

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC129092.1 - phase: 0
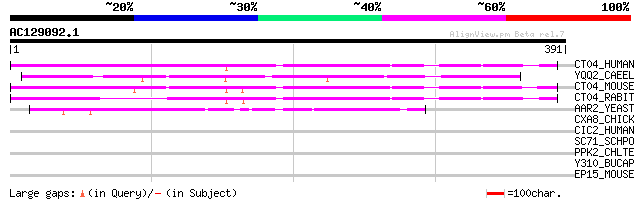
(391 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CT04_HUMAN (Q9Y312) Protein c20orf4 (CGI-23) (PRO0225) 152 2e-36
YQQ2_CAEEL (Q09305) Hypothetical protein F10B5.2 in chromosome II 149 9e-36
CT04_MOUSE (Q9D2V5) Protein c20orf4 homolog 146 8e-35
CT04_RABIT (O77682) Protein c20orf4 homolog 115 2e-25
AAR2_YEAST (P32357) A1 cistron splicing factor AAR2 65 3e-10
CXA8_CHICK (P36381) Gap junction alpha-8 protein (Connexin 45.6)... 33 1.2
CIC2_HUMAN (P54289) Dihydropyridine-sensitive L-type, calcium ch... 31 4.7
SC71_SCHPO (Q9UT02) Protein transport protein sec71 31 6.2
PPK2_CHLTE (O68984) Polyphosphate kinase 2 (EC 2.7.4.1) (Polypho... 31 6.2
Y310_BUCAP (Q8K9M4) Hypothetical metalloprotease BUsg310 precurs... 30 8.1
EP15_MOUSE (P42567) Epidermal growth factor receptor substrate 1... 30 8.1
>CT04_HUMAN (Q9Y312) Protein c20orf4 (CGI-23) (PRO0225)
Length = 384
Score = 152 bits (383), Expect = 2e-36
Identities = 115/400 (28%), Positives = 187/400 (46%), Gaps = 40/400 (10%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD + A L G T++ L++P+ T ID + VGP F+G+KMIPPG HF++YSS +
Sbjct: 6 MDPELAKRLFFEGATVVILNMPKGTEFGIDYNSWEVGPKFRGVKMIPPGIHFLHYSSVDK 65
Query: 61 -DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGPY 119
+ KE P +GFF+ + V +W E + E E + E D+ LGPY
Sbjct: 66 ANPKEVGPRMGFFLSLHQRGLTVLRWSTLREEVDLSPAPESEVEAMRANLQELDQFLGPY 125
Query: 120 NLSHYEDWKRLSDFITKSIIERLEPIGGEVSV-----------ECENDMFRNAPKTPME- 167
+ + W L++FI+++ +E+L+P ++ ++ + +N P+ +E
Sbjct: 126 PYATLKKWISLTNFISEATVEKLQPENRQICAFSDVLPVLSMKHTKDRVGQNLPRCGIEC 185
Query: 168 KALDTQL-KVDNSATSVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLV 226
K+ L ++ G R ++ +P + +G + E+T ++D + LET+L
Sbjct: 186 KSYQEGLARLPEMKPRAGTEIR----FSELPTQMFPEGATPAEITKHSMDLSYALETVLN 241
Query: 227 KDYGGSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIK 286
K + S +LGELQFAF+ ++G EAF WK L++LL +EA L+ I
Sbjct: 242 KQFPSSPQDVLGELQFAFVCFLLGNVYEAFEHWKRLLNLLCR-SEAAMMKHHTLYINLIS 300
Query: 287 VIYYQLKYGLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTR 346
++Y+QL P S D+FL + FFS +D L K
Sbjct: 301 ILYHQL----------GEIPADFFVDIVSQDNFLTSTLQVFFSSAC-SIAVDATLRKKAE 349
Query: 347 KFKKLLESNLGWEFQQNNAVDGLYFDENDEFAPVVEMLDD 386
KF+ L W+F E ++ APVV L +
Sbjct: 350 KFQAHLTKKFRWDFAA----------EPEDCAPVVVELPE 379
>YQQ2_CAEEL (Q09305) Hypothetical protein F10B5.2 in chromosome II
Length = 357
Score = 149 bits (377), Expect = 9e-36
Identities = 108/365 (29%), Positives = 171/365 (46%), Gaps = 36/365 (9%)
Query: 9 LVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTRDGKEFSPM 68
+ +NG LLFL PQ + ID + + G F G+KMIPPG HFVY S + +P
Sbjct: 13 MYRNGAFLLFLGFPQASEFGIDYKSWKTGEKFMGLKMIPPGVHFVYCSIKS------APR 66
Query: 69 IGFFIDAGPSEVIVRKWDQQEERL--VKVSEEEDERYRLAVKNMEFDRQLGPYNLSHYED 126
