

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127019.17 + phase: 0 /pseudo
(170 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
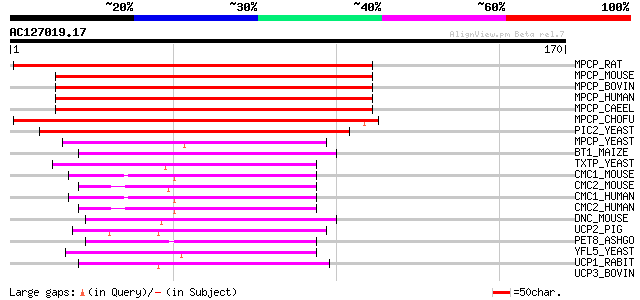
Score E
Sequences producing significant alignments: (bits) Value
MPCP_RAT (P16036) Phosphate carrier protein, mitochondrial precu... 104 1e-22
MPCP_MOUSE (Q8VEM8) Phosphate carrier protein, mitochondrial pre... 101 9e-22
MPCP_BOVIN (P12234) Phosphate carrier protein, mitochondrial pre... 100 1e-21
MPCP_HUMAN (Q00325) Phosphate carrier protein, mitochondrial pre... 100 2e-21
MPCP_CAEEL (P40614) Phosphate carrier protein, mitochondrial pre... 97 1e-20
MPCP_CHOFU (O61703) Phosphate carrier protein, mitochondrial pre... 97 2e-20
PIC2_YEAST (P40035) Mitochondrial phosphate carrier protein 2 (P... 81 1e-15
MPCP_YEAST (P23641) Mitochondrial phosphate carrier protein (Pho... 52 5e-07
BT1_MAIZE (P29518) Brittle-1 protein, chloroplast precursor 52 9e-07
TXTP_YEAST (P38152) Tricarboxylate transport protein (Citrate tr... 50 3e-06
CMC1_MOUSE (Q8BH59) Calcium-binding mitochondrial carrier protei... 48 1e-05
CMC2_MOUSE (Q9QXX4) Calcium-binding mitochondrial carrier protei... 47 2e-05
CMC1_HUMAN (O75746) Calcium-binding mitochondrial carrier protei... 47 2e-05
CMC2_HUMAN (Q9UJS0) Calcium-binding mitochondrial carrier protei... 45 6e-05
DNC_MOUSE (Q9DAM5) Mitochondrial deoxynucleotide carrier 45 1e-04
UCP2_PIG (O97562) Mitochondrial uncoupling protein 2 (UCP 2) 43 3e-04
PET8_ASHGO (O60029) Putative mitochondrial carrier protein PET8 43 3e-04
YFL5_YEAST (P43617) Putative mitochondrial carrier YFR045W 43 4e-04
UCP1_RABIT (P14271) Mitochondrial brown fat uncoupling protein 1... 43 4e-04
UCP3_BOVIN (O77792) Mitochondrial uncoupling protein 3 (UCP 3) 41 0.001
>MPCP_RAT (P16036) Phosphate carrier protein, mitochondrial
precursor (PTP)
Length = 356
Score = 104 bits (259), Expect = 1e-22
Identities = 52/110 (47%), Positives = 68/110 (61%)
Query: 2 AAMEGGISQELTPRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTT 61
AA+EG + + +YYALC GG+LS G TH A PLD++K MQV P KY I + F+
Sbjct: 44 AAVEGYSCEFGSMKYYALCGFGGVLSCGLTHTAVVPLDLVKCRMQVDPQKYKGIFNGFSI 103
Query: 62 LLREQGPSVLWKGWTGKFFGYGAQGGCRFGLYEYFKGVYSNVLVDQHRSL 111
L+E G L KGW GY QG C+FG YE FK +YSN+L +++ L
Sbjct: 104 TLKEDGVRGLAKGWAPTLIGYSMQGLCKFGFYEVFKALYSNILGEENTYL 153
>MPCP_MOUSE (Q8VEM8) Phosphate carrier protein, mitochondrial
