

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC128638.4 - phase: 0
(561 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
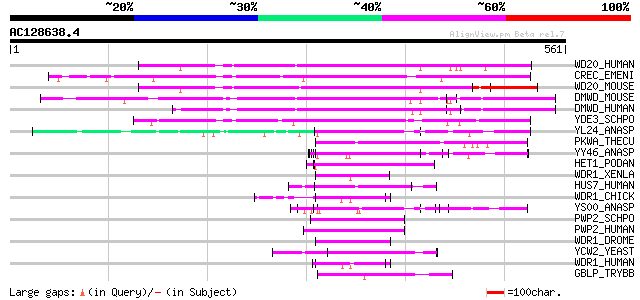
Score E
Sequences producing significant alignments: (bits) Value
WD20_HUMAN (Q8TBZ3) WD-repeat protein 20 (DMR protein) 234 6e-61
CREC_EMENI (Q9P4R5) Catabolite repression protein creC 221 4e-57
WD20_MOUSE (Q9D5R2) WD-repeat protein 20 214 5e-55
DMWD_MOUSE (Q08274) Dystrophia myotonica-containing WD repeat mo... 191 6e-48
DMWD_HUMAN (Q09019) Dystrophia myotonica-containing WD repeat mo... 186 1e-46
YDE3_SCHPO (Q10437) Hypothetical WD-repeat protein C12B10.03 in ... 184 4e-46
YL24_ANASP (Q8YV57) Hypothetical WD-repeat protein all2124 66 3e-10
PKWA_THECU (P49695) Probable serine/threonine-protein kinase pkw... 62 4e-09
YY46_ANASP (Q8YRI1) Hypothetical WD-repeat protein alr3466 60 2e-08
HET1_PODAN (Q00808) Vegetatible incompatibility protein HET-E-1 55 4e-07
WDR1_XENLA (Q9W7F2) WD-repeat protein 1 (Actin interacting prote... 54 1e-06
HUS7_HUMAN (Q9NVX2) WD-repeat protein HUSSY-07 54 1e-06
WDR1_CHICK (O93277) WD-repeat protein 1 (Actin interacting prote... 53 2e-06
YS00_ANASP (Q8YTC2) Hypothetical WD-repeat protein alr2800 52 3e-06
PWP2_SCHPO (Q9C1X1) Periodic tryptophan protein 2 homolog 52 4e-06
PWP2_HUMAN (Q15269) Periodic tryptophan protein 2 homolog 52 4e-06
WDR1_DROME (Q9VU68) Putative actin interacting protein 1 (AIP1) 52 5e-06
YCW2_YEAST (P25382) Hypothetical WD-repeat protein YCR072C 51 7e-06
WDR1_HUMAN (O75083) WD-repeat protein 1 (Actin interacting prote... 51 7e-06
GBLP_TRYBB (Q94775) Guanine nucleotide-binding protein beta subu... 51 7e-06
>WD20_HUMAN (Q8TBZ3) WD-repeat protein 20 (DMR protein)
Length = 569
Score = 234 bits (596), Expect = 6e-61
Identities = 175/530 (33%), Positives = 237/530 (44%), Gaps = 142/530 (26%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WV G + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVHGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S D E
Sbjct: 273 DGKYIVTGGEDDLVTVWSFVDCRVIARGHGHKSWVSVVAFDPYTTSVEEGDPMEFSGSDE 332
Query: 414 -------------------------------TITYRFGSVGQDTQLLLWELEMDEIV--V 440
++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQDLLHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQ 392
Query: 441 PLRR-----------GPPGGS-----PTFG---------------SPTFSAGSQSSHWDN 469
PL R PP GS T G S +AGS+SS D
Sbjct: 393 PLSRARTHTNVMNATSPPAGSNGNSVTTPGNSVPPPLPRSNSLPHSAVSNAGSKSSVMDG 452
Query: 470 AVPLGTLQPA-------------------------------------------------- 479
A+ G + A
Sbjct: 453 AIASGVSKFATLSLHDRKERHHEKDHKRNHSMGHISSKSSDKLNLVTKTKTDPAKTLGTP 512
Query: 480 --PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG
Sbjct: 513 LCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCIVTACQEGFICTWGRPG 562
>CREC_EMENI (Q9P4R5) Catabolite repression protein creC
Length = 630
Score = 221 bits (563), Expect = 4e-57
Identities = 165/540 (30%), Positives = 255/540 (46%), Gaps = 82/540 (15%)
Query: 40 EGRYKLQFD---KTHPSGLLQFNHGKTVSMVTLAHLKEKPAPLTPTASSSSFSASSGVRS 96
EG Y+LQ D T P H ++ L P P PT+ S G+R+
Sbjct: 87 EGTYQLQDDLHLATPPP------HPSEAPIINPNPLATVPNP--PTSGVKLSLISLGLRN 138
Query: 97 ---------AAARLLGGSNGNRALSFVGGNGSSKSNGGASRIGSIGSSSLSSSVANP--- 144
AR G +GN AL+ SK + I SS +S + +
Sbjct: 139 KTAFPSKVQVTARPFG--DGNSALAAAPVKDPSKKRKPKNNIIKSSSSFVSRVIIHEATT 196
Query: 145 ----NFDGKGSYLVFNAGDAILISDLNSQDKD-PIKSIHFSNSNPVCHAFDQDAKDGH-- 197
+ D +G + N A DL+S+ KD P+ I F+ ++ + H ++ K
Sbjct: 197 KRLNDRDPEGLFAFANINRAFQWLDLSSKHKDEPLSKILFTKAHMISHDVNEITKSSAHI 256
Query: 198 DLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCTCISWVPGGDGAFVVA 257
D+++G +GD++ + + +K NK+GI+N+S T I W+PG + F+ A
Sbjct: 257 DVIMGSSAGDIF------WYEPISQKYA---RINKNGIINSSPVTHIKWIPGSENFFIAA 307
Query: 258 HADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYS------------KSNPIARWHI 305
H +G L VY+K K+ A P L +Q+ SV +R+S K+NP+A W +
Sbjct: 308 HENGQLVVYDKEKEDA--LFIPELPEQSAESVKPSRWSLQVLKSVNSKNQKANPVAVWRL 365
Query: 306 CQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYIL 365
I +FS D +LA V DG LR+ DY +E ++ +SYYGG C WS DGKYI+
Sbjct: 366 ANQKITQFAFSPDHRHLAVVLEDGTLRLMDYLQEEVLDVFRSYYGGFTCVCWSPDGKYIV 425
Query: 366 TGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQD 425
TGG+DDLV +WS+ +RK++A +GH+SWVS VAFD + + TYR GSVG D
Sbjct: 426 TGGQDDLVTIWSLPERKIIARCQGHDSWVSAVAFDPW--------RCDERTYRIGSVGDD 477
Query: 426 TQLLLWELEM-----------------DEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWD 468
LLLW+ + ++ P + P + SQ + D
Sbjct: 478 CNLLLWDFSVGMLHRPKVHHQTSARHRTSLIAPSSQQPNRHRADSSGNRMRSDSQRTAAD 537
Query: 469 N-AVPLGTLQ-PAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRP 526
+ + P +Q P S + P+++ V +P+ L F +++++T+ EGHI+ W RP
Sbjct: 538 SESAPDQPVQHPVESRARTALLPPIMSKAVGEDPICWLGFQEDTIMTSSLEGHIRTWDRP 597
>WD20_MOUSE (Q9D5R2) WD-repeat protein 20
Length = 567
Score = 214 bits (545), Expect = 5e-55
Identities = 146/406 (35%), Positives = 201/406 (48%), Gaps = 63/406 (15%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G+ L F+ G + + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQTGNGNRLCFSVGRELYVYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKSNPIA 301
C+ WVPG D F+VAH+ GN+Y Y ++ G + +LK F+V + + SKS
Sbjct: 154 CVKWVPGSDSLFLVAHSSGNMYSYNVEHTYGTTAPHYQLLKQGESFAVHNCK-SKSTRDL 212
Query: 302 RWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDG 361
+W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS DG
Sbjct: 213 KWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSAELHGTMKSYFGGLLCLCWSPDG 272
Query: 362 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE-------- 413
KYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E