IGFF + E++V+KW+ + E +V ++ + +KNM D L PY +Y
Sbjct: 67 IGFFHNFKAGEILVKKWNTESETFEDEEVPTDQISEKKRQLKNM--DSSLAPYPYENYRS 124
Query: 127 WKRLSDFITKSIIERLEPIGGEVS-----VECENDMFRNAPKTPMEKALDTQLKVDNSAT 181
W L+DFIT +ER+ PI G ++ V E + NA K + ++ +N
Sbjct: 125 WYGLTDFITADTVERIHPILGRITSQAELVSLETEFMENAEKEHKDSHFRNRVDRENPV- 183
Query: 182 SVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQL-LE-----TLLVKDYGGSEDL 235
+ + + I ++ + I Q++ L + + ++ +E L++ GG
Sbjct: 184 ---RTRFTDQHGLPIMKIREGYEIRFQDIPPLTVSQNRVGIEYSDRLYRLLRALGGDWKQ 240
Query: 236 LLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIKVIYYQLKYG 295
LL E+Q AF+ + GQ E F QWK ++ L+ C + LF FI+V+++QLK
Sbjct: 241 LLAEMQIAFVCFLQGQVFEGFEQWKRIIH-LMSCCPNSLGSEKELFMSFIRVLFFQLK-- 297
Query: 296 LQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRKFKKLLESN 355
P S D+FL F+ V D + DL K T +FK+ L +
Sbjct: 298 --------ECPTDFFVDIVSRDNFLTTTLSMLFANVRDSAHAGDDLKKKTAQFKQYLTNQ 349
Query: 356 LGWEF 360
W+F
Sbjct: 350 FKWDF 354
>CT04_MOUSE (Q9D2V5) Protein c20orf4 homolog
Length = 384
Score = 146 bits (369), Expect = 8e-35
Identities = 114/402 (28%), Positives = 190/402 (46%), Gaps = 44/402 (10%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD + A +L G T++ L++P+ T ID + VGP F+G+KMIPPG HF+YYSS +
Sbjct: 6 MDPELAKQLFFEGATVVILNMPKGTEFGIDYNSWEVGPKFRGVKMIPPGIHFLYYSSVDK 65
Query: 61 -DGKEFSPMIGFFIDAGPSEVIVRKWD--QQEERLVKVSEEEDERYRLAVKNMEFDRQLG 117
+ +E P +GFF+ + V +W+ Q+E L E E E R + ++ D+ LG
Sbjct: 66 ANPREVGPRMGFFLSLKQRGLTVLRWNAVQEEVDLSPAPEAEVEAMRANLPDL--DQFLG 123
Query: 118 PYNLSHYEDWKRLSDFITKSIIERLEPIGGEVSV-----------ECENDMFRNAP--KT 164
PY + + W L++FI+++ +E+L+P ++ ++ + +N P T
Sbjct: 124 PYPYATLKKWISLTNFISEATMEKLQPESRQICAFSDVLPVLSMKHTKDRVGQNLPLCGT 183
Query: 165 PMEKALDTQLKVDNSATSVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETL 224
+ ++ G R ++ +P + G + E+T ++D + LET+
Sbjct: 184 ECRSYQEGLARLPEMRPRAGTEIR----FSELPTQMFPAGATPAEITRHSMDLSYALETV 239
Query: 225 LVKDYGGSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKF 284
L K + G+ +LGELQFAF+ ++G EAF WK L++LL +E+ L+
Sbjct: 240 LSKQFPGNPQDVLGELQFAFVCFLLGNVYEAFEHWKRLLNLLCR-SESAMGKYHALYISL 298
Query: 285 IKVIYYQLKYGLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKW 344
I ++Y+QL P S D+FL + FFS ++ L K
Sbjct: 299 ISILYHQL----------GEIPADFFVDIVSQDNFLTSTLQVFFSSAC-SIAVEATLRKK 347
Query: 345 TRKFKKLLESNLGWEFQQNNAVDGLYFDENDEFAPVVEMLDD 386
KF+ L W+F E ++ APVV L +
Sbjct: 348 AEKFQAHLTKKFRWDFT----------SEPEDCAPVVVELPE 379
>CT04_RABIT (O77682) Protein c20orf4 homolog
Length = 337
Score = 115 bits (288), Expect = 2e-25
Identities = 101/399 (25%), Positives = 167/399 (41%), Gaps = 85/399 (21%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD + A L G T++ L++P+ T ID + VGP F+G+KMIPPG HF++YS+ +
Sbjct: 6 MDPELAKCLFFEGATVVILNMPKGTEFGIDYNSWEVGPKFRGVKMIPPGIHFLHYSAVDK 65
