 (33%), Positives = 28/56 (49%), Gaps = 9/56 (16%)
Query: 269 TKDEKKCFRCGQKGHLLADCKRGD-------VVCYNCNEEGHISTQCTQPKKDRTG 317
T+ C CG++GH +C D C+ C EEGH+S +C P + R+G
Sbjct: 12 TESSTSCRNCGKEGHYARECPEADSKGDERSTTCFRCGEEGHMSREC--PNEARSG 65
Score = 41.2 bits (95), Expect = 0.021
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 11/50 (22%)
Query: 250 SAEIVCFKCGEKGHKSNVCTK-----------DEKKCFRCGQKGHLLADC 288
S + C+KCG+ GH S C + ++ C++CG+ GH+ DC
Sbjct: 219 SGDRACYKCGKPGHISRECPEAGGSYGGSRGGGDRTCYKCGEAGHISRDC 268
>BYR3_SCHPO (P36627) Cellular nucleic acid binding protein homolog
Length = 179
Score = 62.4 bits (150), Expect = 9e-09
Identities = 28/80 (35%), Positives = 43/80 (53%), Gaps = 8/80 (10%)
Query: 247 RDASAEIVCFKCGEKGHKSNVCTK--DEKKCFRCGQKGHLLADC-----KRGDVVCYNCN 299
R+ + +C+ C + GHK++ CT+ EK C+ CG GHL+ DC R CY C
Sbjct: 30 RECTKGSICYNCNQTGHKASECTEPQQEKTCYACGTAGHLVRDCPSSPNPRQGAECYKCG 89
Query: 300 EEGHISTQCTQPKKDRTGGK 319
GHI+ C + ++GG+
Sbjct: 90 RVGHIARDC-RTNGQQSGGR 108
Score = 59.7 bits (143), Expect = 6e-08
Identities = 24/59 (40%), Positives = 35/59 (58%), Gaps = 3/59 (5%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGD--VVCYNCNEEGHISTQCTQP 311
C+ CG GH++ CT K C+ CG+ GH +C++ +CY CN+ GHI+ CT P
Sbjct: 118 CYACGSYGHQARDCTMGVK-CYSCGKIGHRSFECQQASDGQLCYKCNQPGHIAVNCTSP 175
Score = 58.5 bits (140), Expect = 1e-07
Identities = 19/48 (39%), Positives = 35/48 (72%), Gaps = 1/48 (2%)
Query: 269 TKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKKDRT 316
T+ +C+ CG+ GH +C +G + CYNCN+ GH +++CT+P++++T
Sbjct: 13 TRPGPRCYNCGENGHQARECTKGSI-CYNCNQTGHKASECTEPQQEKT 59
Score = 56.2 bits (134), Expect = 6e-07
Identities = 23/69 (33%), Positives = 34/69 (48%), Gaps = 3/69 (4%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCK--RGDVVCYNCNEEGHISTQCTQPK 312
C+ CGE GH++ CTK C+ C Q GH ++C + + CY C GH+ C
Sbjct: 19 CYNCGENGHQARECTKGSI-CYNCNQTGHKASECTEPQQEKTCYACGTAGHLVRDCPSSP 77
Query: 313 KDRTGGKVF 321
R G + +
Sbjct: 78 NPRQGAECY 86
Score = 43.5 bits (101), Expect = 0.004
Identities = 16/44 (36%), Positives = 26/44 (58%), Gaps = 2/44 (4%)
Query: 247 RDASAEIVCFKCGEKGHKSNVC--TKDEKKCFRCGQKGHLLADC 288
RD + + C+ CG+ GH+S C D + C++C Q GH+ +C
Sbjct: 129 RDCTMGVKCYSCGKIGHRSFECQQASDGQLCYKCNQPGHIAVNC 172
>GLH4_CAEEL (O76743) ATP-dependent RNA helicase glh-4 (EC 3.6.1.-)
(Germline helicase-4)
Length = 1156
Score = 57.0 bits (136), Expect = 4e-07
Identities = 31/83 (37%), Positives = 38/83 (45%), Gaps = 6/83 (7%)
Query: 239 NDERRPNRRDASAEIVCFKCGEKGHKSNVCTKDEKK---CFRCGQKGHLLADCKRGDVV- 294
+D +R N C CGE+GH S C K + C C Q GH +DC + V
Sbjct: 556 SDNQRGNWDGGERPRGCHNCGEEGHISKECDKPKVPRFPCRNCEQLGHFASDCDQPRVPR 615
Query: 295 --CYNCNEEGHISTQCTQPKKDR 315
C NC EGH + C QPK R
Sbjct: 616 GPCRNCGIEGHFAVDCDQPKVPR 638
Score = 45.4 bits (106), Expect = 0.001
Identities = 25/72 (34%), Positives = 32/72 (43%), Gaps = 9/72 (12%)
Query: 255 CFKCGEKGHKSNVCTKDEKK---CFRCGQKGHLLADCKRGDV------VCYNCNEEGHIS 305
C CG +GH + C + + C CGQ+GH DC+ V C C EEGH
Sbjct: 618 CRNCGIEGHFAVDCDQPKVPRGPCRNCGQEGHFAKDCQNERVRMEPTEPCRRCAEEGHWG 677