precursor (PTP)
Length = 357
Score = 101 bits (251), Expect = 9e-22
Identities = 48/97 (49%), Positives = 61/97 (62%)
Query: 15 RYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLWKG 74
+YYALC GG+LS G TH A PLD++K MQV P KY I + F+ L+E G L KG
Sbjct: 58 KYYALCGFGGVLSCGLTHTAVVPLDLVKCRMQVDPQKYKGIFNGFSITLKEDGVRGLAKG 117
Query: 75 WTGKFFGYGAQGGCRFGLYEYFKGVYSNVLVDQHRSL 111
W GY QG C+FG YE FK +YSN+L +++ L
Sbjct: 118 WAPTLIGYSMQGLCKFGFYEVFKALYSNILGEENTYL 154
>MPCP_BOVIN (P12234) Phosphate carrier protein, mitochondrial
precursor (PTP)
Length = 362
Score = 100 bits (250), Expect = 1e-21
Identities = 48/97 (49%), Positives = 64/97 (65%)
Query: 15 RYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLWKG 74
R++ LC +GG++S GTTH A PLD++K MQV P KY SI + F+ L+E G L KG
Sbjct: 63 RFFILCGLGGIISCGTTHTALVPLDLVKCRMQVDPQKYKSIFNGFSVTLKEDGFRGLAKG 122
Query: 75 WTGKFFGYGAQGGCRFGLYEYFKGVYSNVLVDQHRSL 111
W F GY QG C+FG YE FK +YSN+L +++ L
Sbjct: 123 WAPTFIGYSLQGLCKFGFYEVFKVLYSNMLGEENAYL 159
>MPCP_HUMAN (Q00325) Phosphate carrier protein, mitochondrial
precursor (PTP) (OK/SW-cl.48)
Length = 362
Score = 100 bits (248), Expect = 2e-21
Identities = 47/97 (48%), Positives = 63/97 (64%)
Query: 15 RYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLWKG 74
R++ LC +GG++S GTTH A PLD++K MQV P KY I + F+ L+E G L KG
Sbjct: 63 RFFILCGLGGIISCGTTHTALVPLDLVKCRMQVDPQKYKGIFNGFSVTLKEDGVRGLAKG 122
Query: 75 WTGKFFGYGAQGGCRFGLYEYFKGVYSNVLVDQHRSL 111
W F GY QG C+FG YE FK +YSN+L +++ L
Sbjct: 123 WAPTFLGYSMQGLCKFGFYEVFKVLYSNMLGEENTYL 159
>MPCP_CAEEL (P40614) Phosphate carrier protein, mitochondrial
precursor (PTP)
Length = 340
Score = 97.4 bits (241), Expect = 1e-20
Identities = 45/97 (46%), Positives = 63/97 (64%)
Query: 15 RYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLWKG 74
+YYA C +GG+LS G TH A PLD++K +QV+P KY I++ F T + E+G L KG
Sbjct: 41 KYYAYCALGGVLSCGITHTAIVPLDLVKCRIQVNPEKYTGIATGFRTTIAEEGARALVKG 100
Query: 75 WTGKFFGYGAQGGCRFGLYEYFKGVYSNVLVDQHRSL 111
W GY AQG +FG YE FK VY+++L +++ L
Sbjct: 101 WAPTLLGYSAQGLGKFGFYEIFKNVYADMLGEENAYL 137
>MPCP_CHOFU (O61703) Phosphate carrier protein, mitochondrial
precursor (Phosphate transport protein) (PTP)
Length = 349
Score = 96.7 bits (239), Expect = 2e-20
Identities = 50/116 (43%), Positives = 71/116 (61%), Gaps = 4/116 (3%)
Query: 2 AAMEGGISQELTPRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTT 61
