

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127429.2 + phase: 2 /pseudo
(500 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
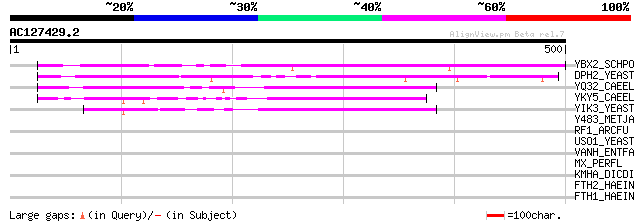
Score E
Sequences producing significant alignments: (bits) Value
YBX2_SCHPO (Q10206) Hypothetical protein C17D1.02 in chromosome II 236 8e-62
DPH2_YEAST (P32461) Diphteria toxin resistance protein 2 173 1e-42
YQ32_CAEEL (Q09454) Hypothetical protein C09G5.2 in chromosome II 141 3e-33
YKY5_CAEEL (P49958) Hypothetical protein C14B1.5 in chromosome III 99 2e-20
YIK3_YEAST (P40487) Hypothetical 48.3 kDa protein in MOB1-SGA1 i... 96 2e-19
Y483_METJA (Q57907) Hypothetical protein MJ0483 39 0.041
RF1_ARCFU (O29048) Peptide chain release factor subunit 1 (Trans... 35 0.59
USO1_YEAST (P25386) Intracellular protein transport protein USO1 32 2.9
VANH_ENTFA (Q47748) D-specific alpha-keto acid dehydrogenase (EC... 31 6.5
MX_PERFL (P20593) Interferon-induced GTP-binding protein Mx (Fra... 31 8.5
KMHA_DICDI (P42527) Myosin heavy chain kinase A (EC 2.7.1.129) (... 31 8.5
FTH2_HAEIN (P45219) Cell division protein ftsH homolog 2 (EC 3.4... 31 8.5
FTH1_HAEIN (P71377) Cell division protein ftsH homolog 1 (EC 3.4... 31 8.5
>YBX2_SCHPO (Q10206) Hypothetical protein C17D1.02 in chromosome II
Length = 503
Score = 236 bits (603), Expect = 8e-62
Identities = 139/486 (28%), Positives = 248/486 (50%), Gaps = 59/486 (12%)
Query: 26 QFPDDLLKDSTRVVSVLRKRLRSLRESDTTGSGDEPDIGLFVMADTAYGSCCVDEVGASH 85
QFPD+ L DS +V S+L + + + ++ADT YGSCCVDEV A H
Sbjct: 57 QFPDEHLADSGKVASILTNLVEA---------------NVQILADTNYGSCCVDEVAAEH 101
Query: 86 INADCVIHYGHTCFSPTTTLPAFFVFGKASIGLADCVESMSQYALTSSKPIMVLYGLEYA 145
++AD ++HYG C SPT+ LP +VFG+ I L + ++ + + I+++ +
Sbjct: 102 MSADAIVHYGRACLSPTSRLPVLYVFGRLPINLHKLEKCLT---IPLDQNILLVSDTRWY 158
Query: 146 HSIQQVRKALLESSTFCTSDPKSEVHFADVPSPYMFPSKDTKKLNDLQEETCGCGDNSSS 205
++ + K+L + + +V + S +++ EE
Sbjct: 159 YAQDSILKSL------------KTLGYQNV-----YESHLKERIEPNLEE---------- 191
Query: 206 DGASGAAYSIGGLTWKLLEGQSMEDYSLFWIGHDNSAFANVVLRFNAC--EIVRYDATKS 263
+ +Y+I G T+ L + S++D +L +IG D+ +++++ + + + +D +
Sbjct: 192 ---ASTSYTIPGRTYSLPKSLSLQDMTLLYIGPDSPTLSSILMSHYSLVNQFLSFDPLSN 248
Query: 264 QMVADLSQQRRILKRRYYLVERAKDANIVGILVGTLGVAGYLHIINQMMGLITAAGKKAY 323
++V + S L+RRY LV+R +DA ++GI++GTLGV YLH++NQ+ +I AGKK Y
Sbjct: 249 KIVEESSFTGAKLRRRYALVQRCRDAGVIGIVIGTLGVHRYLHVLNQLRKMILNAGKKPY 308