Sbjct: 273 KYIVTGGEDDLVTVWSFLDCRVIARGRGHKSWVSVVAFDPYTTSVEESDPMEFSDSDKNF 332
Query: 414 -----------------------------TITYRFGSVGQDTQLLLWELEMDEIV--VPL 442
++TYRFGSVGQDTQL LW+L D + PL
Sbjct: 333 QDLLHFGRDRANSTQSRLSKQNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQPL 392
Query: 443 RRGPPGGS--PTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVP 486
R + G P S GS + N+VP P P +P
Sbjct: 393 SRARAHANVMNATGLPAGSNGSAVTTPGNSVP----PPLPRSNSLP 434
Score = 56.2 bits (134), Expect = 2e-07
Identities = 29/67 (43%), Positives = 42/67 (62%), Gaps = 2/67 (2%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT P M DVP + PL+ ++ E L+ L+F ++ ++TAC+EG I W RPG
Sbjct: 502 DPAKTLGT-SLCPRMEDVPLLEPLICKKIAHERLTVLVFLEDCIVTACQEGFICTWARPG 560
Query: 528 -VAESQP 533
V++ QP
Sbjct: 561 KVSKFQP 567
>DMWD_MOUSE (Q08274) Dystrophia myotonica-containing WD repeat motif
protein (DMR-N9 protein)
Length = 650
Score = 191 bits (484), Expect = 6e-48
Identities = 149/472 (31%), Positives = 220/472 (46%), Gaps = 83/472 (17%)
Query: 32 LKTYFKTPEGRYKL-QFDKTHPSGLLQFNHGKTVSMVTLAHLKEKPAPLTPTASSSSFSA 90
+K+ F+T EG YKL D T SG T A P P P ++ S
Sbjct: 7 IKSQFRTREGFYKLLPGDATRRSG------------PTSAQTPAPPQPTQPPPGPAAASG 54
Query: 91 SSGVRSAAARLLGGSNGNRALSFV--------GGNGSSKSNGGASRIGSIGSSSLSSSVA 142
A++ G AL V +G+ + S +G+ G +
Sbjct: 55 PGAAGPASSPPPAGPGPGPALPAVRLSLVRLGDPDGAGEPPSTPSGLGAGGDRVCFNL-- 112
Query: 143 NPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFSNSNPVCHAFDQ--DAKDGHDLL 200
G L F G S + PI + + P CH F+Q A + LL
Sbjct: 113 -------GRELYFYPGCCRSGSQRSIDLNKPIDKRIYKGTQPTCHDFNQFTAATETISLL 165
Query: 201 IGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCTCISWVPGGDGAFVVAHAD 260
+G +G V + L ++ D K +N++ +++ ++ T + W+P + F+ +HA
Sbjct: 166 VGFSAGQVQYLDLIKK--DTSKL------FNEERLIDKTKVTYLKWLPESESLFLASHAS 217
Query: 261 GNLYVYEKNKDGAGES-SFPILKDQTLFSVAHARYSKS--NPIARWHICQGSINSISFSA 317
G+LY+Y + + +LK F+V +A SK+ NP+A+W + +G +N +FS
Sbjct: 218 GHLYLYNVSHPCTSTPPQYSLLKQGEGFAV-YAAKSKAPRNPLAKWAVGEGPLNEFAFSP 276
Query: 318 DGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWS 377
DG +LA V +DG LRVF + L KSY+GGLLC WS DG+Y++TGGEDDLV VWS
Sbjct: 277 DGRHLACVSQDGCLRVFHFDSMLLRGLMKSYFGGLLCVCWSPDGRYVVTGGEDDLVTVWS 336
Query: 378 MEDRKVVAWGEGHNSWVSGVAFDSYW--------SSPNSNDNGE---------------- 413
+ +VVA G GH SWV+ VAFD Y +S + + +GE
Sbjct: 337 FTEGRVVARGHGHKSWVNAVAFDPYTTRAEEAASASADGDPSGEEEEPEVTSSDTGAPVS 396
Query: 414 ------TITYRFGSVGQDTQLLLWELEMDEIVVP---LRR-----GPPGGSP 451
+ITYRFGS GQDTQ LW+L ++++ P L R G PG +P
Sbjct: 397 PLPKAGSITYRFGSAGQDTQFCLWDL-TEDVLSPHPSLARTRTLPGTPGATP 447
Score = 57.4 bits (137), Expect = 1e-07
Identities = 36/110 (32%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 442 LRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPL 501
+ RG GG+ + + S + S D A LGT P + +VP + PLV ++ E L
Sbjct: 526 ISRGGSGGNSS--NDKLSGPAPRSRLDPAKVLGTAL-CPRIHEVPLLEPLVCKKIAQERL 582
Query: 502 SSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSETLLATSLKEKPSLS 551
+ L+F ++ ++TAC+EG I W RPG A + S PS S
Sbjct: 583 TVLLFLEDCIITACQEGLICTWARPGKAFTDEETEAQAGQASWPRSPSKS 632
>DMWD_HUMAN (Q09019) Dystrophia myotonica-containing WD repeat motif
protein (DMR-N9 protein) (Protein 59) (Fragment)
Length = 553
Score = 186 bits (473), Expect = 1e-46
Identities = 122/339 (35%), Positives = 175/339 (50%), Gaps = 61/339 (17%)
Query: 165 DLNSQDKDPIKSIHFSNSNPVCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGK 222
DLN PI + + P CH F+Q A + LL+G +G V + L ++ D K
Sbjct: 28 DLNK----PIDKRIYKGTQPTCHDFNQFTAATETISLLVGFSAGQVQYLDLIKK--DTSK 81
Query: 223 KIVGAHHYNKDGILNNSRCTCISWVPGGDGAFVVAHADGNLYVYEKNKDGAGES-SFPIL 281
+N++ +++ ++ T + W+P + F+ +HA G+LY+Y + A + +L
Sbjct: 82 L------FNEERLIDKTKVTYLKWLPESESLFLASHASGHLYLYNVSHPCASAPPQYSLL 135
Query: 282 KDQTLFSVAHARYSKS--NPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKE 339
K FSV +A SK+ NP+A+W + +G +N +FS DG +LA V +D LRVF +
Sbjct: 136 KQGEGFSV-YAAKSKAPRNPLAKWAVGEGPLNEFAFSPDGRHLACVSQDACLRVFHFDSM 194
Query: 340 HLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
L KSY+GGLLC WS DG+Y++TGGEDDLV VWS + +VVA G GH SWV+ VAF
Sbjct: 195 LLRGLMKSYFGGLLCVCWSPDGRYVVTGGEDDLVTVWSFTEGRVVARGHGHKSWVNAVAF 254
Query: 400 DSYWSS-----------------------PNSNDNGE-------------TITYRFGSVG 423
DS +++ P + G +ITYRFGS G
Sbjct: 255 DSLYTTRAEEAATAAGADGERSGEEEEEEPEAAGTGSAGGAPLSPLPKAGSITYRFGSAG 314
Query: 424 QDTQLLLWELEMDEIV--VPLRR-----GPPGGSPTFGS 455
QDTQ LW+L D + PL R G PG +P S
Sbjct: 315 QDTQFCLWDLTEDVLYPHPPLARTRTLPGTPGTTPPAAS 353
Score = 57.0 bits (136), Expect = 1e-07
Identities = 38/110 (34%), Positives = 53/110 (47%), Gaps = 2/110 (1%)
Query: 442 LRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPL 501
+ RG GGS + G S S D A LGT P + +VP + PLV ++ E L
Sbjct: 428 ISRGGSGGSGSGGEKP-SGPVPRSRLDPAKVLGTAL-CPRIHEVPLLEPLVCKKIAQERL 485
Query: 502 SSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSETLLATSLKEKPSLS 551
+ L+F ++ ++TAC+EG I W RPG A + S PS S
Sbjct: 486 TVLLFLEDCIITACQEGLICTWPRPGKAFTDEETEAQTGEGSWPRSPSKS 535
>YDE3_SCHPO (Q10437) Hypothetical WD-repeat protein C12B10.03 in
chromosome I
Length = 543
Score = 184 bits (468), Expect = 4e-46
Identities = 138/442 (31%), Positives = 207/442 (46%), Gaps = 64/442 (14%)
Query: 126 ASRIGS--IGSSSLSSSVA---------NPNFDGKGSYLVFNAGDAILISDLNSQDK-DP 173
ASR G+ I + S SS+ P+ GK +++ N + D+++ +
Sbjct: 125 ASRAGAPLINAKSFISSLTFHDNIVRSFQPSIHGK-TFVFANHEKSFYWLDVSAANSHSA 183
Query: 174 IKSIHFSNSNPVCHAFDQDAKD--GHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYN 231
+ + F ++PVCH + K G D++IG +GDV + K + +N
Sbjct: 184 LLKMEFPRASPVCHDINSFTKSPKGLDVIIGFDTGDVL------WYDPINFKYL---RFN 234
Query: 232 KDGILNNSRCTCISWVPGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQT------ 285
