

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127429.4 + phase: 0
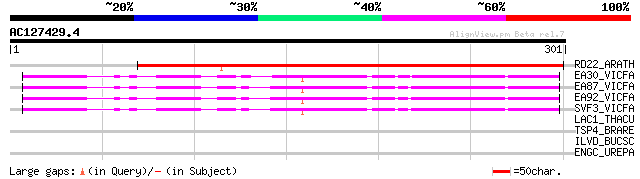
(301 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RD22_ARATH (Q08298) Dehydration-responsive protein RD22 precursor 241 1e-63
EA30_VICFA (P21745) Embryonic abundant protein VF30.1 precursor 105 1e-22
EA87_VICFA (P21746) Embryonic abundant protein USP87 precursor 105 2e-22
EA92_VICFA (P21747) Embryonic abundant protein USP92 precursor 103 5e-22
SVF3_VICFA (P09059) Unknown seed protein 30.1 precursor (VF30.1) 100 6e-21
LAC1_THACU (P56193) Laccase 1 precursor (EC 1.10.3.2) (Benzenedi... 32 2.5
TSP4_BRARE (Q8JGW0) Thrombospondin 4 precursor (Thbs4) 30 9.5
ILVD_BUCSC (Q9RQ52) Dihydroxy-acid dehydratase (EC 4.2.1.9) (DAD) 30 9.5
ENGC_UREPA (Q9PQS5) Probable GTPase engC (EC 3.6.1.-) 30 9.5
>RD22_ARATH (Q08298) Dehydration-responsive protein RD22 precursor
Length = 392
Score = 241 bits (616), Expect = 1e-63
Identities = 123/236 (52%), Positives = 159/236 (67%), Gaps = 5/236 (2%)
Query: 70 YFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHG--PTFLPGDVANSIPF 127
+ Y A E +LHD+ N A+F LEKDL G + N++F +G FLP A ++PF
Sbjct: 156 FVYNYAAKETQLHDDPNAALFFLEKDLVRGKEMNVRFNAEDGYGGKTAFLPRGEAETVPF 215
Query: 128 SSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGN 187
S K L FS++ GS E+E++K TI ECEA + GEEK C TSLESM+DF+ KLG
Sbjct: 216 GSEKFSETLKRFSVEAGSEEAEMMKKTIEECEARKVSGEEKYCATSLESMVDFSVSKLGK 275
Query: 188 -NVDTVSTEVNGESG-LQQYVIAN-GVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYS 244
+V VSTEV ++ +Q+Y IA GVKK+ ++ VVCHK+ YP+AVFYCHK T VY+
Sbjct: 276 YHVRAVSTEVAKKNAPMQKYKIAAAGVKKLSDDKSVVCHKQKYPFAVFYCHKAMMTTVYA 335
Query: 245 VPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFS 300
VPLEG +G R KAVA+CH +TS W+ HLAF+VLKV+ GT PVCH L + VVWFS
Sbjct: 336 VPLEGENGMRAKAVAVCHKNTSAWNPNHLAFKVLKVKPGTVPVCHFLPETHVVWFS 391
Score = 33.1 bits (74), Expect = 0.86
Identities = 14/31 (45%), Positives = 20/31 (64%)
Query: 10 LVATHATLPPELYWKSKLPTTQIPKAITDIL 40
+VA A L PE YW + LP T IP ++ ++L
Sbjct: 16 VVAIAADLTPERYWSTALPNTPIPNSLHNLL 46
>EA30_VICFA (P21745) Embryonic abundant protein VF30.1 precursor
Length = 268
Score = 105 bits (263), Expect = 1e-22
Identities = 89/297 (29%), Positives = 133/297 (43%), Gaps = 60/297 (20%)
Query: 8 LALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTRIVYDAA 67
LA V AT E YW+S P T +PK +D+L P +GKT ++
Sbjct: 14 LAFVGITATSSGEDYWQSIWPNTPLPKTFSDLLIP--------------SGKT----NSL 55
Query: 68 PIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPF 127
PI + + + + E DLH G F + G T G + PF
Sbjct: 56 PI----------KSEELKQYSTLFFEHDLHPGKNFIL--------GNTNSVGSIIR--PF 95
Query: 128 SSNKLENILNYFSIKQGSTESEIVKNTISE-----CEAYGIKGEEKLCVTSLESMIDFTT 182
+ ++ QG T+S + N + C + E K CV+SL+SMID
Sbjct: 96 TKSR-----------QGVTDSIWLANKEKQSLEDFCYSPTAIAEHKHCVSSLKSMIDQVI 144
Query: 183 LKLGNN-VDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATK 241
G+ + +S+ N QYV+ + VKK+G+N V+CH+ N+ VF CH+ T