Query: 61 DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGPYN 120
+ E D+ LGPY
Sbjct: 66 ANLQ----------------------------------------------ELDQFLGPYP 79
Query: 121 LSHYEDWKRLSDFITKSIIERLEPIGGEVSV-----------ECENDMFRNAPK--TPME 167
+ + W L+ FI+++ +ERL+P ++ ++ + +N P+ T +
Sbjct: 80 YATLKKWISLTSFISEATVERLQPESRQICAFSDVLPVLAMKHTKDRVGQNLPQCGTECK 139
Query: 168 KALDTQLKVDNSATSVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVK 227
+ ++ G R ++ +P + G + E+T ++D + LE +L +
Sbjct: 140 SYQEGLARLPEMKPRAGTEIR----FSELPTQMFPAGATPAEITRHSMDLSYALEMVLGR 195
Query: 228 DYGGSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIKV 287
+ GS +LGELQFAF+ ++G EAF WK L++LL +EA L+ I +
Sbjct: 196 QFPGSPQDVLGELQFAFVCFLLGNVYEAFEHWKRLLNLLCR-SEAAMVKHHALYVNLISI 254
Query: 288 IYYQLKYGLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRK 347
+Y+QL P S D+FL + FFS + +D L K K
Sbjct: 255 LYHQL----------GEIPADFFVDIVSQDNFLTSTLQVFFSSAC-STAVDATLRKKAEK 303
Query: 348 FKKLLESNLGWEFQQNNAVDGLYFDENDEFAPVVEMLDD 386
F+ L W+F E ++ APVV L +
Sbjct: 304 FQAHLTKKFRWDFAA----------EPEDCAPVVVELPE 332
>AAR2_YEAST (P32357) A1 cistron splicing factor AAR2
Length = 355
Score = 65.1 bits (157), Expect = 3e-10
Identities = 66/287 (22%), Positives = 125/287 (42%), Gaps = 24/287 (8%)
Query: 15 TLLFLDVPQYTLVAIDTQVFSV--GPTFKGIKMIPPG-THFVYY----SSSTRDGKEFSP 67
T+ F P + ID F+V F GIK IP G H +++ +SS R G F
Sbjct: 3 TVPFTSAPIEVTIGIDQYSFNVKENQPFHGIKDIPIGHVHVIHFQHADNSSMRYGYWFDC 62
Query: 68 MIG-FFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGPYNLSHYED 126
+G F+I P + + + ++++ + + ++ V + D YNL+ +
Sbjct: 63 RMGNFYIQYDPKDGLYKMMEERDGAKFENIVHNFKERQMMVSYPKIDEDDTWYNLTEFVQ 122
Query: 127 WKRLSDFITKSIIERLEPIGGEVSVECENDMFRNAPKTPMEKALDTQLKVDNSATSVGKL 186
++ + K + + ++ EN++ K+ ++KA ++++ N L
Sbjct: 123 MDKIRKIVRKDE-NQFSYVDSSMTTVQENELL----KSSLQKA-GSKMEAKNEDDPAHSL 176
Query: 187 QRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVKDYGGSEDLLLGELQFAFIA 246
YT I + G E+ LDK+ L T++++ + GELQFAF+
Sbjct: 177 N-----YTVINFKSREAIRPGHEMEDF-LDKSYYLNTVMLQGIFKNSSNYFGELQFAFLN 230
Query: 247 LMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIKVIYYQLK 293
M + + LQW +++ L+ P H + K +++YYQ+K
Sbjct: 231 AMFFGNYGSSLQWHAMIELICSSATVPKH----MLDKLDEILYYQIK 273
>CXA8_CHICK (P36381) Gap junction alpha-8 protein (Connexin 45.6)
(Cx45.6)
Length = 399
Score = 33.1 bits (74), Expect = 1.2
Identities = 28/105 (26%), Positives = 45/105 (42%), Gaps = 9/105 (8%)
Query: 20 DVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTRDGKEFSPMIGFF------I 73
+VP+ L AI S P KG K++ +Y T G E SP+ F I
Sbjct: 247 EVPEKPLHAI---AVSSIPKAKGYKLLEEEKPVSHYFPLTEVGVEPSPLPSAFNEFEEKI 303
Query: 74 DAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGP 118
GP E + R +D++ + E E+E+ + + + + Q P
Sbjct: 304 GMGPLEDLSRAFDERLPSYAQAKEPEEEKVKAEEEEEQEEEQQAP 348
>CIC2_HUMAN (P54289) Dihydropyridine-sensitive L-type, calcium
