

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127169.5 + phase: 0 /pseudo
(550 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
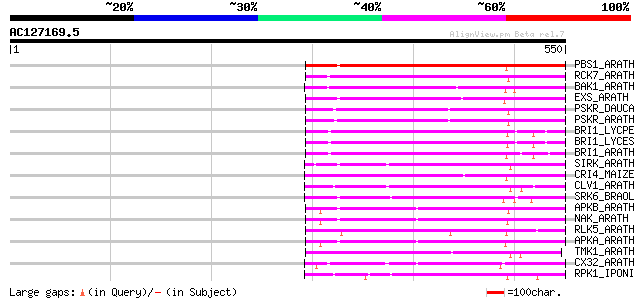
Score E
Sequences producing significant alignments: (bits) Value
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 196 1e-49
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 192 2e-48
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 189 2e-47
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 176 2e-43
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 170 8e-42
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 167 5e-41
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 167 5e-41
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 167 7e-41
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 167 9e-41
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 166 1e-40
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 166 2e-40
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 165 2e-40
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 164 4e-40
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 163 9e-40
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 163 1e-39
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 160 1e-38
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 159 2e-38
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 148 4e-35
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 145 3e-34
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 140 1e-32
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 196 bits (498), Expect = 1e-49
Identities = 103/264 (39%), Positives = 161/264 (60%), Gaps = 9/264 (3%)
Query: 294 GGYSEVYKGDL-CNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
GG+ VYKG L G+ +AVK+L ++ N+E FL+E+ ++ + HPN +L+GYC +
Sbjct: 95 GGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNRE--FLVEVLMLSLLHHPNLVNLIGYCAD 152
Query: 353 NGL-YLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
L++ + G+L LH +LDW +R KIA G A+GL +LH +I+R
Sbjct: 153 GDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKIAAGAAKGLEFLHDKANPPVIYR 212
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D K+SN+LL + P+++DFGLAK P H V GT+GY APE M G + K+D
Sbjct: 213 DFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRVMGTYGYCAPEYAMTGQLTVKSD 272
Query: 472 IFAFGVLLLEIVTGRRPVDS----SKQNILLWAKPLM-ESGNIAELADPRMEGRYDVEQL 526
+++FGV+ LE++TGR+ +DS +QN++ WA+PL + +LADPR++GR+ L
Sbjct: 273 VYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFNDRRKFIKLADPRLKGRFPTRAL 332
Query: 527 YRVVLTASYCVRQTAIWRPAMTEV 550
Y+ + AS C+++ A RP + +V
Sbjct: 333 YQALAVASMCIQEQAATRPLIADV 356
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 192 bits (488), Expect = 2e-48
Identities = 102/264 (38%), Positives = 160/264 (59%), Gaps = 9/264 (3%)
Query: 294 GGYSEVYKGDLCN-GETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
GG+ +V+KG + + +A+K+L D +EF++E+ + HPN L+G+C E
Sbjct: 112 GGFGKVFKGTIEKLDQVVAIKQL--DRNGVQGIREFVVEVLTLSLADHPNLVKLIGFCAE 169