Query: 324 TLVMGKPNPAKLANFPECDVFLYVSCAQTALLDSKDYLAPVITPFEATIAFNRGSQWTGA 383
L +GK NPAKLANF E + F+ ++C + +L+DSK++ P++TPFE A + W
Sbjct: 309 MLAVGKLNPAKLANFQEIECFVLIACGENSLIDSKEFYRPIVTPFELVKALSSDMSWNND 368
Query: 384 YVMGFGDLINL---------PQVQVGNQEEARFSFLKGGYVEDFENQENNVEEEREDLAL 434
+++ F +++ L +V E FS + G +V + +V E D
Sbjct: 369 FILSFDEVLKLSEGKQSKEPSEVLTEESAEPHFSLITGKFVNSTPMRHLDVTLETADAKN 428
Query: 435 ANATEKALQLRDNCNSIIKGGAKSGAEFFANRTYQGLNIPDDNTTPQSFVIGRKGRAAGY 494
+++ +++ R + + G A F ++++ GL+ D+ P G+ G A GY
Sbjct: 429 NDSSSASIEKRGMRSLAVNGVYSPAAAFLQSKSWSGLDSVDEGEGPSKLYEGQSGIAKGY 488
Query: 495 EDENNK 500
E +K
Sbjct: 489 VGEGSK 494
>DPH2_YEAST (P32461) Diphteria toxin resistance protein 2
Length = 534
Score = 173 bits (438), Expect = 1e-42
Identities = 135/491 (27%), Positives = 225/491 (45%), Gaps = 59/491 (12%)
Query: 26 QFPDDLLKDSTRVVSVLRKRLRSLRESDTTGSGDEPDIGLFVMADTAYGSCCVDEVGASH 85
QFPDDL+KDS+ +V +L+ + I +V+ADTAY +CCVDEV A H
Sbjct: 68 QFPDDLIKDSSLIVRLLQSKF------------PHGKIKFWVLADTAYSACCVDEVAAEH 115
Query: 86 INADCVIHYGHTCFSPTTTLPAFFVFGKASIGLADCVESMSQYALTSSKPIMVLYGLEYA 145
++A+ V+H+G C + LP + FG + LA VE+ + S I ++ ++
Sbjct: 116 VHAEVVVHFGDACLNAIQNLPVVYSFGTPFLDLALVVENFQRAFPDLSSKICLMANAPFS 175
Query: 146 HSIQQVRKALLESSTFCTSDPKSEVHFADVPSPYM------FPSKDTKKLNDLQEETCGC 199
+ Q+ +L+ T+ S+V+ + V ++ +D ++ ++ +
Sbjct: 176 KHLSQLYN-ILKGDLHYTNIIYSQVNTSAVEEKFVTILDTFHVPEDVDQVGVFEKNSVLF 234
Query: 200 GDNSSSDGASGAAYSIGGLTWKLLEGQSMEDYSLFWIGHDNSAFANVVLRFNACEIVRYD 259
G + +D S Y + LT + +D L ++ ++ F +V + +D
Sbjct: 235 GQHDKADNISPEDYHLFHLT-------TPQDPRLLYL---STVFQSVHI---------FD 275
Query: 260 ATKSQMVADLSQQRRILKRRYYLVERAKDANIVGILVGTLGVAGYLHIINQMMGLITAAG 319
MV L RRY + A+ A +GILV TL + IN+++ LI
Sbjct: 276 PALPGMVTGPFPS---LMRRYKYMHVARTAGCIGILVNTLSLRNTRETINELVKLIKTRE 332
Query: 320 KKAYTLVMGKPNPAKLANFPECDVFLYVSCAQTALL--DSKDYLAPVITPFEATIAFNRG 377
KK Y V+GKPN AKLANF + D++ + C+Q+ ++ ++ P+ITP+E +A +
Sbjct: 333 KKHYLFVVGKPNVAKLANFEDIDIWCILGCSQSGIIVDQFNEFYKPIITPYELNLALSEE 392
Query: 378 SQWTGAYVMGFGDLINLPQVQVGNQ---------EEARFSFLKGGYVEDFENQENNVEEE 428
WTG +V+ F D I+ + +G Q +E F ++G Y E
Sbjct: 393 VTWTGKWVVDFRDAIDEIEQNLGGQDTISASTTSDEPEFDVVRGRYTSTSRPLRALTHLE 452
Query: 429 REDLALANATEKALQLRDNCN-SIIKGGAKSGAEFFANRTYQGLNIPDDNT----TPQSF 483
E A + K L R + ++IKG + A NR+++GL D+T T
Sbjct: 453 LE--AADDDDSKQLTTRHTASGAVIKGTVSTSASALQNRSWKGLGSDFDSTEVDNTGADI 510
Query: 484 VIGRKGRAAGY 494