Sbjct: 145 SHFGSTKIKAISS--NFAPYQDQYVVED-VKKVGDN-AVMCHRLNFEKVVFNCHQVRDTT 200
Query: 242 VYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
Y V L +DG++ KA+ +CH DT + L ++ L+V GT PVCH + W
Sbjct: 201 AYVVSLVASDGTKTKALTVCHHDTRGMN-PELLYEALEVTPGTVPVCHFIGNKAAAW 256
>EA87_VICFA (P21746) Embryonic abundant protein USP87 precursor
Length = 268
Score = 105 bits (261), Expect = 2e-22
Identities = 89/297 (29%), Positives = 133/297 (43%), Gaps = 60/297 (20%)
Query: 8 LALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTRIVYDAA 67
LA V AT P E YW+S P T +PK +D+L P +GKT ++
Sbjct: 14 LAFVGITATSPREDYWQSIWPNTPLPKTFSDMLIP--------------SGKT----NSL 55
Query: 68 PIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPF 127
PI + + + + E DLH F + G T G + PF
Sbjct: 56 PI----------KSEELKQYSTLFFEHDLHPRKNFIL--------GNTNSVGSIIR--PF 95
Query: 128 SSNKLENILNYFSIKQGSTESEIVKNTISE-----CEAYGIKGEEKLCVTSLESMIDFTT 182
+ + +QG T+S + N + C + E K CV+SL+SMID
Sbjct: 96 TKS-----------RQGVTDSIWLANKEKQSLEDFCYSPTAIAEHKHCVSSLKSMIDQVI 144
Query: 183 LKLGN-NVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATK 241
G+ + +S+ N QYV+ + VKK+G+ N V+CH+ N+ VF CH+ T
Sbjct: 145 SHFGSTKIKAISS--NFAPYQDQYVVED-VKKVGD-NAVMCHRLNFEKVVFNCHQVRDTT 200
Query: 242 VYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
Y V L +DG++ KA+ +CH DT + L ++ L+V GT PVCH + W
Sbjct: 201 AYVVSLVASDGTKTKALTVCHHDTRGMN-PELLYEALEVTLGTVPVCHFIGNKAAAW 256
>EA92_VICFA (P21747) Embryonic abundant protein USP92 precursor
Length = 268
Score = 103 bits (257), Expect = 5e-22
Identities = 88/297 (29%), Positives = 132/297 (43%), Gaps = 60/297 (20%)
Query: 8 LALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTRIVYDAA 67
LA V AT E YW+S P T +PK +D+L P +GKT ++
Sbjct: 14 LAFVGITATSSEEDYWQSIWPNTPLPKTFSDLLIP--------------SGKT----NSL 55
Query: 68 PIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPF 127
PI + + + + E DLH F + G T G + PF
Sbjct: 56 PI----------KSEELKQYSTLFFEHDLHPRKNFIL--------GNTNSVGSIIR--PF 95
Query: 128 SSNKLENILNYFSIKQGSTESEIVKNTISE-----CEAYGIKGEEKLCVTSLESMIDFTT 182
+ + +QG T+S + N + C + E K CV+SL+SMID
Sbjct: 96 TKS-----------RQGVTDSIWLANKEKQSFEDFCYSPTAIAEHKHCVSSLKSMIDQVI 144
Query: 183 LKLGN-NVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATK 241
G+ + +S+ N QYV+ + VKK+G+ N V+CH+ N+ VF CH+ T
Sbjct: 145 SHFGSTKIKAISS--NFAPYQDQYVVED-VKKVGD-NAVMCHRLNFEKVVFNCHQVRETT 200
Query: 242 VYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
Y V L +DG++ KA+ +CH DT + L ++ L+V GT PVCH + W
Sbjct: 201 AYVVSLVASDGTKTKALTVCHHDTRGMN-PELLYEALEVTPGTVPVCHFIGNKAAAW 256
>SVF3_VICFA (P09059) Unknown seed protein 30.1 precursor (VF30.1)
Length = 268
Score = 100 bits (248), Expect = 6e-21
Identities = 87/297 (29%), Positives = 131/297 (43%), Gaps = 60/297 (20%)
Query: 8 LALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTRIVYDAA 67
LA V AT E YW+S P T +PK +D+ P +GKT ++
Sbjct: 14 LAFVGITATSSGEDYWQSIWPNTPLPKTFSDLSIP--------------SGKT----NSL 55