channel alpha-2/delta subunits precursor
Length = 1091
Score = 31.2 bits (69), Expect = 4.7
Identities = 24/102 (23%), Positives = 43/102 (41%), Gaps = 6/102 (5%)
Query: 260 KSLVSLLLGCTEAPFHTRTRLFTKFIKVIYYQLKYGLQKDRKDNTG------PLLLDDSW 313
K+LVSL L + + R +V+YY K L ++ D+ P+ ++D+
Sbjct: 95 KALVSLALEAEKVQAAHQWREDFASNEVVYYNAKDDLDPEKNDSEPGSQRIKPVFIEDAN 154
Query: 314 FSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRKFKKLLESN 355
F H + + +GS I + L WT ++ + N
Sbjct: 155 FGRQISYQHAAVHIPTDIYEGSTIVLNELNWTSALDEVFKKN 196
>SC71_SCHPO (Q9UT02) Protein transport protein sec71
Length = 1811
Score = 30.8 bits (68), Expect = 6.2
Identities = 20/72 (27%), Positives = 33/72 (45%), Gaps = 3/72 (4%)
Query: 125 EDWKRLSDFITKSIIERLEPIGGEVSVECENDMFRNAPKTPMEKALDTQLKVDNSATSVG 184
+D LSD + E ++ E+ ++ E D N P+ P L + N+ +VG
Sbjct: 860 DDGANLSDSFLTEVYEEIQK--NEIVLKDEQDPTSNFPEIPGTSNLSFAANISNALATVG 917
Query: 185 K-LQRKGCYYTS 195
+ LQR+ Y S
Sbjct: 918 RDLQREAYYMAS 929
>PPK2_CHLTE (O68984) Polyphosphate kinase 2 (EC 2.7.4.1)
(Polyphosphoric acid kinase 2) (ATP-polyphosphate
phosphotransferase 2)
Length = 714
Score = 30.8 bits (68), Expect = 6.2
Identities = 16/47 (34%), Positives = 29/47 (61%), Gaps = 1/47 (2%)
Query: 183 VGKL-QRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVKD 228
+G+L QR C++ I +K KGI ++SL++++ QLL+ K+
Sbjct: 106 IGQLRQRNACFFDDILPALKRKGIEFVSVSSLSVEQQQLLQHYFRKE 152
>Y310_BUCAP (Q8K9M4) Hypothetical metalloprotease BUsg310 precursor
(EC 3.4.24.-)
Length = 415
Score = 30.4 bits (67), Expect = 8.1
Identities = 21/82 (25%), Positives = 38/82 (45%), Gaps = 3/82 (3%)
Query: 46 IPPGTHFVYYSSSTRDGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRL 105
+P GT + SS +F+ + GF+I ++ ++ LVKV ++ + ++
Sbjct: 298 MPQGTPVIATSSGKIIKAQFNKIAGFYISLKNKNYYTTRYMHLKKILVKVGQKIKKGEKI 357
Query: 106 AVKNMEFDRQLGPYNLSHYEDW 127
A+ R GP+ HYE W
Sbjct: 358 ALSG-NTGRTTGPH--LHYEIW 376
>EP15_MOUSE (P42567) Epidermal growth factor receptor substrate 15
(Protein Eps15) (AF-1p protein)
Length = 897
Score = 30.4 bits (67), Expect = 8.1
Identities = 18/59 (30%), Positives = 31/59 (52%), Gaps = 6/59 (10%)
Query: 55 YSSSTRDGKEFSPMIGFFIDAGPSEVIVRKWD------QQEERLVKVSEEEDERYRLAV 107
++SST KE P A PSE + +W ++E+RL +++++E E LA+
Sbjct: 829 FTSSTTTNKEADPSNFANFSAYPSEEDMIEWAKRESEREEEQRLARLNQQEQEDLELAI 887
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,489,717
Number of Sequences: 164201
Number of extensions: 1986706
Number of successful extensions: 4826
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 4808
Number of HSP's gapped (non-prelim): 15
length of query: 391
length of database: 59,974,054
effective HSP length: 112
effective length of query: 279
effective length of database: 41,583,542
effective search space: 11601808218
effective search space used: 11601808218
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC129092.1