K+G LN+S T I WV G D F+V+ +G L +Y+K + E I+ +
Sbjct: 235 KNGQLNSSSVTAIKWVAGKDSQFLVSFRNGWLVLYDKYRH---EQPLHIVVPEKNLKSLY 291
Query: 286 ---------LFSVAHARYSKSNPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDY 336
L S+ H K NP+A + + IN FS D YLA V G L++FD+
Sbjct: 292 LSSPGTFNILISINHRDDRKLNPVACYAFSKSPINGFCFSPDYQYLALVSERGTLKLFDF 351
Query: 337 TKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSG 396
KEH++ SY+ GL C WS DGK+I GG+DDLV ++S RK+VA +GH SWV+
Sbjct: 352 VKEHVLDVFHSYFAGLTCVTWSPDGKFIAIGGKDDLVSIYSFPLRKLVARCQGHKSWVTD 411
Query: 397 VAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWELEMDEIVVP------LRRGPPGGS 450
V FD+ W + N YR SVG D +LLLW+ + I P +
Sbjct: 412 VIFDA-WRCDDDN-------YRIASVGLDRKLLLWDFSVSAIHRPKSAVYYVNHHSNNSK 463
Query: 451 PTF------GSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSL 504
P G T + +S++ N T+ P S +P ISP+ + V PLSS+
Sbjct: 464 PAISDFDDVGDLTMGSEIDNSNYVNGDI--TIHPTLSRSLIPVISPITIYDVDDSPLSSV 521
Query: 505 IFTQESVLTACREGHIKIWTRP 526
F + ++T G I+ W RP
Sbjct: 522 FFDPDCMITCATNGRIRTWQRP 543
>YL24_ANASP (Q8YV57) Hypothetical WD-repeat protein all2124
Length = 1683
Score = 65.9 bits (159), Expect = 3e-10
Identities = 52/223 (23%), Positives = 100/223 (44%), Gaps = 32/223 (14%)
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
++N++ FS DG LA+ D ++++D T L+ + G++ +S DG+ I G
Sbjct: 1157 TVNNVYFSPDGKNLASASSDHSIKLWDTTSGQLLMTLTGHSAGVITVRFSPDGQTIAAGS 1216
Query: 369 EDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQL 428
ED V++W +D K++ GH WV+ ++F + +G+T+ S D +
Sbjct: 1217 EDKTVKLWHRQDGKLLKTLNGHQDWVNSLSF---------SPDGKTL----ASASADKTI 1263
Query: 429 LLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQ---SSHWDNAVPLGTLQPAPSMRDV 485
LW + ++V L+ G + + FS+ + S+ DN + L
Sbjct: 1264 KLWRIADGKLVKTLK----GHNDSVWDVNFSSDGKAIASASRDNTIKLWNRHGI------ 1313
Query: 486 PKISPLVAHRVHTEPLSSLIFTQES--VLTACREGHIKIWTRP 526
L H+ + ++ F +S + +A + I++W RP
Sbjct: 1314 ----ELETFTGHSGGVYAVNFLPDSNIIASASLDNTIRLWQRP 1352
Score = 45.8 bits (107), Expect = 3e-04
Identities = 77/428 (17%), Positives = 159/428 (36%), Gaps = 59/428 (13%)
Query: 24 TTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNHGKTVSMVTLAHLKEKPAPLTPTA 83
T +Q + + +P+G+ H L G+ + +T +T
Sbjct: 1150 TITGHEQTVNNVYFSPDGKNLASASSDHSIKLWDTTSGQLLMTLT----GHSAGVITVRF 1205
Query: 84 SSSSFSASSGVRSAAARLLGGSNGNRALSFVGGNGSSKSNGGASRIGSIGSSSLSSSVAN 143
S + ++G +L +G + NG + S+ S ++A+
Sbjct: 1206 SPDGQTIAAGSEDKTVKLWHRQDGKLLKTL---------NGHQDWVNSLSFSPDGKTLAS 1256
Query: 144 PNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFSNSNPVCHAFDQDAK----DGHDL 199
+ D + ++ D L+ L + D + ++FS+ + +D + H +
Sbjct: 1257 ASADK--TIKLWRIADGKLVKTLKGHN-DSVWDVNFSSDGKAIASASRDNTIKLWNRHGI 1313
Query: 200 LIGLF---SGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCTCISWVPGGDGAFVV 256
+ F SG VY+V+ L D I A N + + + + G G + V
Sbjct: 1314 ELETFTGHSGGVYAVNF---LPD-SNIIASASLDNTIRLWQRPLISPLEVLAGNSGVYAV 1369
Query: 257 -----------AHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAH-------ARYSKSN 298
A ADGN+ ++ ++DG+ + P ++ ++ ++ A +
Sbjct: 1370 SFLHDGSIIATAGADGNIQLWH-SQDGSLLKTLP--GNKAIYGISFTPQGDLIASANADK 1426
Query: 299 PIARWHICQGS-----------INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKS 347
+ W + G +N ++FS DG LA+ RD +++++ + K
Sbjct: 1427 TVKIWRVRDGKALKTLIGHDNEVNKVNFSPDGKTLASASRDNTVKLWNVSDGKFKKTLKG 1486
Query: 348 YYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPN 407
+ + ++S DGK I + D +++W ++ HN V V F+ S
Sbjct: 1487 HTDEVFWVSFSPDGKIIASASADKTIRLWDSFSGNLIKSLPAHNDLVYSVNFNPDGSMLA 1546
Query: 408 SNDNGETI 415
S +T+
Sbjct: 1547 STSADKTV 1554
Score = 43.5 bits (101), Expect = 0.001
Identities = 49/217 (22%), Positives = 90/217 (40%), Gaps = 32/217 (14%)
Query: 312 SISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDD 371
SIS S DG +A+ D ++++ L + + ++S DG+ I +GG D
Sbjct: 1077 SISISRDGQTIASGSLDKTIKLWS-RDGRLFRTLNGHEDAVYSVSFSPDGQTIASGGSDK 1135
Query: 372 LVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLW 431
+++W D ++ GH V+ V F SP+ + S D + LW
Sbjct: 1136 TIKLWQTSDGTLLKTITGHEQTVNNVYF-----SPDGKN--------LASASSDHSIKLW 1182
Query: 432 ELEMDEIVVPLRRGPPGGSPTFGSP---TFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKI 488
+ ++++ L G SP T +AGS+ D V L Q ++ +
Sbjct: 1183 DTTSGQLLMTLTGHSAGVITVRFSPDGQTIAAGSE----DKTVKLWHRQDGKLLKTL--- 1235
Query: 489 SPLVAHRVHTEPLSSLIFTQE--SVLTACREGHIKIW 523
H + ++SL F+ + ++ +A + IK+W
Sbjct: 1236 ------NGHQDWVNSLSFSPDGKTLASASADKTIKLW 1266
Score = 41.2 bits (95), Expect = 0.007
Identities = 30/154 (19%), Positives = 63/154 (40%), Gaps = 24/154 (15%)
Query: 292 ARYSKSNPIARWHICQGSINS-----------ISFSADGAYLATVGRDGYLRVFDYTKEH 340
A S+ N + W++ G +SFS DG +A+ D +R++D +
Sbjct: 1462 ASASRDNTVKLWNVSDGKFKKTLKGHTDEVFWVSFSPDGKIIASASADKTIRLWDSFSGN 1521
Query: 341 LVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFD 400
L+ ++ + ++ DG + + D V++W D ++ GH++ V +F
Sbjct: 1522 LIKSLPAHNDLVYSVNFNPDGSMLASTSADKTVKLWRSHDGHLLHTFSGHSNVVYSSSF- 1580
Query: 401 SYWSSPNSNDNGETITYRFGSVGQDTQLLLWELE 434
SP+ S +D + +W+++
Sbjct: 1581 ----SPDGR--------YIASASEDKTVKIWQID 1602
Score = 41.2 bits (95), Expect = 0.007
Identities = 19/73 (26%), Positives = 39/73 (53%), Gaps = 1/73 (1%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
+ S SFS DG Y+A+ D ++++ HL+ + G++ +S DGK +++G
Sbjct: 1575 VYSSSFSPDGRYIASASEDKTVKIWQIDG-HLLTTLPQHQAGVMSAIFSPDGKTLISGSL 1633
Query: 370 DDLVQVWSMEDRK 382
D ++W + ++
Sbjct: 1634 DTTTKIWRFDSQQ 1646
Score = 32.3 bits (72), Expect = 3.4
Identities = 23/82 (28%), Positives = 39/82 (47%), Gaps = 14/82 (17%)
Query: 351 GLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSND 410
G++ + S DG+ I +G D +++WS D ++ GH V V+F +