Query: 306 TQCTQPKKDRTG 317
+C KD G
Sbjct: 678 YECPTRPKDLQG 689
Score = 36.6 bits (83), Expect = 0.52
Identities = 18/47 (38%), Positives = 25/47 (52%), Gaps = 2/47 (4%)
Query: 295 CYNCNEEGHISTQCTQPKKDRTGGKVFALTG--TQTTNEDRLIRGTC 339
C+NC EEGHIS +C +PK R + G ++ R+ RG C
Sbjct: 572 CHNCGEEGHISKECDKPKVPRFPCRNCEQLGHFASDCDQPRVPRGPC 618
>YL92_SCHPO (Q9HFF2) Hypothetical protein C683.02c in chromosome I
Length = 218
Score = 56.2 bits (134), Expect = 6e-07
Identities = 30/91 (32%), Positives = 41/91 (44%), Gaps = 12/91 (13%)
Query: 254 VCFKCGEKGHKSNVCTKDE----KKCFRCGQKGHLLADCKRGDV-------VCYNCNEEG 302
+CF+CG K H N C+K KCF C + GHL C++ C C+
Sbjct: 101 ICFRCGSKEHSLNACSKKGPLKFAKCFICHENGHLSGQCEQNPKGLYPKGGCCKFCSSVH 160
Query: 303 HISTQCTQPKKDRTG-GKVFALTGTQTTNED 332
H++ C Q KD G V + GT +ED
Sbjct: 161 HLAKDCDQVNKDDVSFGHVVGVAGTTGADED 191
Score = 38.1 bits (87), Expect = 0.18
Identities = 29/110 (26%), Positives = 43/110 (38%), Gaps = 37/110 (33%)
Query: 231 ADKGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADC-- 288
A G + DER+ +R + + N +D K CF C Q+GH++ DC
Sbjct: 45 ASFGSSKRYDERQKKKRSEYRRL---------RRINQRNRD-KFCFACRQQGHIVQDCPE 94
Query: 289 ----------------------KRGDV---VCYNCNEEGHISTQCTQPKK 313
K+G + C+ C+E GH+S QC Q K
Sbjct: 95 AKDNVSICFRCGSKEHSLNACSKKGPLKFAKCFICHENGHLSGQCEQNPK 144
>GLH1_CAEEL (P34689) ATP-dependent RNA helicase glh-1 (EC 3.6.1.-)
(Germline helicase-1)
Length = 763
Score = 56.2 bits (134), Expect = 6e-07
Identities = 34/130 (26%), Positives = 45/130 (34%), Gaps = 39/130 (30%)
Query: 244 PNRRDASAEIVCFKCGEKGHKSNVCTKDEK------------------------------ 273
P R VC+ C + GH S CT++ K
Sbjct: 174 PEPRKEREPRVCYNCQQPGHTSRECTEERKPREGRTGGFGGGAGFGNNGGNDGFGGDGGF 233
Query: 274 ---------KCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKKDRTGGKVFALT 324
KCF C +GH A+C C+NC E+GH S +C P K R G +
Sbjct: 234 GGGEERGPMKCFNCKGEGHRSAECPEPPRGCFNCGEQGHRSNECPNPAKPREGVEGEGPK 293
Query: 325 GTQTTNEDRL 334
T ED +
Sbjct: 294 ATYVPVEDNM 303
Score = 54.3 bits (129), Expect = 2e-06
Identities = 28/68 (41%), Positives = 36/68 (52%), Gaps = 11/68 (16%)
Query: 259 GEKGHKSNVCTKDEKKCFRCGQKGHLLADC-----KRGDVVCYNCNEEGHISTQCTQ--- 310
GE GH + CF C Q GH +DC +R VCYNC + GH S +CT+
Sbjct: 147 GEGGHGGG---ERNNNCFNCQQPGHRSSDCPEPRKEREPRVCYNCQQPGHTSRECTEERK 203
Query: 311 PKKDRTGG 318
P++ RTGG
Sbjct: 204 PREGRTGG 211
Score = 42.7 bits (99), Expect = 0.007
Identities = 26/95 (27%), Positives = 40/95 (41%), Gaps = 6/95 (6%)
Query: 198 CRICEEDTKAHYKVMSERRGKGRQSRPKPYSAPADKGKQRLNDERRPNRRDASAE----I 253
C C++ + ER K R+ R + A G ND + E +
Sbjct: 185 CYNCQQPGHTSRECTEER--KPREGRTGGFGGGAGFGNNGGNDGFGGDGGFGGGEERGPM 242
Query: 254 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADC 288
CF C +GH+S C + + CF CG++GH +C
Sbjct: 243 KCFNCKGEGHRSAECPEPPRGCFNCGEQGHRSNEC 277
>GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-)
(Germline helicase-2)
Length = 974
Score = 55.1 bits (131), Expect = 1e-06
Identities = 31/118 (26%), Positives = 42/118 (35%), Gaps = 37/118 (31%)