G G A GY
Sbjct: 511 EEGISGVARGY 521
>YQ32_CAEEL (Q09454) Hypothetical protein C09G5.2 in chromosome II
Length = 476
Score = 141 bits (356), Expect = 3e-33
Identities = 106/368 (28%), Positives = 171/368 (45%), Gaps = 56/368 (15%)
Query: 26 QFPDDLLKDSTRVVSVLRKRLRSLRESDTTGSGDEPDIGLFVMADTAYGSCCVDEVGASH 85
QFPD LL S +V ++ R RS T FV+ADT+Y SCCVDEV A+H
Sbjct: 62 QFPDSLLPYSEKVTKLIESRFRSESAKKT-----------FVLADTSYRSCCVDEVAAAH 110
Query: 86 INADCVIHYGHTCFS-PTTTLPAFFVFGKASIGLADC-VESMSQYALTSSKPIMVLYGLE 143
+ ++H+G C S PT + +V G I + +E + +++ I++L
Sbjct: 111 ADCTALVHFGEACHSAPTDKIDVKYVLGSMPIFIDQFRMEFQNVADQLTAEHIVLLMDSC 170
Query: 144 YAHSIQQVRKALLESSTFCTSDPKSEVHFADVPSPYMFPSKDTKKLND----LQEETCGC 199
++H ++V + E V + + PS+D K N L E C
Sbjct: 171 FSHEQEKVAAVIKE-----VLPGNRHVECS------LLPSEDVLKQNRQNIYLGREIPSC 219
Query: 200 GDNSSSDGASGAAYSIGGLTWKLLEGQSMEDYSLFWIGHDNSAFANV-VLRFNACEIV-R 257
N+ + L + G NS + +L + +C V
Sbjct: 220 LRNN-------------------------QPTDLIFCGFPNSPLLPIWLLSYPSCSTVSH 254
Query: 258 YDATKSQMVADLSQQRRILKRRYYLVERAKDANIVGILVGTLGVAGYLHIINQMMGLITA 317
++ + + ++ R+L++R +LVE+ KDA+ VG++VG++GV + + +M +
Sbjct: 255 FNPINKTIQHESTRSSRLLRKRLFLVEKLKDADTVGLVVGSVGVDKHREAVKRMREMCKK 314
Query: 318 AGKKAYTLVMGKPNPAKLANF-PECDVFLYVSCAQTALLDSKDYLAPVITPFEATIAFNR 376
AGKK Y + +GK N KL+NF + DVF+ +SC +LDS DY PV++ FEA IA N
Sbjct: 315 AGKKIYVISVGKINVPKLSNFSTDIDVFVLLSCPFGVVLDSSDYFRPVVSYFEAEIALNP 374
Query: 377 GSQWTGAY 384
W +
Sbjct: 375 AKTWAADF 382
>YKY5_CAEEL (P49958) Hypothetical protein C14B1.5 in chromosome III
Length = 396
Score = 99.4 bits (246), Expect = 2e-20
Identities = 83/356 (23%), Positives = 150/356 (41%), Gaps = 66/356 (18%)
Query: 26 QFPDDLLKDSTRVVSVLRKRLRSLRESDTTGSGDEPDIGLFVMADTAYGSCCVDEVGASH 85
QFP+ L+ + + +L K DT +M D YG+CCVD+ A
Sbjct: 54 QFPEGLIMYACVIADILEK----YTGCDTV-----------IMGDVTYGACCVDDYTAKS 98
Query: 86 INADCVIHYGHTCFSP---TTTLPAFFVFGKASIGLA---DCVESMSQYALTSSKPIMVL 139
+ D ++HYGH+C P T + +VF I L+ DCV+ Q K ++V+
Sbjct: 99 MGCDLLVHYGHSCLVPIQNTDGIAMLYVFVNIHINLSHLIDCVKENFQ-----GKRLVVV 153
Query: 140 YGLEYAHSIQQVRKALLESSTFCTSDPKSEVHFADVPSPYMFPSKDTKKLNDLQEETCGC 199
+++ S+Q +R +TF D + D+P K L+ E GC
Sbjct: 154 STVQFIPSLQTLR------TTFNKDDSSIRI---DIPQ--------CKPLSP--GEVLGC 194
Query: 200 GDNSSSDGASGAAYSIGGLTWKLLEGQSMEDYSLFWIGHDNSAFANVVLRFNACEIVRYD 259
+S + Y ++ ++G ++++ E +YD
Sbjct: 195 ----TSPRLDASKYD-----------------AIVYLGDGRFHLESIMIHNPEIEAFQYD 233