Sbjct: 1074 GVISISISRDGQTIASGSLDKTIKLWS-RDGRLFRTLNGHEDAVYSVSF---------SP 1123
Query: 411 NGETITYRFGSVGQDTQLLLWE 432
+G+TI S G D + LW+
Sbjct: 1124 DGQTI----ASGGSDKTIKLWQ 1141
>PKWA_THECU (P49695) Probable serine/threonine-protein kinase pkwA
(EC 2.7.1.37)
Length = 742
Score = 62.0 bits (149), Expect = 4e-09
Identities = 66/239 (27%), Positives = 103/239 (42%), Gaps = 30/239 (12%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYT--KEHLVCGGKSYYGGLLCCAWSMDGKYILTG 367
+ +++FS DGA LA+ D +R++D +E V G ++Y +L A+S DG + +G
Sbjct: 504 VRAVAFSPDGALLASGSDDATVRLWDVAAAEERAVFEGHTHY--VLDIAFSPDGSMVASG 561
Query: 368 GEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQ 427
D ++W++ A +GH +V VAF S S TI + G++
Sbjct: 562 SRDGTARLWNVATGTEHAVLKGHTDYVYAVAFSPDGSMVASGSRDGTIRLWDVATGKERD 621
Query: 428 LLLWELEMDEIVVPLRRGPPGGSPTFGSPTF-----SAGSQSSH-------WDNAV---P 472
+L E VV L P G GS + A ++ H W AV P
Sbjct: 622 VLQAPAEN---VVSLAFSPDGSMLVHGSDSTVHLWDVASGEALHTFEGHTDWVRAVAFSP 678
Query: 473 LGTLQPAPS------MRDVPKISPLVAHRVHTEPLSSLIFTQE--SVLTACREGHIKIW 523
G L + S + DV HTEP+ S+ F E ++ +A +G I+IW
Sbjct: 679 DGALLASGSDDRTIRLWDVAAQEEHTTLEGHTEPVHSVAFHPEGTTLASASEDGTIRIW 737
Score = 33.9 bits (76), Expect = 1.2
Identities = 16/44 (36%), Positives = 21/44 (47%)
Query: 356 AWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
A+S G + G D L+ VW + + EGH WV VAF
Sbjct: 466 AFSPGGSLLAGGSGDKLIHVWDVASGDELHTLEGHTDWVRAVAF 509
>YY46_ANASP (Q8YRI1) Hypothetical WD-repeat protein alr3466
Length = 1526
Score = 59.7 bits (143), Expect = 2e-08
Identities = 52/229 (22%), Positives = 96/229 (41%), Gaps = 34/229 (14%)
Query: 304 HICQGS---INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMD 360
+I QG +NS+ F+ DG+ LA+ D +R+++ +C + + + ++ D
Sbjct: 1194 YILQGHTSWVNSVVFNPDGSTLASGSSDQTVRLWEINSSKCLCTFQGHTSWVNSVVFNPD 1253
Query: 361 GKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFG 420
G + +G D V++W + K + +GH +WV+ VAF N +G + G
Sbjct: 1254 GSMLASGSSDKTVRLWDISSSKCLHTFQGHTNWVNSVAF---------NPDGSMLASGSG 1304
Query: 421 SVGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGS---QSSHWDNAVPLGTLQ 477
D + LWE+ + + + G + S TFS S D V L ++
Sbjct: 1305 ----DQTVRLWEISSSKCLHTFQ----GHTSWVSSVTFSPDGTMLASGSDDQTVRLWSIS 1356
Query: 478 PAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGH--IKIWT 524
L HT + S+IF+ + + A G +++W+
Sbjct: 1357 SGEC---------LYTFLGHTNWVGSVIFSPDGAILASGSGDQTVRLWS 1396
Score = 58.2 bits (139), Expect = 6e-08
Identities = 29/106 (27%), Positives = 56/106 (52%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
+ S+ FS DGA LA+ G D +R++D + + + + Y + +S +G + G
Sbjct: 1077 VRSVVFSPDGAMLASGGDDQIVRLWDISSGNCLYTLQGYTSWVRFLVFSPNGVTLANGSS 1136
Query: 370 DDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETI 415
D +V++W + +K + +GH +WV+ VAF ++ S +T+
Sbjct: 1137 DQIVRLWDISSKKCLYTLQGHTNWVNAVAFSPDGATLASGSGDQTV 1182
Score = 53.9 bits (128), Expect = 1e-06
Identities = 38/132 (28%), Positives = 61/132 (45%), Gaps = 17/132 (12%)
Query: 308 GSINSISFSADGAYLATVGRDGYLRVFDYT--KEHLVCGGKSYYGGLLCCAWSMDGKYIL 365
GS+ +++FS DG AT G +R ++ KE L C G + + + +S DGK +
Sbjct: 865 GSVLTVAFSPDGKLFATGDSGGIVRFWEAATGKELLTCKGHNSWVNSV--GFSQDGKMLA 922
Query: 366 TGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQD 425
+G +D V++W + + + +GH S V V F SPNS S D
Sbjct: 923 SGSDDQTVRLWDISSGQCLKTFKGHTSRVRSVVF-----SPNS--------LMLASGSSD 969
Query: 426 TQLLLWELEMDE 437
+ LW++ E
Sbjct: 970 QTVRLWDISSGE 981
Score = 53.1 bits (126), Expect = 2e-06
Identities = 45/216 (20%), Positives = 89/216 (40%), Gaps = 25/216 (11%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
+ + FS +G LA D +R++D + + + + + + A+S DG + +G
Sbjct: 1119 VRFLVFSPNGVTLANGSSDQIVRLWDISSKKCLYTLQGHTNWVNAVAFSPDGATLASGSG 1178
Query: 370 DDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLL 429
D V++W + K + +GH SWV+ V F N +G T+ S D +
Sbjct: 1179 DQTVRLWDISSSKCLYILQGHTSWVNSVVF---------NPDGSTL----ASGSSDQTVR 1225
Query: 430 LWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKIS 489
LWE+ + + + G + S F+ + + G+ + D+
Sbjct: 1226 LWEINSSKCLCTFQ----GHTSWVNSVVFNPDG------SMLASGSSDKTVRLWDISSSK 1275
Query: 490 PLVAHRVHTEPLSSLIFTQESVLTACREGH--IKIW 523
L + HT ++S+ F + + A G +++W
Sbjct: 1276 CLHTFQGHTNWVNSVAFNPDGSMLASGSGDQTVRLW 1311
Score = 52.4 bits (124), Expect = 3e-06
Identities = 37/144 (25%), Positives = 71/144 (48%), Gaps = 16/144 (11%)
Query: 303 WHICQGS---INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
++I QG + S+ FS+DGA LA+ D +R++D + + + + + + +S
Sbjct: 1025 FYIFQGHTSCVRSVVFSSDGAMLASGSDDQTVRLWDISSGNCLYTLQGHTSCVRSVVFSP 1084
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRF 419
DG + +GG+D +V++W + + +G+ SWV + F SPN +T
Sbjct: 1085 DGAMLASGGDDQIVRLWDISSGNCLYTLQGYTSWVRFLVF-----SPNG------VTLAN 1133
Query: 420 GSVGQDTQLLLWELEMDEIVVPLR 443
GS D + LW++ + + L+
Sbjct: 1134 GS--SDQIVRLWDISSKKCLYTLQ 1155
Score = 50.8 bits (120), Expect = 9e-06
Identities = 31/118 (26%), Positives = 59/118 (49%), Gaps = 9/118 (7%)
Query: 304 HICQGS---INSISFSADGAYLATVGRDGYLRVFDYTKEHLV---CGGKSYYGGLLCCAW 357
H QG ++S++FS DG LA+ D +R++ + + G ++ G ++ +
Sbjct: 1320 HTFQGHTSWVSSVTFSPDGTMLASGSDDQTVRLWSISSGECLYTFLGHTNWVGSVI---F 1376
Query: 358 SMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETI 415
S DG + +G D V++WS+ K + +GHN+WV + F + S + +T+
Sbjct: 1377 SPDGAILASGSGDQTVRLWSISSGKCLYTLQGHNNWVGSIVFSPDGTLLASGSDDQTV 1434
Score = 48.5 bits (114), Expect = 5e-05
Identities = 28/113 (24%), Positives = 53/113 (46%), Gaps = 3/113 (2%)
Query: 306 CQGS---INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGK 362
C+G +NS+ FS DG LA+ D +R++D + + K + + +S +
Sbjct: 902 CKGHNSWVNSVGFSQDGKMLASGSDDQTVRLWDISSGQCLKTFKGHTSRVRSVVFSPNSL 961