Query: 353 NGL-YLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
L++ Y G+L LH LDW R KIA G ARGL YLH +I+R
Sbjct: 170 GDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMKIAAGAARGLEYLHDRMTPPVIYR 229
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D+K SN+LLG DY+P+++DFGLAK P+ H V GT+GY AP+ M G + K+D
Sbjct: 230 DLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSD 289
Query: 472 IFAFGVLLLEIVTGRRPVDSSK----QNILLWAKPLM-ESGNIAELADPRMEGRYDVEQL 526
I++FGV+LLE++TGR+ +D++K QN++ WA+PL + N ++ DP ++G+Y V L
Sbjct: 290 IYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPLFKDRRNFPKMVDPLLQGQYPVRGL 349
Query: 527 YRVVLTASYCVRQTAIWRPAMTEV 550
Y+ + ++ CV++ RP +++V
Sbjct: 350 YQALAISAMCVQEQPTMRPVVSDV 373
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 189 bits (480), Expect = 2e-47
Identities = 103/265 (38%), Positives = 158/265 (58%), Gaps = 9/265 (3%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
RGG+ +VYKG L +G +AVKRL K+ + E +F E+ +I H N L G+C
Sbjct: 297 RGGFGKVYKGRLADGTLVAVKRL-KEERTQGGELQFQTEVEMISMAVHRNLLRLRGFCMT 355
Query: 353 -NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
L++ Y NG++++ L ++ LDWP R +IA+G ARGL YLH C +IIHR
Sbjct: 356 PTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIALGSARGLAYLHDHCDPKIIHR 415
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D+KA+N+LL ++E + DFGLAK + K T H V GT G++APE G EKTD
Sbjct: 416 DVKAANILLDEEFEAVVGDFGLAKLMDYKDT-HVTTAVRGTIGHIAPEYLSTGKSSEKTD 474
Query: 472 IFAFGVLLLEIVTGRRPVD----SSKQNILL--WAKPLMESGNIAELADPRMEGRYDVEQ 525
+F +GV+LLE++TG+R D ++ +++L W K L++ + L D ++G Y E+
Sbjct: 475 VFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKKLEALVDVDLQGNYKDEE 534
Query: 526 LYRVVLTASYCVRQTAIWRPAMTEV 550
+ +++ A C + + + RP M+EV
Sbjct: 535 VEQLIQVALLCTQSSPMERPKMSEV 559
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 176 bits (445), Expect = 2e-43
Identities = 102/263 (38%), Positives = 146/263 (54%), Gaps = 9/263 (3%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYC-FE 352
GG+ VYK L +T+AVK+L++ N+E F+ E+ +G V HPN SLLGYC F
Sbjct: 926 GGFGTVYKACLPGEKTVAVKKLSEAKTQGNRE--FMAEMETLGKVKHPNLVSLLGYCSFS 983
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L++ Y NG+L L LDW R KIA+G ARGL +LH IIHRD
Sbjct: 984 EEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARGLAFLHHGFIPHIIHRD 1043
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKASN+LL D+EP++ DFGLA+ + +H + + + GTFGY+ PE K D+
Sbjct: 1044 IKASNILLDGDFEPKVADFGLARLISACESHVSTV-IAGTFGYIPPEYGQSARATTKGDV 1102
Query: 473 FAFGVLLLEIVTGRRPV-----DSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLY 527
++FGV+LLE+VTG+ P +S N++ WA + G ++ DP +
Sbjct: 1103 YSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQGKAVDVIDPLLVSVALKNSQL 1162
Query: 528 RVVLTASYCVRQTAIWRPAMTEV 550
R++ A C+ +T RP M +V
Sbjct: 1163 RLLQIAMLCLAETPAKRPNMLDV 1185
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 170 bits (431), Expect = 8e-42
Identities = 95/262 (36%), Positives = 145/262 (55%), Gaps = 8/262 (3%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYC-FE 352