Query: 260 ATKSQMVADLSQQRRILKRRYYLVERAKDANIVGILVGTLGVAGYLHIINQMMGLITAAG 319
++ + + K R +E A+ G++ GTLG G L ++ ++ + G
Sbjct: 234 PYSRKLTREFYDHDLMRKNRIGSIEIARKCTTFGLIQGTLGRQGNLKVVEELEAQLERKG 293
Query: 320 KKAYTLVMGKPNPAKLANFPECDVFLYVSCAQTALLDSKDYLAPVITPFEATIAFN 375
KK +++ + P KLA FPE D ++ V+C + ++ + P++ PFE +A +
Sbjct: 294 KKFLRVLLSEIFPEKLAMFPEVDCWVQVACPRLSIDWGTQFPKPLLYPFELAVALD 349
>YIK3_YEAST (P40487) Hypothetical 48.3 kDa protein in MOB1-SGA1
intergenic region
Length = 425
Score = 95.9 bits (237), Expect = 2e-19
Identities = 69/321 (21%), Positives = 138/321 (42%), Gaps = 44/321 (13%)
Query: 67 VMADTAYGSCCVDEVGASHINADCVIHYGHTCFSP--TTTLPAFFVFGKASIGLADCVES 124
VM D +YG+CC+D+ A ++ D ++HY H+C P T + +VF +I +++
Sbjct: 124 VMGDVSYGACCIDDFTARALDCDFIVHYAHSCLVPIDVTKIKVLYVFVTINIQEDHIIKT 183
Query: 125 MSQYALTSSKPIMVLYGLEYAHSIQQVRKALLESSTFCTSDPKSEVHFADVPSPYMFPSK 184
+ + S+ I +++ ++ VR LL E H + P + P
Sbjct: 184 LQKNFPKGSR-IATFGTIQFNPAVHSVRDKLLND----------EEHMLYIIPPQIKPLS 232
Query: 185 DTKKLNDLQEETCGCGDNSSSDGASGAAYSIGGLTWKLLEGQSMEDY-SLFWIGHDNSAF 243
+ E GC E E Y ++ +IG
Sbjct: 233 --------RGEVLGCTS----------------------ERLDKEQYDAMVFIGDGRFHL 262
Query: 244 ANVVLRFNACEIVRYDATKSQMVADLSQQRRILKRRYYLVERAKDANIVGILVGTLGVAG 303
+ ++ +YD + + Q+++++ R +E A+ + G+++G LG G
Sbjct: 263 ESAMIHNPEIPAFKYDPYNRKFTREGYDQKQLVEVRAEAIEVARKGKVFGLILGALGRQG 322
Query: 304 YLHIINQMMGLITAAGKKAYTLVMGKPNPAKLANFPECDVFLYVSCAQTALLDSKDYLAP 363
L+ + + + AAGK +++ + P KLA F + DVF+ V+C + ++ + P
Sbjct: 323 NLNTVKNLEKNLIAAGKTVVKIILSEVFPQKLAMFDQIDVFVQVACPRLSIDWGYAFNKP 382
Query: 364 VITPFEATIAFNRGSQWTGAY 384
++TP+EA++ + ++ Y
Sbjct: 383 LLTPYEASVLLKKDVMFSEKY 403
>Y483_METJA (Q57907) Hypothetical protein MJ0483
Length = 340
Score = 38.5 bits (88), Expect = 0.041
Identities = 17/58 (29%), Positives = 32/58 (54%), Gaps = 6/58 (10%)
Query: 62 DIGLFVMADTAYGSCCVDEVGASHINADCVIHYGHTCFS------PTTTLPAFFVFGK 113
+I +++ +T +G+C + + ++N D +IHYGH S T +PA+ +F K
Sbjct: 69 NIEIYLWGNTCFGACDLIDNHVKNLNVDLIIHYGHEKLSYANPEIKTLFIPAYHIFNK 126
>RF1_ARCFU (O29048) Peptide chain release factor subunit 1
(Translation termination factor aRF1)
Length = 407
Score = 34.7 bits (78), Expect = 0.59
Identities = 51/235 (21%), Positives = 88/235 (36%), Gaps = 48/235 (20%)
Query: 254 EIVRYDATKSQMVADLSQQRRILKRRYYLVERAKDANIVGILVGTLGVAGYLHIINQMM- 312
E++ + +AD+S Q R E ++ +NI T +AG I+N++
Sbjct: 25 ELITLYIPPDKNIADVSNQLRS--------ELSQASNIKSKQTRTNVLAGIEAILNRLKH 76