Query: 363 YILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETI 415
+ +G D V++W + + + +GH WV VAF+ S + +T+
Sbjct: 962 MLASGSSDQTVRLWDISSGECLYIFQGHTGWVYSVAFNLDGSMLATGSGDQTV 1014
Score = 47.0 bits (110), Expect = 1e-04
Identities = 26/106 (24%), Positives = 51/106 (47%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
+ S+ FS + LA+ D +R++D + + + + G + A+++DG + TG
Sbjct: 951 VRSVVFSPNSLMLASGSSDQTVRLWDISSGECLYIFQGHTGWVYSVAFNLDGSMLATGSG 1010
Query: 370 DDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETI 415
D V++W + + +GH S V V F S + S + +T+
Sbjct: 1011 DQTVRLWDISSSQCFYIFQGHTSCVRSVVFSSDGAMLASGSDDQTV 1056
Score = 46.2 bits (108), Expect = 2e-04
Identities = 32/136 (23%), Positives = 59/136 (42%), Gaps = 13/136 (9%)
Query: 308 GSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTG 367
G + S++F+ DG+ LAT D +R++D + + + + +S DG + +G
Sbjct: 991 GWVYSVAFNLDGSMLATGSGDQTVRLWDISSSQCFYIFQGHTSCVRSVVFSSDGAMLASG 1050
Query: 368 GEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQ 427
+D V++W + + +GH S V V F SP+ S G D
Sbjct: 1051 SDDQTVRLWDISSGNCLYTLQGHTSCVRSVVF-----SPDG--------AMLASGGDDQI 1097
Query: 428 LLLWELEMDEIVVPLR 443
+ LW++ + L+
Sbjct: 1098 VRLWDISSGNCLYTLQ 1113
Score = 44.7 bits (104), Expect = 7e-04
Identities = 34/137 (24%), Positives = 64/137 (45%), Gaps = 19/137 (13%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKE---HLVCGGKSYYGGLLCCAWSMDGKYILT 366
+ S+ FS DGA LA+ D +R++ + + + G ++ G ++ +S DG + +
Sbjct: 1371 VGSVIFSPDGAILASGSGDQTVRLWSISSGKCLYTLQGHNNWVGSIV---FSPDGTLLAS 1427
Query: 367 GGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDT 426
G +D V++W++ + + GH + V VAF S S + ETI
Sbjct: 1428 GSDDQTVRLWNISSGECLYTLHGHINSVRSVAFSSDGLILASGSDDETIK---------- 1477
Query: 427 QLLLWELEMDEIVVPLR 443
LW+++ E + L+
Sbjct: 1478 ---LWDVKTGECIKTLK 1491
>HET1_PODAN (Q00808) Vegetatible incompatibility protein HET-E-1
Length = 1356
Score = 55.5 bits (132), Expect = 4e-07
Identities = 33/122 (27%), Positives = 55/122 (45%)
Query: 308 GSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTG 367
G ++S++FS DG +A+ DG ++++D + + G + A+S DG+ + +G
Sbjct: 1136 GWVHSVAFSPDGQRVASGSIDGTIKIWDAASGTCTQTLEGHGGWVQSVAFSPDGQRVASG 1195
Query: 368 GEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQ 427
D +++W EGH WV VAF S + TI + G TQ
Sbjct: 1196 SSDKTIKIWDTASGTCTQTLEGHGGWVQSVAFSPDGQRVASGSSDNTIKIWDTASGTCTQ 1255
Query: 428 LL 429
L
Sbjct: 1256 TL 1257
Score = 51.2 bits (121), Expect = 7e-06
Identities = 31/121 (25%), Positives = 52/121 (42%)
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
S+ S++FS DG +A+ D ++++D + + G + A+S DG+ + +G
Sbjct: 969 SVLSVAFSPDGQRVASGSGDKTIKIWDTASGTCTQTLEGHGGSVWSVAFSPDGQRVASGS 1028
Query: 369 EDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQL 428
+D +++W EGH WV V F S + TI G TQ
Sbjct: 1029 DDKTIKIWDTASGTCTQTLEGHGGWVQSVVFSPDGQRVASGSDDHTIKIWDAVSGTCTQT 1088
Query: 429 L 429
L
Sbjct: 1089 L 1089
Score = 48.1 bits (113), Expect = 6e-05
Identities = 34/136 (25%), Positives = 57/136 (41%), Gaps = 7/136 (5%)
Query: 301 ARWHICQ-------GSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLL 353
A W+ C S+ S++FSADG +A+ D ++++D + + G +
Sbjct: 828 AEWNACTQTLEGHGSSVLSVAFSADGQRVASGSDDKTIKIWDTASGTGTQTLEGHGGSVW 887
Query: 354 CCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE 413
A+S D + + +G +D +++W EGH V VAF S +
Sbjct: 888 SVAFSPDRERVASGSDDKTIKIWDAASGTCTQTLEGHGGRVQSVAFSPDGQRVASGSDDH 947
Query: 414 TITYRFGSVGQDTQLL 429
TI + G TQ L
Sbjct: 948 TIKIWDAASGTCTQTL 963
Score = 46.6 bits (109), Expect = 2e-04
Identities = 32/122 (26%), Positives = 54/122 (44%)
Query: 308 GSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTG 367
GS+ S++FS D +A+ D ++++D + + G + A+S DG+ + +G
Sbjct: 884 GSVWSVAFSPDRERVASGSDDKTIKIWDAASGTCTQTLEGHGGRVQSVAFSPDGQRVASG 943
Query: 368 GEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQ 427
+D +++W EGH S V VAF S +TI + G TQ
Sbjct: 944 SDDHTIKIWDAASGTCTQTLEGHGSSVLSVAFSPDGQRVASGSGDKTIKIWDTASGTCTQ 1003
Query: 428 LL 429
L
Sbjct: 1004 TL 1005
Score = 46.2 bits (108), Expect = 2e-04
Identities = 31/122 (25%), Positives = 50/122 (40%)
Query: 308 GSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTG 367
G + S+ FS DG +A+ D ++++D + + + A+S DG+ + +G
Sbjct: 1052 GWVQSVVFSPDGQRVASGSDDHTIKIWDAVSGTCTQTLEGHGDSVWSVAFSPDGQRVASG 1111
Query: 368 GEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQ 427
D +++W EGH WV VAF S TI + G TQ
Sbjct: 1112 SIDGTIKIWDAASGTCTQTLEGHGGWVHSVAFSPDGQRVASGSIDGTIKIWDAASGTCTQ 1171
Query: 428 LL 429
L
Sbjct: 1172 TL 1173
Score = 44.3 bits (103), Expect = 9e-04
Identities = 31/122 (25%), Positives = 52/122 (42%)
Query: 308 GSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTG 367
GS+ S++FS DG +A+ D ++++D + + G + +S DG+ + +G
Sbjct: 1010 GSVWSVAFSPDGQRVASGSDDKTIKIWDTASGTCTQTLEGHGGWVQSVVFSPDGQRVASG 1069
Query: 368 GEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQ 427
+D +++W EGH V VAF S TI + G TQ
Sbjct: 1070 SDDHTIKIWDAVSGTCTQTLEGHGDSVWSVAFSPDGQRVASGSIDGTIKIWDAASGTCTQ 1129
Query: 428 LL 429
L
Sbjct: 1130 TL 1131
>WDR1_XENLA (Q9W7F2) WD-repeat protein 1 (Actin interacting protein
1) (XAIP1)
Length = 608
Score = 53.5 bits (127), Expect = 1e-06
Identities = 25/78 (32%), Positives = 41/78 (52%), Gaps = 3/78 (3%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVC---GGKSYYGGLLCCAWSMDGKYILT 366
+N + FS DG+ LA+ G DG + ++D VC G K++ GG+ +WS DG +L+
Sbjct: 192 VNCVRFSPDGSKLASAGADGQIFLYDGKTGEKVCSLGGSKAHDGGIYAVSWSPDGTQLLS 251
Query: 367 GGEDDLVQVWSMEDRKVV 384
D ++W + V
Sbjct: 252 ASGDKTTKIWDVAANSAV 269
Score = 35.0 bits (79), Expect = 0.52
Identities = 31/122 (25%), Positives = 50/122 (40%), Gaps = 29/122 (23%)
Query: 250 GDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKSNPIARWHICQGS 309