Query: 68 PIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPF 127
PI + + + + E DLH F + G T G + PF
Sbjct: 56 PI----------KSEELKQYSTLFFEHDLHPRKNFIL--------GNTNSVGSIIR--PF 95
Query: 128 SSNKLENILNYFSIKQGSTESEIVKNTISE-----CEAYGIKGEEKLCVTSLESMIDFTT 182
+ + +QG T+S + N + C + E K CV+SL+SMID
Sbjct: 96 TKS-----------RQGVTDSIWLANKEKQSLEDFCYSPTAIAEHKHCVSSLKSMIDQVI 144
Query: 183 LKLGN-NVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATK 241
G+ + +S+ N QYV+ + VKK+G+ N V+CH+ N+ VF CH+ T
Sbjct: 145 SHFGSTKIKAISS--NFAPYQDQYVVED-VKKVGD-NAVMCHRLNFEKVVFNCHQVRDTT 200
Query: 242 VYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
Y V L +DG++ KA+ +CH DT + L ++ L+V GT PVCH + W
Sbjct: 201 AYVVSLVASDGTKTKALTVCHHDTRGMN-PELLYEALEVTLGTVPVCHFIGNKAAAW 256
>LAC1_THACU (P56193) Laccase 1 precursor (EC 1.10.3.2)
(Benzenediol:oxygen oxidoreductase) (Urishiol oxidase)
(Diphenol oxidase)
Length = 576
Score = 31.6 bits (70), Expect = 2.5
Identities = 21/74 (28%), Positives = 37/74 (49%), Gaps = 8/74 (10%)
Query: 123 NSIPFSSNKLENILNYFSIKQGSTESEIVK--NTI----SECEAYGIKGEEKLCVTSLES 176
N IP+ S K+ +L + + G TES+ K +T+ ++C + IKG + +T
Sbjct: 415 NGIPYESPKIPTLLKILTDEDGVTESDFTKEEHTVILPKNKCIEFNIKGNSGIPITHPVH 474
Query: 177 MIDFT--TLKLGNN 188
+ T ++ GNN
Sbjct: 475 LHGHTWDVVQFGNN 488
>TSP4_BRARE (Q8JGW0) Thrombospondin 4 precursor (Thbs4)
Length = 949
Score = 29.6 bits (65), Expect = 9.5
Identities = 15/38 (39%), Positives = 24/38 (62%), Gaps = 5/38 (13%)
Query: 130 NKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEE 167
N+L+++L Q E+ ++NTISEC+A G+ G E
Sbjct: 238 NELKDVLI-----QQVKETSFLRNTISECQACGLSGAE 270
>ILVD_BUCSC (Q9RQ52) Dihydroxy-acid dehydratase (EC 4.2.1.9) (DAD)
Length = 613
Score = 29.6 bits (65), Expect = 9.5
Identities = 16/30 (53%), Positives = 19/30 (63%), Gaps = 1/30 (3%)
Query: 42 PMDDGKISVDGK-ISVNGKTRIVYDAAPIY 70
PM+ G+I VDGK I +N IVY A P Y
Sbjct: 147 PMESGRIVVDGKEIKINLVDAIVYGANPNY 176
>ENGC_UREPA (Q9PQS5) Probable GTPase engC (EC 3.6.1.-)
Length = 302
Score = 29.6 bits (65), Expect = 9.5
Identities = 25/96 (26%), Positives = 44/96 (45%), Gaps = 10/96 (10%)
Query: 201 GLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAI 260
GL +Y +AN K N L++ +K+N YAVF K + + ++K+ +
Sbjct: 113 GLSKYDLANNHLKQKINKLILDYKKN-NYAVFILTKENDILLLK--------EKIKSKTL 163
Query: 261 CHSDTSQWSLKHLAFQV-LKVQQGTFPVCHILQQGQ 295
C + S L ++ +QQ T + H L +G+
Sbjct: 164 CLAGNSGVGKSTLINKLDPSIQQRTQEISHFLNRGK 199
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,989,018
Number of Sequences: 164201
Number of extensions: 1502595
Number of successful extensions: 3359
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 3339
Number of HSP's gapped (non-prelim): 13
length of query: 301
length of database: 59,974,054
effective HSP length: 110
effective length of query: 191
effective length of database: 41,911,944
effective search space: 8005181304
effective search space used: 8005181304
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC127429.4