Query: 313 -------------GLITAAGKKAY-TLVMGKPNPAKLANFPECDVFLYVSCAQTALLDSK 358
G++ GK+ + T ++ P P L + CD Y+ + L + K
Sbjct: 77 FRKPPENGMVIISGVVNINGKEKHITEIIEPPEPVPLYKY-HCDSKFYLDPLKEMLTEKK 135
Query: 359 DYLAPVITPFEATIAFNRGSQ-----WTGAYVMGFGDLINLPQVQVGNQEEARFSFLKGG 413
Y V+ EAT+ +G + W + V G + + G Q RF L+
Sbjct: 136 LYGLIVLDRREATVGLLKGKRIEVLDWDTSMVPG--------KHRQGGQSSVRFERLREI 187
Query: 414 YVEDFENQENNVEEEREDLALANATEKALQLRDNCNSIIKGGAKSGAEFFANRTY 468
+ +F + + A+E L +D I+ GG E F Y
Sbjct: 188 AIHEFYKKVGEM-----------ASEALLPYKDKLIGILIGGPSPTKEEFYEGEY 231
>USO1_YEAST (P25386) Intracellular protein transport protein USO1
Length = 1790
Score = 32.3 bits (72), Expect = 2.9
Identities = 21/64 (32%), Positives = 36/64 (55%), Gaps = 2/64 (3%)
Query: 122 VESMSQYALTSSKPIMVLYGLEYAHSI-QQVRK-ALLESSTFCTSDPKSEVHFADVPSPY 179
V + Q AL S +MV+ L + H+I ++VR ALL ++ S+ +++ F+ + PY
Sbjct: 319 VTTKHQNALLDSSVLMVVLRLAFFHNIPKKVRPVALLTAANMVRSNEHAQLEFSKIDVPY 378
Query: 180 MFPS 183
PS
Sbjct: 379 FDPS 382
>VANH_ENTFA (Q47748) D-specific alpha-keto acid dehydrogenase (EC
1.1.1.-) (Vancomycin B-type resistance protein vanHB)
Length = 323
Score = 31.2 bits (69), Expect = 6.5
Identities = 38/156 (24%), Positives = 66/156 (41%), Gaps = 11/156 (7%)
Query: 207 GASGAAYSIGGLTWKL--LEGQSMEDYSLFWIGHDNSAFANVV-LRFNACEIVRYDATKS 263
GA +++ ++L + G+ + D ++ IG + A V LR C ++ YD ++
Sbjct: 122 GAKSTIHAVAQQNFRLDCVRGKELRDMTVGVIGTGHIGQAVVKRLRGFGCRVLAYDNSR- 180
Query: 264 QMVADLSQQRRILKRRYYLVER----AKDANIVGIL-VGTLGVAGYLHIINQMMGLITAA 318
++ AD Q +LK + A +++G +G + +L IN G +
Sbjct: 181 KIEADYVQLDELLKNSDIVTLHVPLCADTRHLIGQRQIGEMKQGAFL--INTGRGALVDT 238
Query: 319 GKKAYTLVMGKPNPAKLANFPECDVFLYVSCAQTAL 354
G L GK A L D F+Y C+Q L
Sbjct: 239 GSLVEALGSGKLGGAALDVLEGEDQFVYTDCSQKVL 274
>MX_PERFL (P20593) Interferon-induced GTP-binding protein Mx
(Fragment)
Length = 301
Score = 30.8 bits (68), Expect = 8.5
Identities = 26/102 (25%), Positives = 39/102 (37%), Gaps = 11/102 (10%)
Query: 105 LPAFFVFGKASIGLADCVESMSQYALTSSKPIMVLYGLEYAHSIQQVRKALLESSTFCTS 164
LPA V G S G + +E++S AL I+ LE +Q +
Sbjct: 35 LPAIAVIGDQSSGKSSVLEALSGVALPRGSGIVTRCPLELKMKRRQEGEEWY-------- 86
Query: 165 DPKSEVHFADVPSPYMFPSKDTKKLNDLQEETCGCGDNSSSD 206
++ + D P+ KK+ D Q+E G G S D
Sbjct: 87 ---GKISYQDCEKEIEDPADVEKKIRDAQDEMAGVGVGISDD 125
>KMHA_DICDI (P42527) Myosin heavy chain kinase A (EC 2.7.1.129)
(MHCK A)
Length = 1146