AA G + + +Y+ALC +GG+LS G TH A PLD++K +QV KY ++ + F
Sbjct: 34 AAPVGDSCEFGSTKYFALCGLGGILSCGITHTAVVPLDLVKCRLQVDADKYKNVVNGFRV 93
Query: 62 LLREQGPSVLWKGWTGKFFGYGAQGGCRFGLYEYFKGVYSNVLVDQ----HRSLVY 113
+RE+G L KGW F GY QG C+FGLYE FK Y+N+L ++ +R+ VY
Sbjct: 94 SVREEGLRGLAKGWAPTFIGYSLQGLCKFGLYEVFKVQYNNMLDEETAYTYRTFVY 149
>PIC2_YEAST (P40035) Mitochondrial phosphate carrier protein 2
(Phosphate transport protein 2) (PTP 2) (mPic 2) (Pi
carrier isoform 2)
Length = 300
Score = 80.9 bits (198), Expect = 1e-15
Identities = 38/95 (40%), Positives = 58/95 (61%)
Query: 10 QELTPRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPS 69
Q T +YA CT+GG+++ G TH + TPLD++K +QV+P Y S F ++ +G
Sbjct: 11 QLYTKEFYATCTLGGIIACGPTHSSITPLDLVKCRLQVNPKLYTSNLQGFRKIIANEGWK 70
Query: 70 VLWKGWTGKFFGYGAQGGCRFGLYEYFKGVYSNVL 104
++ G+ F GY QG ++G YEYFK +YS+ L
Sbjct: 71 KVYTGFGATFVGYSLQGAGKYGGYEYFKHLYSSWL 105
>MPCP_YEAST (P23641) Mitochondrial phosphate carrier protein
(Phosphate transport protein) (PTP) (mPic 1)
(Mitochondrial import receptor) (p32)
Length = 311
Score = 52.4 bits (124), Expect = 5e-07
Identities = 26/82 (31%), Positives = 40/82 (48%), Gaps = 1/82 (1%)
Query: 17 YALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKY-YSISSCFTTLLREQGPSVLWKGW 75
Y + G + G+TH + P+DV+K +Q+ P Y + F ++ +G L G+
Sbjct: 17 YMKFALAGAIGCGSTHSSMVPIDVVKTRIQLEPTVYNKGMVGSFKQIIAGEGAGALLTGF 76
Query: 76 TGKFFGYGAQGGCRFGLYEYFK 97
GY QG +FG YE FK
Sbjct: 77 GPTLLGYSIQGAFKFGGYEVFK 98
>BT1_MAIZE (P29518) Brittle-1 protein, chloroplast precursor
Length = 436
Score = 51.6 bits (122), Expect = 9e-07
Identities = 23/79 (29%), Positives = 41/79 (51%)
Query: 22 IGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLWKGWTGKFFG 81
+ G L+ + L T P++++K + + Y +++ F +LR++GPS L++G T G
Sbjct: 234 VAGALAGFASTLCTYPMELIKTRVTIEKDVYDNVAHAFVKILRDEGPSELYRGLTPSLIG 293
Query: 82 YGAQGGCRFGLYEYFKGVY 100
C F YE K +Y
Sbjct: 294 VVPYAACNFYAYETLKRLY 312
>TXTP_YEAST (P38152) Tricarboxylate transport protein (Citrate
transport protein) (CTP)
Length = 299
Score = 50.1 bits (118), Expect = 3e-06
Identities = 29/82 (35%), Positives = 39/82 (47%), Gaps = 1/82 (1%)
Query: 14 PRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQ-VHPIKYYSISSCFTTLLREQGPSVLW 72
P L + G S T +T PLD +K MQ + KY S +CF T+ +E+G W
Sbjct: 211 PLSSGLTFLVGAFSGIVTVYSTMPLDTVKTRMQSLDSTKYSSTMNCFATIFKEEGLKTFW 270
Query: 73 KGWTGKFFGYGAQGGCRFGLYE 94
KG T + GG F +YE