GG+ VYK L +G +A+KRL+ D ++EF E+ + HPN LLGYC ++
Sbjct: 752 GGFGLVYKATLPDGTKVAIKRLSGDTGQ--MDREFQAEVETLSRAQHPNLVHLLGYCNYK 809
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
N LI++Y NG+L LH SLDW R +IA G A GL YLH+ C+ I+HRD
Sbjct: 810 NDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGAAEGLAYLHQSCEPHILHRD 869
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IK+SN+LL + + DFGLA+ L + H + GT GY+ PE + K D+
Sbjct: 870 IKSSNILLSDTFVAHLADFGLAR-LILPYDTHVTTDLVGTLGYIPPEYGQASVATYKGDV 928
Query: 473 FAFGVLLLEIVTGRRPVDSSK----QNILLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
++FGV+LLE++TGRRP+D K ++++ W + +E+ DP + + E++
Sbjct: 929 YSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRESEIFDPFIYDKDHAEEMLL 988
Query: 529 VVLTASYCVRQTAIWRPAMTEV 550
V+ A C+ + RP ++
Sbjct: 989 VLEIACRCLGENPKTRPTTQQL 1010
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 167 bits (424), Expect = 5e-41
Identities = 91/262 (34%), Positives = 148/262 (55%), Gaps = 8/262 (3%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCF-E 352
GG+ VYK L +G+ +A+K+L+ D E+EF E+ + HPN L G+CF +
Sbjct: 743 GGFGMVYKATLPDGKKVAIKKLSGDCGQI--EREFEAEVETLSRAQHPNLVLLRGFCFYK 800
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
N LI++Y +NG+L LH L W R +IA G A+GL YLH+ C I+HRD
Sbjct: 801 NDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGAAKGLLYLHEGCDPHILHRD 860
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IK+SN+LL ++ + DFGLA+ L + + H + GT GY+ PE + K D+
Sbjct: 861 IKSSNILLDENFNSHLADFGLAR-LMSPYETHVSTDLVGTLGYIPPEYGQASVATYKGDV 919
Query: 473 FAFGVLLLEIVTGRRPVDSSK----QNILLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
++FGV+LLE++T +RPVD K ++++ W + +E+ DP + + + ++++R
Sbjct: 920 YSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKMKHESRASEVFDPLIYSKENDKEMFR 979
Query: 529 VVLTASYCVRQTAIWRPAMTEV 550
V+ A C+ + RP ++
Sbjct: 980 VLEIACLCLSENPKQRPTTQQL 1001
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 167 bits (424), Expect = 5e-41
Identities = 98/264 (37%), Positives = 150/264 (56%), Gaps = 11/264 (4%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYC-FE 352
GG+ +VYK L +G +A+K+L + ++EF E+ IG + H N LLGYC
Sbjct: 897 GGFGDVYKAQLKDGSVVAIKKLI--HVSGQGDREFTAEMETIGKIKHRNLVPLLGYCKVG 954
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L++ Y + G+L LH K G L+WP R KIAIG ARGL +LH C IIHRD
Sbjct: 955 EERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARRKIAIGAARGLAFLHHNCIPHIIHRD 1014
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
+K+SNVLL + E +++DFG+A+ + TH +V + GT GY+ PE + K D+
Sbjct: 1015 MKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDV 1074
Query: 473 FAFGVLLLEIVTGRRPVDSS---KQNILLWAKPLMESGNIAELADPRM---EGRYDVEQL 526
+++GV+LLE++TG++P DS+ N++ W K L G I ++ D + + ++E L
Sbjct: 1075 YSYGVVLLELLTGKQPTDSADFGDNNLVGWVK-LHAKGKITDVFDRELLKEDASIEIELL 1133
Query: 527 YRVVLTASYCVRQTAIWRPAMTEV 550
+ + A C+ RP M +V
Sbjct: 1134 QHLKV-ACACLDDRHWKRPTMIQV 1156
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 167 bits (423), Expect = 7e-41