Score = 30.8 bits (68), Expect = 8.5
Identities = 14/57 (24%), Positives = 30/57 (52%)
Query: 425 VEEEREDLALANATEKALQLRDNCNSIIKGGAKSGAEFFANRTYQGLNIPDDNTTPQ 481
VE++++ LA T++ + + + ++KGG G + ++ G NI +T+ Q
Sbjct: 142 VEQQQQQLAKRLITQQIQEKKSTSSPLVKGGISGGGGSGGDDSFDGANISSMSTSKQ 198
>FTH2_HAEIN (P45219) Cell division protein ftsH homolog 2 (EC
3.4.24.-)
Length = 381
Score = 30.8 bits (68), Expect = 8.5
Identities = 40/178 (22%), Positives = 71/178 (39%), Gaps = 13/178 (7%)
Query: 257 RYDATKSQMVADLSQQRRILKRRYYLVERAKDANIVGILVGTLGVAG--YLHIINQMMGL 314
R+D + D+ + +ILK V A+D + + + GT G +G +++N+
Sbjct: 57 RFDRQVVVGLPDVKGREQILKVHMRKVSVAQDVDAMTLARGTPGYSGADLANLVNEAALF 116
Query: 315 ITAAGKKAYTLVMGKPNPAKLANFPECDVFLYVSCAQ--TALLDS----KDYLAPVITPF 368
K+ T++ + K+ PE + + TA ++ YL P P
Sbjct: 117 AARVNKRTVTMLEFEKAKDKINMGPERRTMIMTDKQKESTAYHEAGHAIVGYLVPEHDPV 176
Query: 369 EATIAFNRGSQWTGAYVMGFGDLINLPQVQVGNQEEARFSFLKGG-YVEDFENQENNV 425
RG + + GD I++ Q Q+ E++ S L G ED E N+
Sbjct: 177 HKVTIIPRGRALGVTFFLPEGDQISISQKQL----ESKLSTLYAGRLAEDLIYGEENI 230
>FTH1_HAEIN (P71377) Cell division protein ftsH homolog 1 (EC
3.4.24.-)
Length = 635
Score = 30.8 bits (68), Expect = 8.5
Identities = 40/178 (22%), Positives = 71/178 (39%), Gaps = 13/178 (7%)
Query: 257 RYDATKSQMVADLSQQRRILKRRYYLVERAKDANIVGILVGTLGVAG--YLHIINQMMGL 314
R+D + D+ + +ILK V A+D + + + GT G +G +++N+
Sbjct: 311 RFDRQVVVGLPDVKGREQILKVHMRKVSVAQDVDAMTLARGTPGYSGADLANLVNEAALF 370
Query: 315 ITAAGKKAYTLVMGKPNPAKLANFPECDVFLYVSCAQ--TALLDS----KDYLAPVITPF 368
K+ T++ + K+ PE + + TA ++ YL P P
Sbjct: 371 AARVNKRTVTMLEFEKAKDKINMGPERRTMIMTDKQKESTAYHEAGHAIVGYLVPEHDPV 430
Query: 369 EATIAFNRGSQWTGAYVMGFGDLINLPQVQVGNQEEARFSFLKGG-YVEDFENQENNV 425
RG + + GD I++ Q Q+ E++ S L G ED E N+
Sbjct: 431 HKVTIIPRGRALGVTFFLPEGDQISISQKQL----ESKLSTLYAGRLAEDLIYGEENI 484
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 59,295,777
Number of Sequences: 164201
Number of extensions: 2529577
Number of successful extensions: 5675
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 5654
Number of HSP's gapped (non-prelim): 18
length of query: 500
length of database: 59,974,054
effective HSP length: 114
effective length of query: 386
effective length of database: 41,255,140
effective search space: 15924484040
effective search space used: 15924484040
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 68 (30.8 bits)
Medicago: description of AC127429.2