Sbjct: 271 KGATPRLGRLVLSGGIVFTIYE 292
>CMC1_MOUSE (Q8BH59) Calcium-binding mitochondrial carrier protein
Aralar1 (Mitochondrial aspartate glutamate carrier 1)
(Solute carrier family 25, member 12)
Length = 677
Score = 47.8 bits (112), Expect = 1e-05
Identities = 28/80 (35%), Positives = 35/80 (43%), Gaps = 5/80 (6%)
Query: 19 LCTIGGMLSAGTTHLATTPLDVLKVNMQVHP----IKYYSISSCFTTLLREQGPSVLWKG 74
L T G + L T P DV+K +QV Y + CF +LRE+GPS WKG
Sbjct: 521 LLTAGALAGVPAASLVT-PADVIKTRLQVAARAGQTTYSGVVDCFRKILREEGPSAFWKG 579
Query: 75 WTGKFFGYGAQGGCRFGLYE 94
+ F Q G YE
Sbjct: 580 TAARVFRSSPQFGVTLVTYE 599
>CMC2_MOUSE (Q9QXX4) Calcium-binding mitochondrial carrier protein
Aralar2 (Mitochondrial aspartate glutamate carrier 2)
(Solute carrier family 25, member 13) (Citrin)
Length = 676
Score = 47.4 bits (111), Expect = 2e-05
Identities = 28/77 (36%), Positives = 36/77 (46%), Gaps = 8/77 (10%)
Query: 22 IGGMLSAGTTHLATTPLDVLKVNMQV----HPIKYYSISSCFTTLLREQGPSVLWKGWTG 77
I GM +A TP DV+K +QV Y ++ CF +LRE+GP LWKG
Sbjct: 530 IAGMPAASLV----TPADVIKTRLQVAARAGQTTYNGVTDCFRKILREEGPKALWKGVAA 585
Query: 78 KFFGYGAQGGCRFGLYE 94
+ F Q G YE
Sbjct: 586 RVFRSSPQFGVTLLTYE 602
>CMC1_HUMAN (O75746) Calcium-binding mitochondrial carrier protein
Aralar1 (Mitochondrial aspartate glutamate carrier 1)
(Solute carrier family 25, member 12)
Length = 678
Score = 47.4 bits (111), Expect = 2e-05
Identities = 28/80 (35%), Positives = 34/80 (42%), Gaps = 5/80 (6%)
Query: 19 LCTIGGMLSAGTTHLATTPLDVLKVNMQVHP----IKYYSISSCFTTLLREQGPSVLWKG 74
L G M L T P DV+K +QV Y + CF +LRE+GPS WKG
Sbjct: 521 LLAAGAMAGVPAASLVT-PADVIKTRLQVAARAGQTTYSGVIDCFRKILREEGPSAFWKG 579
Query: 75 WTGKFFGYGAQGGCRFGLYE 94
+ F Q G YE
Sbjct: 580 TAARVFRSSPQFGVTLAHYE 599
>CMC2_HUMAN (Q9UJS0) Calcium-binding mitochondrial carrier protein
Aralar2 (Mitochondrial aspartate glutamate carrier 2)
(Solute carrier family 25, member 13) (Citrin)
Length = 675
Score = 45.4 bits (106), Expect = 6e-05
Identities = 28/77 (36%), Positives = 35/77 (45%), Gaps = 8/77 (10%)
Query: 22 IGGMLSAGTTHLATTPLDVLKVNMQVHP----IKYYSISSCFTTLLREQGPSVLWKGWTG 77
I GM +A TP DV+K +QV Y + CF +LRE+GP LWKG
Sbjct: 529 IAGMPAASLV----TPADVIKTRLQVAARAGQTTYSGVIDCFRKILREEGPKALWKGAGA 584
Query: 78 KFFGYGAQGGCRFGLYE 94
+ F Q G YE
Sbjct: 585 RVFRSSPQFGVTLLTYE 601
>DNC_MOUSE (Q9DAM5) Mitochondrial deoxynucleotide carrier
Length = 318
Score = 44.7 bits (104), Expect = 1e-04