G G V ADG +++Y + LKD+ K+ P +G+
Sbjct: 456 GSGTVAVGGADGKVHLYSIQGNS--------LKDE----------GKTLP------AKGA 491
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYG---GLLCCAWSMDGKYILT 366
+ +++S DGA+LA + + VF + SYYG L AWS D ++ +
Sbjct: 492 VTDLAYSHDGAFLAVTDANKVVTVFSVADGY--SEKNSYYGHHAKALSVAWSPDNEHFAS 549
Query: 367 GG 368
G
Sbjct: 550 SG 551
Score = 31.2 bits (69), Expect = 7.5
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 2/57 (3%)
Query: 315 FSADGAYLATVGRDGYLRVFDYT-KEHLV-CGGKSYYGGLLCCAWSMDGKYILTGGE 369
++ G Y+A+ G LR++D T KEHL+ + + G + AW+ D K I GE
Sbjct: 66 YAPSGFYIASGDTSGKLRIWDTTQKEHLLKYEYQPFAGKIKDIAWTEDSKRIAVVGE 122
>HUS7_HUMAN (Q9NVX2) WD-repeat protein HUSSY-07
Length = 485
Score = 53.5 bits (127), Expect = 1e-06
Identities = 29/101 (28%), Positives = 56/101 (54%), Gaps = 5/101 (4%)
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKE--HLVCGGKSYYGGLLCCAWSMDGKYILT 366
++ S++FS G YLA+ D +R +D + E H C G ++ +L +WS DGK + +
Sbjct: 116 AVISVAFSPTGKYLASGSGDTTVRFWDLSTETPHFTCKGHRHW--VLSISWSPDGKKLAS 173
Query: 367 GGEDDLVQVWSMEDRKVVAWG-EGHNSWVSGVAFDSYWSSP 406
G ++ + +W K V GH+ W++G++++ ++P
Sbjct: 174 GCKNGQILLWDPSTGKQVGRTLAGHSKWITGLSWEPLHANP 214
Score = 48.5 bits (114), Expect = 5e-05
Identities = 38/149 (25%), Positives = 63/149 (41%), Gaps = 15/149 (10%)
Query: 283 DQTLFSVAHARYSKSNPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLV 342
D TLF + A K P+ R Q IN + FS D +A+ D ++++D +
Sbjct: 350 DFTLFLWSPAEDKK--PLTRMTGHQALINQVLFSPDSRIVASASFDKSIKLWDGRTGKYL 407
Query: 343 CGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSY 402
+ + + AWS D + +++G D ++VW ++ +K+ GH V V
Sbjct: 408 ASLRGHVAAVYQIAWSADSRLLVSGSSDSTLKVWDVKAQKLAMDLPGHADEVYAVD---- 463
Query: 403 WSSPNSNDNGETITYRFGSVGQDTQLLLW 431
WS R S G+D L +W
Sbjct: 464 WSPDGQ---------RVASGGKDKCLRIW 483
Score = 40.0 bits (92), Expect = 0.016
Identities = 22/84 (26%), Positives = 42/84 (49%), Gaps = 7/84 (8%)
Query: 321 YLATVGRDGYLRVFDYTK---EHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWS 377
Y+A+ +DG +R++D T E ++ G + + C W DG + + +D ++VW
Sbjct: 218 YVASSSKDGSVRIWDTTAGRCERILTG---HTQSVTCLRWGGDG-LLYSASQDRTIKVWR 273
Query: 378 MEDRKVVAWGEGHNSWVSGVAFDS 401
D + +GH WV+ +A +
Sbjct: 274 AHDGVLCRTLQGHGHWVNTMALST 297
>WDR1_CHICK (O93277) WD-repeat protein 1 (Actin interacting protein
1) (AIP1)
Length = 609
Score = 52.8 bits (125), Expect = 2e-06
Identities = 25/79 (31%), Positives = 40/79 (49%), Gaps = 3/79 (3%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVC---GGKSYYGGLLCCAWSMDGKYILT 366
+N + FS DG AT DG + ++D VC GGK++ GG+ +WS D +L+
Sbjct: 194 VNCVRFSPDGNRFATASADGQIFIYDGKTGEKVCALGGGKAHDGGIYAISWSPDSSQLLS 253
Query: 367 GGEDDLVQVWSMEDRKVVA 385
D ++W + VV+
Sbjct: 254 ASGDKTAKIWDVGANSVVS 272
Score = 49.3 bits (116), Expect = 3e-05
Identities = 40/136 (29%), Positives = 64/136 (46%), Gaps = 30/136 (22%)
Query: 248 PGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKSNPIARWHICQ 307
PGG G+ V DGN+ +Y + G S D+TL + +
Sbjct: 457 PGG-GSVAVGGTDGNVRLY--SIQGTSLKS----DDKTLEA------------------K 491
Query: 308 GSINSISFSADGAYLATVGRDGYLRVF---DYTKEHLVCGGKSYYGGLLCCAWSMDGKYI 364
G + +++S DGA+LA + + VF D EH V G ++ ++C AWS D ++
Sbjct: 492 GPVTDLAYSHDGAFLAVCDANKVVTVFSVPDGYVEHNVFYG--HHAKVVCIAWSPDNEHF 549
Query: 365 LTGGEDDLVQVWSMED 380
+GG D +V VW++ D
Sbjct: 550 ASGGMDMMVYVWTVSD 565
Score = 33.1 bits (74), Expect = 2.0
Identities = 30/104 (28%), Positives = 48/104 (45%), Gaps = 7/104 (6%)
Query: 315 FSADGAYLATVGRDGYLRVFDYT-KEHLV-CGGKSYYGGLLCCAWSMDGKYILTGGE--D 370
++ G Y+A+ G LR++D T KEHL+ + + G + AW+ D K I GE +
Sbjct: 68 YAPSGFYIASGDVSGKLRIWDTTQKEHLLKYEYQPFAGKIKDLAWTEDSKRIAVVGEGRE 127
Query: 371 DLVQVWSMEDRKVVAWGEGHNSWVSGVAFDS---YWSSPNSNDN 411
V+ + V GHN ++ V Y + S+DN
Sbjct: 128 KFGAVFLWDSGSSVGEITGHNKVINSVDIKQTRPYRLATGSDDN 171
>YS00_ANASP (Q8YTC2) Hypothetical WD-repeat protein alr2800
Length = 1258
Score = 52.4 bits (124), Expect = 3e-06
Identities = 39/129 (30%), Positives = 65/129 (50%), Gaps = 17/129 (13%)
Query: 308 GSINSISFSADGAYLATVGRDGYLRVFDYT--KEHLVCGGKSYYGGLLCCAWSMDGKYIL 365
G+I S +FS +G LAT D ++RV++ K L+C G S + + +S DG+ +
Sbjct: 643 GNILSAAFSPEGQLLATCDTDCHVRVWEVKSGKLLLICRGHSNWVRFV--VFSPDGEILA 700
Query: 366 TGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQD 425
+ G D+ V++WS+ D + GH V VAF + +GET+ S D
Sbjct: 701 SCGADENVKLWSVRDGVCIKTLTGHEHEVFSVAF---------HPDGETL----ASASGD 747
Query: 426 TQLLLWELE 434
+ LW+++
Sbjct: 748 KTIKLWDIQ 756
Score = 51.2 bits (121), Expect = 7e-06
Identities = 44/167 (26%), Positives = 78/167 (46%), Gaps = 24/167 (14%)
Query: 285 TLFSVAHARYSK-------SNPIARW----HICQGSIN-------SISFSADGAYLATVG 326
+++S+A++ SK I W HIC +++ S++FS DG LA V
Sbjct: 854 SVYSIAYSPDSKILVSGSGDRTIKLWDCQTHICIKTLHGHTNEVCSVAFSPDGQTLACVS 913
Query: 327 RDGYLRVFDYTKEHLVCGGKSYYGGL---LCCAWSMDGKYILTGGEDDLVQVWSMEDRKV 383
D +R+++ + K++YG L A+S D + + +G D V++W + K
Sbjct: 914 LDQSVRLWNCRTGQCL---KAWYGNTDWALPVAFSPDRQILASGSNDKTVKLWDWQTGKY 970
Query: 384 VAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLL 430
++ EGH ++ G+AF + S ++ S GQ Q+LL
Sbjct: 971 ISSLEGHTDFIYGIAFSPDSQTLASASTDSSVRLWNISTGQCFQILL 1017
Score = 47.8 bits (112), Expect = 8e-05
Identities = 50/222 (22%), Positives = 93/222 (41%), Gaps = 33/222 (14%)
Query: 308 GSINSISFSADGAYLATVGRDGYLRVFDY-TKEHLVCGGKSYYG---GLLCCAWSMDGKY 363
G + S++FSADG LA+ D +++++Y T E L K+Y G + A+S D K
Sbjct: 811 GWVRSVAFSADGQTLASGSGDRTIKIWNYHTGECL----KTYIGHTNSVYSIAYSPDSKI 866
Query: 364 ILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVG 423
+++G D +++W + + GH + V VAF + +G+T+ V
Sbjct: 867 LVSGSGDRTIKLWDCQTHICIKTLHGHTNEVCSVAF---------SPDGQTL----ACVS 913
Query: 424 QDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMR 483