Identities = 98/264 (37%), Positives = 150/264 (56%), Gaps = 11/264 (4%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYC-FE 352
GG+ +VYK L +G +A+K+L + ++EF E+ IG + H N LLGYC
Sbjct: 897 GGFGDVYKAQLKDGSVVAIKKLI--HVSGQGDREFTAEMETIGKIKHRNLVPLLGYCKVG 954
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L++ Y + G+L LH K G L+WP R KIAIG ARGL +LH C IIHRD
Sbjct: 955 EERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARRKIAIGAARGLAFLHHNCIPHIIHRD 1014
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
+K+SNVLL + E +++DFG+A+ + TH +V + GT GY+ PE + K D+
Sbjct: 1015 MKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDV 1074
Query: 473 FAFGVLLLEIVTGRRPVDSS---KQNILLWAKPLMESGNIAELADPRM---EGRYDVEQL 526
+++GV+LLE++TG++P DS+ N++ W K L G I ++ D + + ++E L
Sbjct: 1075 YSYGVVLLELLTGKQPTDSADFGDNNLVGWVK-LHAKGKITDVFDRELLKEDASIEIELL 1133
Query: 527 YRVVLTASYCVRQTAIWRPAMTEV 550
+ + A C+ RP M +V
Sbjct: 1134 QHLKV-ACACLDDRHWKRPTMIQV 1156
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 167 bits (422), Expect = 9e-41
Identities = 97/264 (36%), Positives = 153/264 (57%), Gaps = 11/264 (4%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYC-FE 352
GG+ +VYK L +G +A+K+L + ++EF+ E+ IG + H N LLGYC
Sbjct: 892 GGFGDVYKAILKDGSAVAIKKLI--HVSGQGDREFMAEMETIGKIKHRNLVPLLGYCKVG 949
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
+ L++ + + G+L LH KAG L+W R KIAIG ARGL +LH C IIHRD
Sbjct: 950 DERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIAIGSARGLAFLHHNCSPHIIHRD 1009
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
+K+SNVLL + E +++DFG+A+ + TH +V + GT GY+ PE + K D+
Sbjct: 1010 MKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDV 1069
Query: 473 FAFGVLLLEIVTGRRPVDS---SKQNILLWAKPLMESGNIAELADPRM---EGRYDVEQL 526
+++GV+LLE++TG+RP DS N++ W K + I+++ DP + + ++E L
Sbjct: 1070 YSYGVVLLELLTGKRPTDSPDFGDNNLVGWVKQHAKL-RISDVFDPELMKEDPALEIELL 1128
Query: 527 YRVVLTASYCVRQTAIWRPAMTEV 550
+ + + C+ A RP M +V
Sbjct: 1129 QHLKVAVA-CLDDRAWRRPTMVQV 1151
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 166 bits (420), Expect = 1e-40
Identities = 96/262 (36%), Positives = 152/262 (57%), Gaps = 9/262 (3%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG+ +VY G + NGE +AVK L++++ KE F E+ ++ V H N SL+GYC E
Sbjct: 582 KGGFGKVYHG-VINGEQVAVKVLSEESAQGYKE--FRAEVDLLMRVHHTNLTSLVGYCNE 638
Query: 353 -NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
N + LI+ Y N NL L G K L W R KI++ A+GL YLH CK I+HR
Sbjct: 639 INHMVLIYEYMANENLGDYLAG--KRSFILSWEERLKISLDAAQGLEYLHNGCKPPIVHR 696
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D+K +N+LL + ++ DFGL++ + + V G+ GYL PE + ++EK+D
Sbjct: 697 DVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTVVAGSIGYLDPEYYSTRQMNEKSD 756
Query: 472 IFAFGVLLLEIVTGRRPVDSSKQ---NILLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
+++ GV+LLE++TG+ + SSK +I + ++ +G+I + D R+ RYDV ++
Sbjct: 757 VYSLGVVLLEVITGQPAIASSKTEKVHISDHVRSILANGDIRGIVDQRLRERYDVGSAWK 816
Query: 529 VVLTASYCVRQTAIWRPAMTEV 550
+ A C T+ RP M++V