Identities = 26/79 (32%), Positives = 36/79 (44%), Gaps = 2/79 (2%)
Query: 24 GMLSAGTTHLATTPLDVLKVNM--QVHPIKYYSISSCFTTLLREQGPSVLWKGWTGKFFG 81
G LSAGT L P+DVL+ + Q P Y ++ T+ + +GP V +KG T
Sbjct: 125 GGLSAGTATLTVHPVDVLRTRLAAQGEPKIYNNLREAIRTMYKTEGPFVFYKGLTPTVIA 184
Query: 82 YGAQGGCRFGLYEYFKGVY 100
G +F Y K Y
Sbjct: 185 IFPYAGLQFSCYRSLKRAY 203
Score = 38.9 bits (89), Expect = 0.006
Identities = 23/75 (30%), Positives = 33/75 (43%), Gaps = 12/75 (16%)
Query: 22 IGGMLSAGTTHLATTPLDVLKVNMQVH---------PIKYYSISSCFTTLLREQGPSVLW 72
+ G +S T +PLDV+K+ Q+ KY+ I +L+E+GP W
Sbjct: 20 VAGSVSGFVTRALISPLDVIKIRFQLQIERLCPSDPNAKYHGIFQAAKQILQEEGPRAFW 79
Query: 73 KGWTGK---FFGYGA 84
KG GYGA
Sbjct: 80 KGHVPAQILSIGYGA 94
>UCP2_PIG (O97562) Mitochondrial uncoupling protein 2 (UCP 2)
Length = 309
Score = 43.1 bits (100), Expect = 3e-04
Identities = 26/80 (32%), Positives = 38/80 (47%), Gaps = 2/80 (2%)
Query: 20 CTIGGMLSAG-TTHLATTPLDVLKVN-MQVHPIKYYSISSCFTTLLREQGPSVLWKGWTG 77
C AG T + +P+DV+K M P +Y S C T+L+++GP +KG+T
Sbjct: 216 CHFTSAFGAGFCTTVIASPVDVVKTRYMNSAPGQYSSAGHCALTMLQKEGPRAFYKGFTP 275
Query: 78 KFFGYGAQGGCRFGLYEYFK 97
F G+ F YE K
Sbjct: 276 SFLRLGSWNVVMFVTYEQLK 295
Score = 32.3 bits (72), Expect = 0.54
Identities = 23/63 (36%), Positives = 29/63 (45%), Gaps = 10/63 (15%)
Query: 22 IGGMLSAGTTHLATT-----PLDVLKVNMQVHPI-----KYYSISSCFTTLLREQGPSVL 71
IG L AG+T A P DV+KV Q +Y S + T+ RE+G L
Sbjct: 116 IGSRLLAGSTTGALAVAVAQPTDVVKVRFQAQARAGGGRRYRSTVDAYKTIAREEGLRGL 175
Query: 72 WKG 74
WKG
Sbjct: 176 WKG 178
>PET8_ASHGO (O60029) Putative mitochondrial carrier protein PET8
Length = 271
Score = 43.1 bits (100), Expect = 3e-04
Identities = 26/71 (36%), Positives = 35/71 (48%), Gaps = 1/71 (1%)
Query: 24 GMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLWKGWTGKFFGYG 83
G L+ G ATTPLDVLK M +H + + TL RE+G V ++G +
Sbjct: 196 GSLAGGIAAAATTPLDVLKTRMMLHE-RRVPMLHLARTLFREEGARVFFRGIGPRTMWIS 254
Query: 84 AQGGCRFGLYE 94
A G G+YE
Sbjct: 255 AGGAIFLGVYE 265
>YFL5_YEAST (P43617) Putative mitochondrial carrier YFR045W
Length = 285
Score = 42.7 bits (99), Expect = 4e-04
Identities = 24/79 (30%), Positives = 37/79 (46%), Gaps = 2/79 (2%)
Query: 18 ALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIK--YYSISSCFTTLLREQGPSVLWKGW 75
A I G+ ++ T T P+DV+K M K Y + +C + ++G + WKG
Sbjct: 195 ASSVITGLATSFTLVAMTQPIDVVKTRMMSQNAKTEYKNTLNCMYRIFVQEGMATFWKGS 254
Query: 76 TGKFFGYGAQGGCRFGLYE 94