D + LW + + P SP + S+ D V L Q +
Sbjct: 914 LDQSVRLWNCRTGQCLKAWYGNTDWALPVAFSPDRQILASGSN-DKTVKLWDWQTGKYIS 972
Query: 484 DVPKISPLVAHRVHTEPLSSLIFTQES--VLTACREGHIKIW 523
+ HT+ + + F+ +S + +A + +++W
Sbjct: 973 SL---------EGHTDFIYGIAFSPDSQTLASASTDSSVRLW 1005
Score = 46.2 bits (108), Expect = 2e-04
Identities = 38/163 (23%), Positives = 65/163 (39%), Gaps = 24/163 (14%)
Query: 292 ARYSKSNPIARWHICQGS-----------INSISFSADGAYLATVGRDGYLRVFDYTKEH 340
A S + W C G + S FS +G +AT D ++++D+ +
Sbjct: 1078 ASASADQSVRLWDCCTGRCVGILRGHSNRVYSAIFSPNGEIIATCSTDQTVKIWDWQQGK 1137
Query: 341 LVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFD 400
+ + + A+S DGK + + D V++W + K GH VS VAF
Sbjct: 1138 CLKTLTGHTNWVFDIAFSPDGKILASASHDQTVRIWDVNTGKCHHICIGHTHLVSSVAF- 1196
Query: 401 SYWSSPNSNDNGETITYRFGSVGQDTQLLLWELEMDEIVVPLR 443
+ +GE + S QD + +W ++ E + LR
Sbjct: 1197 --------SPDGEVV----ASGSQDQTVRIWNVKTGECLQILR 1227
Score = 45.1 bits (105), Expect = 5e-04
Identities = 25/104 (24%), Positives = 46/104 (44%)
Query: 312 SISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDD 371
S++F DG LA+ D ++++D + + + C A+S DG + + D
Sbjct: 731 SVAFHPDGETLASASGDKTIKLWDIQDGTCLQTLTGHTDWVRCVAFSPDGNTLASSAADH 790
Query: 372 LVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETI 415
+++W + K + + H WV VAF + + S TI
Sbjct: 791 TIKLWDVSQGKCLRTLKSHTGWVRSVAFSADGQTLASGSGDRTI 834
Score = 43.5 bits (101), Expect = 0.001
Identities = 22/90 (24%), Positives = 48/90 (52%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
+ ++FS DG LA+ D ++++D ++ + KS+ G + A+S DG+ + +G
Sbjct: 771 VRCVAFSPDGNTLASSAADHTIKLWDVSQGKCLRTLKSHTGWVRSVAFSADGQTLASGSG 830
Query: 370 DDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
D +++W+ + + GH + V +A+
Sbjct: 831 DRTIKIWNYHTGECLKTYIGHTNSVYSIAY 860
Score = 34.7 bits (78), Expect = 0.68
Identities = 31/162 (19%), Positives = 63/162 (38%), Gaps = 24/162 (14%)
Query: 292 ARYSKSNPIARWHICQGS-----------INSISFSADGAYLATVGRDGYLRVFDYTKEH 340
A S + + W+I G + ++ F G +AT D +++++ +
Sbjct: 994 ASASTDSSVRLWNISTGQCFQILLEHTDWVYAVVFHPQGKIIATGSADCTVKLWNISTGQ 1053
Query: 341 LVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFD 400
+ + +L AWS DG+ + + D V++W + V GH++ V F
Sbjct: 1054 CLKTLSEHSDKILGMAWSPDGQLLASASADQSVRLWDCCTGRCVGILRGHSNRVYSAIF- 1112
Query: 401 SYWSSPNSNDNGETITYRFGSVGQDTQLLLWELEMDEIVVPL 442
+ NGE I + D + +W+ + + + L
Sbjct: 1113 --------SPNGEII----ATCSTDQTVKIWDWQQGKCLKTL 1142
>PWP2_SCHPO (Q9C1X1) Periodic tryptophan protein 2 homolog
Length = 854
Score = 52.0 bits (123), Expect = 4e-06
Identities = 29/96 (30%), Positives = 52/96 (53%), Gaps = 1/96 (1%)
Query: 305 ICQGSINSISFSADGAYLAT-VGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKY 363
I Q +I++++ ++ G ++A + G L V+++ E V +S+Y L +S DG+
Sbjct: 294 ITQSNIDTLTVNSTGDWIAIGSSKLGQLLVWEWQSESYVLKQQSHYDALSTLQYSSDGQR 353
Query: 364 ILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
I+TG +D ++VW M + H S VSG+ F
Sbjct: 354 IITGADDGKIKVWDMNSGFCIVTFTQHTSAVSGLCF 389
Score = 42.7 bits (99), Expect = 0.002
Identities = 25/105 (23%), Positives = 50/105 (46%)
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILT 366
+G ++S+SF++ G+ LA+ D +R++D + +L A+ DGK +
Sbjct: 467 EGPVSSLSFNSSGSLLASGSWDKTVRIWDIFSRSGIVEPLPIPSDVLSLAFHPDGKEVCV 526
Query: 367 GGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDN 411
D + W++++ K + +G G FD ++ NS+ N
Sbjct: 527 ASLDGQLTFWNVQEGKQTSLIDGRKDLSGGRRFDDARTAENSSLN 571
>PWP2_HUMAN (Q15269) Periodic tryptophan protein 2 homolog
Length = 919
Score = 52.0 bits (123), Expect = 4e-06
Identities = 32/103 (31%), Positives = 54/103 (52%), Gaps = 1/103 (0%)
Query: 298 NPIARWHICQGSINSISFSADGAYLAT-VGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCA 356
N I I SI S++ ++ G ++A G L V+++ E V + ++ ++ A
Sbjct: 321 NLIHSLSISDQSIASVAINSSGDWIAFGCSGLGQLLVWEWQSESYVLKQQGHFNSMVALA 380
Query: 357 WSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
+S DG+YI+TGG+D V+VW+ H+S V+GV F
Sbjct: 381 YSPDGQYIVTGGDDGKVKVWNTLSGFCFVTFTEHSSGVTGVTF 423
Score = 38.9 bits (89), Expect = 0.036
Identities = 43/187 (22%), Positives = 72/187 (37%), Gaps = 41/187 (21%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYG----GLLCCAWSMDGKYIL 365
+ ++F+A G + T DG +R FD H +++ C A G+ +
Sbjct: 418 VTGVTFTATGYVVVTSSMDGTVRAFDL---HRYRNFRTFTSPRPTQFSCVAVDASGEIVS 474
Query: 366 TGGEDDL-VQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQ 424
G +D + VWSM+ +++ GH +SG+ F+ S S
Sbjct: 475 AGAQDSFEIFVWSMQTGRLLDVLSGHEGPISGLCFNPMKSV-------------LASASW 521
Query: 425 DTQLLLWE------------LEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWD--NA 470
D + LW+ L D + V R P G+ + SQ + WD NA
Sbjct: 522 DKTVRLWDMFDSWRTKETLALTSDALAVTFR---PDGAEL---AVATLNSQITFWDPENA 575
Query: 471 VPLGTLQ 477
V G+++
Sbjct: 576 VQTGSIE 582
Score = 33.1 bits (74), Expect = 2.0
Identities = 40/151 (26%), Positives = 61/151 (39%), Gaps = 23/151 (15%)
Query: 307 QGSINSISFSADG-AYLATVGRDGYLRVFDYTKE--HLVCGGKSYYG---GLLCCAWSMD 360
+GS++S+SFS DG ++ T G + K + K+Y+G C W+ D
Sbjct: 96 KGSVHSVSFSPDGRKFVVTKGNIAQMYHAPGKKREFNAFVLDKTYFGPYDETTCIDWTDD 155
Query: 361 GKYILTGGEDDLVQVWSME--DRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYR 418
+ + G +D V+ E D + GH + F+S NS D
Sbjct: 156 SRCFVVGSKDMSTWVFGAERWDNLIYYALGGHKDAIVACFFES-----NSLD-------- 202
Query: 419 FGSVGQDTQLLLWELEMDEIVVPLRRGPPGG 449
S+ QD L +W + D LR PP G
Sbjct: 203 LYSLSQDGVLCMW--QCDTPPEGLRLKPPAG 231
>WDR1_DROME (Q9VU68) Putative actin interacting protein 1 (AIP1)
Length = 608
Score = 51.6 bits (122), Expect = 5e-06
Identities = 24/78 (30%), Positives = 44/78 (55%), Gaps = 2/78 (2%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLV--CGGKSYYGGLLCCAWSMDGKYILTG 367