Query: 254 VCFKCGEKGHKSNVCTKDEK-------------------------------------KCF 276
VC+ C + GH S C ++ K KCF
Sbjct: 397 VCYNCQQPGHNSRDCPEERKPREGRNGFTSGFGGGNDGGFGGGNAEGFGNNEERGPMKCF 456
Query: 277 RCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKKDRTGGKVFALTGTQTTNEDRL 334
C +GH A+C C+NC E+GH S +C P K R G + T ED +
Sbjct: 457 NCKGEGHRSAECPEPPRGCFNCGEQGHRSNECPNPAKPREGAEGEGPKATYVPVEDNM 514
Score = 50.1 bits (118), Expect = 5e-05
Identities = 27/101 (26%), Positives = 39/101 (37%), Gaps = 42/101 (41%)
Query: 255 CFKCGEKGHKSNVCTKDEKK-----CFRCGQKGHLLADC--------------------- 288
CF C + GH+SN C + +K+ C+ C Q GH DC
Sbjct: 373 CFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHNSRDCPEERKPREGRNGFTSGFGGGN 432
Query: 289 ----------------KRGDVVCYNCNEEGHISTQCTQPKK 313
+RG + C+NC EGH S +C +P +
Sbjct: 433 DGGFGGGNAEGFGNNEERGPMKCFNCKGEGHRSAECPEPPR 473
Score = 47.0 bits (110), Expect = 4e-04
Identities = 23/67 (34%), Positives = 31/67 (45%), Gaps = 15/67 (22%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKKD 314
CF C + GH+SN C + +K +R VCYNC + GH S C + +K
Sbjct: 259 CFNCQQPGHRSNDCPEPKK---------------EREPRVCYNCQQPGHNSRDCPEERKP 303
Query: 315 RTGGKVF 321
R G F
Sbjct: 304 REGRNGF 310
Score = 47.0 bits (110), Expect = 4e-04
Identities = 24/67 (35%), Positives = 32/67 (46%), Gaps = 12/67 (17%)
Query: 260 EKGHKSNVCTKDEKKCFRCGQKGHLLADC-----KRGDVVCYNCNEEGHISTQCTQPKKD 314
++G ++N CF C Q GH DC +R VCYNC + GH S C + +K
Sbjct: 365 DRGERNN-------NCFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHNSRDCPEERKP 417
Query: 315 RTGGKVF 321
R G F
Sbjct: 418 REGRNGF 424
>GAG_VILV2 (P23425) Gag polyprotein [Contains: Core protein p16;
Core protein p25; Core protein p14]
Length = 442
Score = 51.6 bits (122), Expect = 2e-05
Identities = 23/65 (35%), Positives = 38/65 (58%), Gaps = 9/65 (13%)
Query: 259 GEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKK---DR 315
G+ GHK +KC+ CG+ GHL C++G ++C++C + GH+ C Q K+ +R
Sbjct: 376 GKAGHKGV-----NQKCYNCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKQQGNNR 429
Query: 316 TGGKV 320
G +V
Sbjct: 430 RGPRV 434
Score = 37.7 bits (86), Expect = 0.23
Identities = 13/36 (36%), Positives = 22/36 (61%), Gaps = 1/36 (2%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKR 290
C+ CG+ GH + C + C CG++GH+ DC++
Sbjct: 387 CYNCGKPGHLARQC-RQGIICHHCGKRGHMQKDCRQ 421
>GAG_VILV1 (P23424) Gag polyprotein [Contains: Core protein p16;
Core protein p25; Core protein p14]
Length = 442
Score = 51.6 bits (122), Expect = 2e-05
Identities = 23/65 (35%), Positives = 38/65 (58%), Gaps = 9/65 (13%)
Query: 259 GEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKK---DR 315
G+ GHK +KC+ CG+ GHL C++G ++C++C + GH+ C Q K+ +R
Sbjct: 376 GKAGHKGV-----NQKCYNCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKQQGNNR 429
Query: 316 TGGKV 320
G +V
Sbjct: 430 RGPRV 434
Score = 37.7 bits (86), Expect = 0.23
Identities = 13/36 (36%), Positives = 22/36 (61%), Gaps = 1/36 (2%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKR 290