Sbjct: 817 MSEIALACTEHTSAQRPTMSQV 838
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 166 bits (419), Expect = 2e-40
Identities = 97/263 (36%), Positives = 152/263 (56%), Gaps = 6/263 (2%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+G +S V+KG L +G +AVKR K + KEF EL ++ + H + +LLGYC +
Sbjct: 513 KGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHNELDLLSRLNHAHLLNLLGYCED 572
Query: 353 NG-LYLIFNYSQNGNLSTALHGYG-KAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIH 410
L++ + +G+L LHG L+W R IA+ ARG+ YLH +IH
Sbjct: 573 GSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARRVTIAVQAARGIEYLHGYACPPVIH 632
Query: 411 RDIKASNVLLGPDYEPQITDFGLAKWLP-NKWTHHAVIPVEGTFGYLAPEVFMHGIVDEK 469
RDIK+SN+L+ D+ ++ DFGL+ P + T + +P GT GYL PE + + K
Sbjct: 633 RDIKSSNILIDEDHNARVADFGLSILGPADSGTPLSELPA-GTLGYLDPEYYRLHYLTTK 691
Query: 470 TDIFAFGVLLLEIVTGRRPVDS--SKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLY 527
+D+++FGV+LLEI++GR+ +D + NI+ WA PL+++G+I + DP + D+E L
Sbjct: 692 SDVYSFGVVLLEILSGRKAIDMQFEEGNIVEWAVPLIKAGDIFAILDPVLSPPSDLEALK 751
Query: 528 RVVLTASYCVRQTAIWRPAMTEV 550
++ A CVR RP+M +V
Sbjct: 752 KIASVACKCVRMRGKDRPSMDKV 774
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 165 bits (418), Expect = 2e-40
Identities = 93/268 (34%), Positives = 145/268 (53%), Gaps = 14/268 (5%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG VY+G + N +A+KRL + F E+ +G + H + LLGY
Sbjct: 700 KGGAGIVYRGSMPNNVDVAIKRLVGRGTG-RSDHGFTAEIQTLGRIRHRHIVRLLGYVAN 758
Query: 353 NGL-YLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
L++ Y NG+L LHG G L W R+++A+ A+GL YLH C I+HR
Sbjct: 759 KDTNLLLYEYMPNGSLGELLHG--SKGGHLQWETRHRVAVEAAKGLCYLHHDCSPLILHR 816
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D+K++N+LL D+E + DFGLAK+L + + + G++GY+APE VDEK+D
Sbjct: 817 DVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPEYAYTLKVDEKSD 876
Query: 472 IFAFGVLLLEIVTGRRPVDSSKQ--NILLWAKPLME-------SGNIAELADPRMEGRYD 522
+++FGV+LLE++ G++PV + +I+ W + E + + + DPR+ G Y
Sbjct: 877 VYSFGVVLLELIAGKKPVGEFGEGVDIVRWVRNTEEEITQPSDAAIVVAIVDPRLTG-YP 935
Query: 523 VEQLYRVVLTASYCVRQTAIWRPAMTEV 550
+ + V A CV + A RP M EV
Sbjct: 936 LTSVIHVFKIAMMCVEEEAAARPTMREV 963
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 164 bits (416), Expect = 4e-40
Identities = 101/272 (37%), Positives = 157/272 (57%), Gaps = 19/272 (6%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG+ VYKG L +G+ IAVKRL+K + E F+ E+ +I + H N +LG C E
Sbjct: 536 QGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDE--FMNEVTLIARLQHINLVQVLGCCIE 593
Query: 353 NG-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
LI+ Y +N +L + L G + + L+W R+ I GVARGL YLH+ + RIIHR
Sbjct: 594 GDEKMLIYEYLENLSLDSYLFGKTRR-SKLNWNERFDITNGVARGLLYLHQDSRFRIIHR 652
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D+K SN+LL + P+I+DFG+A+ T + V GT+GY++PE M+GI EK+D
Sbjct: 653 DLKVSNILLDKNMIPKISDFGMARIFERDETEANTMKVVGTYGYMSPEYAMYGIFSEKSD 712
Query: 472 IFAFGVLLLEIVTGRRP---VDSSKQNILL---WAKPLMESGNIAELADP-------RME 518
+F+FGV++LEIV+G++ + +N LL W++ + G E+ DP