+ ++ +S DG + A+ G DG + ++D T LV G ++ GG+ AW D +LT
Sbjct: 198 VQAVRYSPDGKFFASAGFDGKVFLYDGTSSELVGEFGSPAHKGGVYALAWKPDSTQLLTC 257
Query: 368 GEDDLVQVWSMEDRKVVA 385
D ++W++E R++V+
Sbjct: 258 SGDKTCRLWTVESRELVS 275
>YCW2_YEAST (P25382) Hypothetical WD-repeat protein YCR072C
Length = 515
Score = 51.2 bits (121), Expect = 7e-06
Identities = 44/169 (26%), Positives = 72/169 (42%), Gaps = 18/169 (10%)
Query: 266 YEK--NKDGAGESSFPILKDQ-TLFSVAHARYSKSNPIARWHICQGSINSISFSADGAYL 322
YEK K+G E D T+F + +K PIAR Q +N ++FS DG Y+
Sbjct: 360 YEKICKKNGNSEEMMVTASDDYTMFLWNPLKSTK--PIARMTGHQKLVNHVAFSPDGRYI 417
Query: 323 ATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRK 382
+ D ++++D + + + + AWS D + +++ +D ++VW + RK
Sbjct: 418 VSASFDNSIKLWDGRDGKFISTFRGHVASVYQVAWSSDCRLLVSCSKDTTLKVWDVRTRK 477
Query: 383 VVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLW 431
+ GH V V WS R S G+D + LW
Sbjct: 478 LSVDLPGHKDEVYTVD----WSVDGK---------RVCSGGKDKMVRLW 513
Score = 47.0 bits (110), Expect = 1e-04
Identities = 26/113 (23%), Positives = 54/113 (47%), Gaps = 9/113 (7%)
Query: 322 LATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDR 381
+ T D R++D + + K +Y +LC +WS DG+ I TG D+ +++W +
Sbjct: 159 MVTGAGDNTARIWDCDTQTPMHTLKGHYNWVLCVSWSPDGEVIATGSMDNTIRLWDPKSG 218
Query: 382 KVVAWG-EGHNSWVSGVAFDS-YWSSPNSNDNGETITYRFGSVGQDTQLLLWE 432
+ + GH+ W++ ++++ + P S R S +D + +W+
Sbjct: 219 QCLGDALRGHSKWITSLSWEPIHLVKPGSKP-------RLASSSKDGTIKIWD 264
Score = 42.4 bits (98), Expect = 0.003
Identities = 21/76 (27%), Positives = 41/76 (53%), Gaps = 2/76 (2%)
Query: 302 RWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDG 361
R H+ S+ +++S+D L + +D L+V+D L + + WS+DG
Sbjct: 441 RGHVA--SVYQVAWSSDCRLLVSCSKDTTLKVWDVRTRKLSVDLPGHKDEVYTVDWSVDG 498
Query: 362 KYILTGGEDDLVQVWS 377
K + +GG+D +V++W+
Sbjct: 499 KRVCSGGKDKMVRLWT 514
>WDR1_HUMAN (O75083) WD-repeat protein 1 (Actin interacting protein
1) (AIP1) (NORI-1)
Length = 606
Score = 51.2 bits (121), Expect = 7e-06
Identities = 24/79 (30%), Positives = 40/79 (50%), Gaps = 3/79 (3%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVC---GGKSYYGGLLCCAWSMDGKYILT 366
+N + FS DG AT DG + ++D VC G K++ GG+ +WS D ++L+
Sbjct: 192 VNCVRFSPDGNRFATASADGQIYIYDGKTGEKVCALGGSKAHDGGIYAISWSPDSTHLLS 251
Query: 367 GGEDDLVQVWSMEDRKVVA 385
D ++W + VV+
Sbjct: 252 ASGDKTSKIWDVSVNSVVS 270
Score = 48.9 bits (115), Expect = 3e-05
Identities = 23/78 (29%), Positives = 44/78 (55%), Gaps = 7/78 (8%)
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFD----YTKEHLVCGGKSYYGGLLCCAWSMDGK 362
+G + +++S DGA+LA + VF Y++ ++ G ++ ++C AWS D +
Sbjct: 489 KGPVTDVAYSHDGAFLAVCDASKVVTVFSVADGYSENNVFYG---HHAKIVCLAWSPDNE 545
Query: 363 YILTGGEDDLVQVWSMED 380
+ +GG D +V VW++ D
Sbjct: 546 HFASGGMDMMVYVWTLSD 563
Score = 33.1 bits (74), Expect = 2.0
Identities = 30/104 (28%), Positives = 48/104 (45%), Gaps = 7/104 (6%)
Query: 315 FSADGAYLATVGRDGYLRVFDYT-KEHLV-CGGKSYYGGLLCCAWSMDGKYILTGGE--D 370
++ G Y+A+ G LR++D T KEHL+ + + G + AW+ D K I GE +
Sbjct: 66 YAPSGFYIASGDVSGKLRIWDTTQKEHLLKYEYQPFAGKIKDIAWTEDSKRIAVVGEGRE 125
Query: 371 DLVQVWSMEDRKVVAWGEGHNSWVSGVAFDS---YWSSPNSNDN 411
V+ + V GHN ++ V Y + S+DN
Sbjct: 126 KFGAVFLWDSGSSVGEITGHNKVINSVDIKQSRPYRLATGSDDN 169
>GBLP_TRYBB (Q94775) Guanine nucleotide-binding protein beta
subunit-like protein (Activated protein kinase C
receptor homolog) (Track)
Length = 318
Score = 51.2 bits (121), Expect = 7e-06
Identities = 37/139 (26%), Positives = 64/139 (45%), Gaps = 16/139 (11%)
Query: 312 SISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAW---SMDGKYILTGG 368
S++FS D + + GRD LRV++ E + + + + C S+D I++GG
Sbjct: 114 SVAFSPDNRQIVSGGRDNALRVWNVKGECMHTLSRGAHTDWVSCVRFSPSLDAPVIVSGG 173
Query: 369 EDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQL 428
D+LV+VW + ++V +GH ++V+ V S S+D +D
Sbjct: 174 WDNLVKVWDLATGRLVTDLKGHTNYVTSVTVSPDGSLCASSD-------------KDGVA 220
Query: 429 LLWELEMDEIVVPLRRGPP 447
LW+L E + + G P
Sbjct: 221 RLWDLTKGEALSEMAAGAP 239
Score = 42.7 bits (99), Expect = 0.002
Identities = 30/135 (22%), Positives = 59/135 (43%), Gaps = 12/135 (8%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
++ ++ S +G + + D LR+++ + +L A+S D + I++GG
Sbjct: 70 VSDVALSNNGNFAVSASWDHSLRLWNLQNGQCQYKFLGHTKDVLSVAFSPDNRQIVSGGR 129
Query: 370 DDLVQVWSMEDRKVVAWGEG-HNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQL 428
D+ ++VW+++ + G H WVS V F +P S G D +
Sbjct: 130 DNALRVWNVKGECMHTLSRGAHTDWVSCVRFSPSLDAP-----------VIVSGGWDNLV 178
Query: 429 LLWELEMDEIVVPLR 443
+W+L +V L+
Sbjct: 179 KVWDLATGRLVTDLK 193
Score = 37.4 bits (85), Expect = 0.10
Identities = 20/78 (25%), Positives = 37/78 (46%), Gaps = 8/78 (10%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVC-------GGKSYYGGLLCCAWSMDGK 362
IN I FS + ++ G +R+FD + ++ G K + AWS DG
Sbjct: 240 INQICFSPNRYWMCAATEKG-IRIFDLENKDIIVELAPEHQGSKKIVPECVSIAWSADGS 298
Query: 363 YILTGGEDDLVQVWSMED 380
+ +G D++++VW + +
Sbjct: 299 TLYSGYTDNVIRVWGVSE 316
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 73,885,573
Number of Sequences: 164201
Number of extensions: 3559453
Number of successful extensions: 12123
Number of sequences better than 10.0: 456
Number of HSP's better than 10.0 without gapping: 200
Number of HSP's successfully gapped in prelim test: 260
Number of HSP's that attempted gapping in prelim test: 9505
Number of HSP's gapped (non-prelim): 1999
length of query: 561
length of database: 59,974,054
effective HSP length: 115
effective length of query: 446
effective length of database: 41,090,939
effective search space: 18326558794
effective search space used: 18326558794
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 68 (30.8 bits)
Medicago: description of AC128638.4