C+ CG+ GH + C + C CG++GH+ DC++
Sbjct: 387 CYNCGKPGHLARQC-RQGIICHHCGKRGHMQKDCRQ 421
>GAG_VILV (P03352) Gag polyprotein [Contains: Core protein p16; Core
protein p25; Core protein p14]
Length = 442
Score = 51.6 bits (122), Expect = 2e-05
Identities = 23/65 (35%), Positives = 38/65 (58%), Gaps = 9/65 (13%)
Query: 259 GEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKK---DR 315
G+ GHK +KC+ CG+ GHL C++G ++C++C + GH+ C Q K+ +R
Sbjct: 376 GKAGHKGV-----NQKCYNCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKQQGNNR 429
Query: 316 TGGKV 320
G +V
Sbjct: 430 RGPRV 434
Score = 37.7 bits (86), Expect = 0.23
Identities = 13/36 (36%), Positives = 22/36 (61%), Gaps = 1/36 (2%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKR 290
C+ CG+ GH + C + C CG++GH+ DC++
Sbjct: 387 CYNCGKPGHLARQC-RQGIICHHCGKRGHMQKDCRQ 421
>GAG_VILVK (P35955) Gag polyprotein [Contains: Core protein p16;
Core protein p25; Core protein p14]
Length = 442
Score = 50.1 bits (118), Expect = 5e-05
Identities = 23/67 (34%), Positives = 38/67 (56%), Gaps = 12/67 (17%)
Query: 257 KCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKK--- 313
K G+KG +KC+ CG+ GHL C++G ++C++C + GH+ C Q K+
Sbjct: 377 KAGQKGVN--------QKCYNCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKQQGN 427
Query: 314 DRTGGKV 320
+R G +V
Sbjct: 428 NRRGPRV 434
Score = 37.7 bits (86), Expect = 0.23
Identities = 13/36 (36%), Positives = 22/36 (61%), Gaps = 1/36 (2%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKR 290
C+ CG+ GH + C + C CG++GH+ DC++
Sbjct: 387 CYNCGKPGHLARQC-RQGIICHHCGKRGHMQKDCRQ 421
>GAG_SIVAI (Q02843) Gag polyprotein [Contains: Core protein p17;
Core protein p24; Core protein p15]
Length = 513
Score = 49.3 bits (116), Expect = 8e-05
Identities = 19/49 (38%), Positives = 27/49 (54%), Gaps = 1/49 (2%)
Query: 244 PNRRDASAEIVCFKCGEKGHKSNVCTKDEK-KCFRCGQKGHLLADCKRG 291
P ++ + CF CG+ GH C + KCF+CG+ GH+ DCK G
Sbjct: 381 PQKKGPRGPLKCFNCGKFGHMQRECKAPRQIKCFKCGKIGHMAKDCKNG 429
Score = 39.7 bits (91), Expect = 0.061
Identities = 14/36 (38%), Positives = 22/36 (60%), Gaps = 1/36 (2%)
Query: 274 KCFRCGQKGHLLADCKRG-DVVCYNCNEEGHISTQC 308
KCF CG+ GH+ +CK + C+ C + GH++ C
Sbjct: 391 KCFNCGKFGHMQRECKAPRQIKCFKCGKIGHMAKDC 426
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.351 0.154 0.543
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 147,883,042
Number of Sequences: 164201
Number of extensions: 5711670
Number of successful extensions: 23741
Number of sequences better than 10.0: 112
Number of HSP's better than 10.0 without gapping: 48
Number of HSP's successfully gapped in prelim test: 65
Number of HSP's that attempted gapping in prelim test: 23109
Number of HSP's gapped (non-prelim): 401
length of query: 1454
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1331
effective length of database: 39,777,331
effective search space: 52943627561
effective search space used: 52943627561
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 72 (32.3 bits)
Medicago: description of AC130807.5