Sbjct: 713 VFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSR--WKEGRALEIVDPVIVDSLSSQP 770
Query: 519 GRYDVEQLYRVVLTASYCVRQTAIWRPAMTEV 550
+ +++ + + CV++ A RPAM+ V
Sbjct: 771 SIFQPQEVLKCIQIGLLCVQELAEHRPAMSSV 802
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 163 bits (413), Expect = 9e-40
Identities = 94/273 (34%), Positives = 150/273 (54%), Gaps = 19/273 (6%)
Query: 294 GGYSEVYKGDLCN----------GETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNT 343
GG+ V+KG + G IAVK+L +D ++E +L E+ +G HPN
Sbjct: 78 GGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQGHQE--WLAEVNYLGQFSHPNL 135
Query: 344 ASLLGYCFENG-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHK 402
L+GYC E+ L++ + G+L L G L W +R K+A+G A+GL +LH
Sbjct: 136 VKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQPLSWTLRLKVALGAAKGLAFLHN 195
Query: 403 CCKHRIIHRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFM 462
+ +I+RD K SN+LL +Y +++DFGLAK P H + GT+GY APE
Sbjct: 196 A-ETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTGDKSHVSTRIMGTYGYAAPEYLA 254
Query: 463 HGIVDEKTDIFAFGVLLLEIVTGRRPVDSSK----QNILLWAKPLM-ESGNIAELADPRM 517
G + K+D++++GV+LLE+++GRR VD ++ Q ++ WA+PL+ + + D R+
Sbjct: 255 TGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQKLVEWARPLLANKRKLFRVIDNRL 314
Query: 518 EGRYDVEQLYRVVLTASYCVRQTAIWRPAMTEV 550
+ +Y +E+ +V A C+ RP M EV
Sbjct: 315 QDQYSMEEACKVATLALRCLTFEIKLRPNMNEV 347
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 163 bits (412), Expect = 1e-39
Identities = 95/273 (34%), Positives = 152/273 (54%), Gaps = 19/273 (6%)
Query: 294 GGYSEVYKGDLCN----------GETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNT 343
GG+ V+KG + G IAVKRL ++ ++E +L E+ +G + HPN
Sbjct: 77 GGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEGFQGHRE--WLAEINYLGQLDHPNL 134
Query: 344 ASLLGYCFENG-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHK 402
L+GYC E L++ + G+L L G L W R ++A+G ARGL +LH
Sbjct: 135 VKLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTFYQPLSWNTRVRMALGAARGLAFLHN 194
Query: 403 CCKHRIIHRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFM 462
+ ++I+RD KASN+LL +Y +++DFGLA+ P H V GT GY APE
Sbjct: 195 A-QPQVIYRDFKASNILLDSNYNAKLSDFGLARDGPMGDNSHVSTRVMGTQGYAAPEYLA 253
Query: 463 HGIVDEKTDIFAFGVLLLEIVTGRRPVDSS----KQNILLWAKP-LMESGNIAELADPRM 517
G + K+D+++FGV+LLE+++GRR +D + + N++ WA+P L + + DPR+
Sbjct: 254 TGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVGEHNLVDWARPYLTNKRRLLRVMDPRL 313
Query: 518 EGRYDVEQLYRVVLTASYCVRQTAIWRPAMTEV 550
+G+Y + + ++ + A C+ A RP M E+
Sbjct: 314 QGQYSLTRALKIAVLALDCISIDAKSRPTMNEI 346
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 160 bits (404), Expect = 1e-38
Identities = 94/270 (34%), Positives = 148/270 (54%), Gaps = 14/270 (5%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKE--------FLMELGIIGHVCHPNTAS 345
G +VYK +L GE +AVK+L K K + E F E+ +G + H +
Sbjct: 692 GSSGKVYKVELRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAEVETLGTIRHKSIVR 751
Query: 346 LLGYCFENGL-YLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCC 404
L C L++ Y NG+L+ LHG K G L WP R +IA+ A GL YLH C
Sbjct: 752 LWCCCSSGDCKLLVYEYMPNGSLADVLHGDRKGGVVLGWPERLRIALDAAEGLSYLHHDC 811
Query: 405 KHRIIHRDIKASNVLLGPDYEPQITDFGLAK--WLPNKWTHHAVIPVEGTFGYLAPEVFM 462
I+HRD+K+SN+LL DY ++ DFG+AK + T A+ + G+ GY+APE
Sbjct: 812 VPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAMSGIAGSCGYIAPEYVY 871
Query: 463 HGIVDEKTDIFAFGVLLLEIVTGRRPVDS--SKQNILLWAKPLMESGNIAELADPRMEGR 520
V+EK+DI++FGV+LLE+VTG++P DS +++ W ++ + + DP+++ +
Sbjct: 872 TLRVNEKSDIYSFGVVLLELVTGKQPTDSELGDKDMAKWVCTALDKCGLEPVIDPKLDLK 931
Query: 521 YDVEQLYRVVLTASYCVRQTAIWRPAMTEV 550
+ E++ +V+ C + RP+M +V
Sbjct: 932 FK-EEISKVIHIGLLCTSPLPLNRPSMRKV 960
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 159 bits (401), Expect = 2e-38
Identities = 97/273 (35%), Positives = 151/273 (54%), Gaps = 19/273 (6%)
Query: 294 GGYSEVYKGDLCN----------GETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNT 343
GG+ V+KG + G IAVK+L +D ++E +L E+ +G H +
Sbjct: 77 GGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQGHQE--WLAEVNYLGQFSHRHL 134
Query: 344 ASLLGYCFENG-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHK 402
L+GYC E+ L++ + G+L L G L W +R K+A+G A+GL +LH
Sbjct: 135 VKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQPLSWKLRLKVALGAAKGLAFLHS 194
Query: 403 CCKHRIIHRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFM 462
+ R+I+RD K SN+LL +Y +++DFGLAK P H V GT GY APE
Sbjct: 195 S-ETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKSHVSTRVMGTHGYAAPEYLA 253
Query: 463 HGIVDEKTDIFAFGVLLLEIVTGRRPVD----SSKQNILLWAKP-LMESGNIAELADPRM 517
G + K+D+++FGV+LLE+++GRR VD S ++N++ WAKP L+ I + D R+
Sbjct: 254 TGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEWAKPYLVNKRKIFRVIDNRL 313
Query: 518 EGRYDVEQLYRVVLTASYCVRQTAIWRPAMTEV 550
+ +Y +E+ +V + C+ RP M+EV
Sbjct: 314 QDQYSMEEACKVATLSLRCLTTEIKLRPNMSEV 346
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 148 bits (373), Expect = 4e-35
Identities = 90/263 (34%), Positives = 139/263 (52%), Gaps = 10/263 (3%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE- 352
GG+ VYKG+L +G IAVKR+ EF E+ ++ V H + +LLGYC +
Sbjct: 597 GGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSEIAVLTKVRHRHLVTLLGYCLDG 656
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAG-NSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
N L++ Y G LS L + + G L W R +A+ VARG+ YLH IHR
Sbjct: 657 NEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLALDVARGVEYLHGLAHQSFIHR 716
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D+K SN+LLG D ++ DFGL + P + + GTFGYLAPE + G V K D
Sbjct: 717 DLKPSNILLGDDMRAKVADFGLVRLAP-EGKGSIETRIAGTFGYLAPEYAVTGRVTTKVD 775
Query: 472 IFAFGVLLLEIVTGRRPVDSSKQ----NILLWAKPLM--ESGNIAELADPRME-GRYDVE 524
+++FGV+L+E++TGR+ +D S+ +++ W K + + + + D ++ +
Sbjct: 776 VYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMYINKEASFKKAIDTTIDLDEETLA 835
Query: 525 QLYRVVLTASYCVRQTAIWRPAM 547
++ V A +C + RP M
Sbjct: 836 SVHTVAELAGHCCAREPYQRPDM 858