+F G GG F +YE
Sbjct: 255 IFRFMKVGISGGLTFTVYE 273
>UCP1_RABIT (P14271) Mitochondrial brown fat uncoupling protein 1
(UCP 1) (Thermogenin)
Length = 306
Score = 42.7 bits (99), Expect = 4e-04
Identities = 21/78 (26%), Positives = 41/78 (51%), Gaps = 1/78 (1%)
Query: 22 IGGMLSAGTTHLATTPLDVLKVN-MQVHPIKYYSISSCFTTLLREQGPSVLWKGWTGKFF 80
+ +++ T L ++P+DV+K + P +Y S+ +C T+ ++GP+ +KG+ F
Sbjct: 216 VSALIAGFCTTLLSSPVDVVKTRFINSPPGQYASVPNCAMTMFTKEGPTAFFKGFVPSFL 275
Query: 81 GYGAQGGCRFGLYEYFKG 98
G+ F +E KG
Sbjct: 276 RLGSWNVIMFVCFEKLKG 293
Score = 32.0 bits (71), Expect = 0.70
Identities = 21/76 (27%), Positives = 32/76 (41%), Gaps = 7/76 (9%)
Query: 26 LSAGTTHLATTPLDVLKVNMQVHP-------IKYYSISSCFTTLLREQGPSVLWKGWTGK 78
++A + T PLD KV Q+ I+Y + TTL + +GP L+ G
Sbjct: 22 VAACLADVITFPLDTAKVRQQIQGEFPITSGIRYKGVLGTITTLAKTEGPLKLYSGLPAG 81
Query: 79 FFGYGAQGGCRFGLYE 94
+ R GLY+
Sbjct: 82 LQRQISFASLRIGLYD 97
>UCP3_BOVIN (O77792) Mitochondrial uncoupling protein 3 (UCP 3)
Length = 311
Score = 41.2 bits (95), Expect = 0.001
Identities = 21/66 (31%), Positives = 34/66 (50%), Gaps = 1/66 (1%)
Query: 33 LATTPLDVLKVN-MQVHPIKYYSISSCFTTLLREQGPSVLWKGWTGKFFGYGAQGGCRFG 91
L +P+DV+K M P +Y+S C ++ ++GP+ +KG+T F G+ F
Sbjct: 232 LVASPVDVVKTRYMNSPPGQYHSPFDCMLKMVTQEGPTAFYKGFTPSFLRLGSWNVVMFV 291
Query: 92 LYEYFK 97
YE K
Sbjct: 292 TYEQMK 297
Score = 31.6 bits (70), Expect = 0.92
Identities = 22/77 (28%), Positives = 29/77 (37%), Gaps = 12/77 (15%)
Query: 5 EGGISQELTPRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPI-------KYYSISS 57
+G + R A CT G M P DV+K+ Q KY
Sbjct: 109 KGSDHSSIITRILAGCTTGAMAVT-----CAQPTDVVKIRFQASMHTGLGGNRKYSGTMD 163
Query: 58 CFTTLLREQGPSVLWKG 74
+ T+ RE+G LWKG
Sbjct: 164 AYRTIAREEGVRGLWKG 180
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.352 0.156 0.557
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,465,488
Number of Sequences: 164201
Number of extensions: 646376
Number of successful extensions: 2834
Number of sequences better than 10.0: 113
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 2622
Number of HSP's gapped (non-prelim): 231
length of query: 170
length of database: 59,974,054
effective HSP length: 102
effective length of query: 68
effective length of database: 43,225,552
effective search space: 2939337536
effective search space used: 2939337536
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC127019.17