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 145 bits (365), Expect = 3e-34
Identities = 91/276 (32%), Positives = 148/276 (52%), Gaps = 26/276 (9%)
Query: 293 RGGYSEVYKG----------DLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPN 342
+GG+ +VY+G + +G +A+KRL +++ E+ E+ +G + H N
Sbjct: 94 QGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRL--NSESVQGFAEWRSEVNFLGMLSHRN 151
Query: 343 TASLLGYCFENG-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLH 401
LLGYC E+ L L++ + G+L + L + + W +R KI IG ARGL +LH
Sbjct: 152 LVKLLGYCREDKELLLVYEFMPKGSLESHLF---RRNDPFPWDLRIKIVIGAARGLAFLH 208
Query: 402 KCCKHRIIHRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVF 461
+ +I+RD KASN+LL +Y+ +++DFGLAK P H + GT+GY APE
Sbjct: 209 SL-QREVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADEKSHVTTRIMGTYGYAAPEYM 267
Query: 462 MHGIVDEKTDIFAFGVLLLEIVTG------RRPVDSSKQNILLWAKP-LMESGNIAELAD 514
G + K+D+FAFGV+LLEI+TG +RP +++++ W +P L + ++ D
Sbjct: 268 ATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRP--RGQESLVDWLRPELSNKHRVKQIMD 325
Query: 515 PRMEGRYDVEQLYRVVLTASYCVRQTAIWRPAMTEV 550
++G+Y + + C+ RP M EV
Sbjct: 326 KGIKGQYTTKVATEMARITLSCIEPDPKNRPHMKEV 361
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 140 bits (352), Expect = 1e-32
Identities = 90/268 (33%), Positives = 139/268 (51%), Gaps = 13/268 (4%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCF- 351
+G + +YK L + AVK+L N + E+ IG V H N L +
Sbjct: 824 KGAHGTIYKATLSPDKVYAVKKLVFTGIK-NGSVSMVREIETIGKVRHRNLIKLEEFWLR 882
Query: 352 -ENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIH 410
E GL +++ Y +NG+L LH LDW R+ IA+G A GL YLH C I+H
Sbjct: 883 KEYGL-ILYTYMENGSLHDILHETNPP-KPLDWSTRHNIAVGTAHGLAYLHFDCDPAIVH 940
Query: 411 RDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKT 470
RDIK N+LL D EP I+DFG+AK L T V+GT GY+APE + ++
Sbjct: 941 RDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAPENAFTTVKSRES 1000
Query: 471 DIFAFGVLLLEIVTGRRPVDSS---KQNILLWAKPL-MESGNIAELADPRMEGRY----D 522
D++++GV+LLE++T ++ +D S + +I+ W + + ++G I ++ DP +
Sbjct: 1001 DVYSYGVVLLELITRKKALDPSFNGETDIVGWVRSVWTQTGEIQKIVDPSLLDELIDSSV 1060
Query: 523 VEQLYRVVLTASYCVRQTAIWRPAMTEV 550
+EQ+ + A C + RP M +V
Sbjct: 1061 MEQVTEALSLALRCAEKEVDKRPTMRDV 1088
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.337 0.149 0.491
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 62,619,897
Number of Sequences: 164201
Number of extensions: 2613832
Number of successful extensions: 11483
Number of sequences better than 10.0: 1616
Number of HSP's better than 10.0 without gapping: 974
Number of HSP's successfully gapped in prelim test: 642
Number of HSP's that attempted gapping in prelim test: 8529
Number of HSP's gapped (non-prelim): 1827
length of query: 550
length of database: 59,974,054
effective HSP length: 115
effective length of query: 435
effective length of database: 41,090,939
effective search space: 17874558465
effective search space used: 17874558465
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC127169.5


