

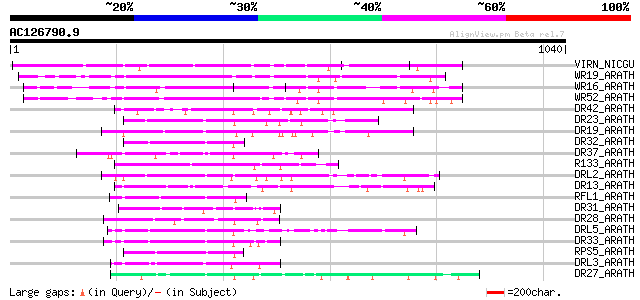
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126790.9 + phase: 0 /pseudo
(1040 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 426 e-118
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 264 8e-70
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 236 3e-61
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 211 6e-54
DR42_ARATH (Q9FKZ1) Probable disease resistance protein At5g66900 105 8e-22
DR23_ARATH (O82484) Putative disease resistance protein At4g10780 86 4e-16
DR19_ARATH (Q9C8T9) Putative disease resistance protein At1g63350 86 5e-16
DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730 81 1e-14
DR37_ARATH (Q9LVT4) Probable disease resistance protein At5g47250 80 3e-14
R133_ARATH (Q9STE7) Putative disease resistance RPP13-like prote... 79 5e-14
DRL2_ARATH (P60839) Probable disease resistance protein At1g12290 79 6e-14
DR13_ARATH (Q9XIF0) Putative disease resistance protein At1g59780 79 8e-14
RFL1_ARATH (Q8L3R3) Disease resistance protein RFL1 (RPS5-like p... 77 2e-13
DR31_ARATH (Q9FLB4) Putative disease resistance protein At5g05400 76 4e-13
DR28_ARATH (O81825) Putative disease resistance protein At4g27220 76 4e-13
DRL5_ARATH (Q9C8K0) Probable disease resistance protein At1g51480 75 1e-12
DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740 74 2e-12
RPS5_ARATH (O64973) Disease resistance protein RPS5 (Resistance ... 74 2e-12
DRL3_ARATH (Q9LMP6) Probable disease resistance protein At1g15890 74 3e-12
DR27_ARATH (Q9T048) Putative disease resistance protein At4g27190 74 3e-12
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 426 bits (1095), Expect = e-118
Identities = 272/759 (35%), Positives = 429/759 (55%), Gaps = 45/759 (5%)
Query: 6 SSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLK 65
+S+S S + Y VFLSFRG DTR FT +LY+ L DKGI TF D L+ G I L K
Sbjct: 2 ASSSSSSRWSYDVFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIPGELCK 61
Query: 66 AIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYG 125
AIEES+ I VFS NYA+S +CL+ELV I+ C + V+P+F+ V+P+ VR++K S+
Sbjct: 62 AIEESQFAIVVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQKESFA 121
Query: 126 EALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVKYISNKIS 185
+A EHE ++++D +E +Q W+ AL++AANL G D+ + I +IV IS+K+
Sbjct: 122 KAFEEHETKYKDD---VEGIQRWRIALNEAANLKGSCDNRDKTDADCIRQIVDQISSKLC 178
Query: 186 RQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFV---- 241
+ L VG+ + +++++SLL+ G +GV ++GI+G+GG+GK+T+AR I++ +
Sbjct: 179 KISLSYLQNIVGIDTHLEKIESLLEIGI-NGVRIMGIWGMGGVGKTTIARAIFDTLLGRM 237
Query: 242 --ADQFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRKK 299
+ QF+G+CFL D++EN + + LQ LL + + ++ +G + RL KK
Sbjct: 238 DSSYQFDGACFLKDIKEN--KRGMHSLQNALLSELLREKANYNNEEDGKHQMASRLRSKK 295
Query: 300 ILLILDDVDNLKQ-LHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEA 358
+L++LDD+DN L LAG LDWFG GSR+IITTR+K L+ + I + V L + E+
Sbjct: 296 VLIVLDDIDNKDHYLEYLAGDLDWFGNGSRIIITTRDKHLIEKNDI--IYEVTALPDHES 353
Query: 359 LELLRWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDR 418
++L + AF + +E + V YA GLPL L+V GS L + +WK ++
Sbjct: 354 IQLFKQHAFGKEVPNENFEKLSLEVVNYAKGLPLALKVWGSLLHNLRLTEWKSAIEHMKN 413
Query: 419 IPNKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHC---ITHHLG 475
I LK+SYD LE ++Q +FLDIAC +G E +D + + C + L
Sbjct: 414 NSYSGIIDKLKISYDGLEPKQQEMFLDIACFLRG----EEKDYILQILESCHIGAEYGLR 469
Query: 476 VLAGKSLVKISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVL 535
+L KSLV IS Y N V++HDLI+DMGK +V + K+PGERSRLW +++ V+
Sbjct: 470 ILIDKSLVFISEY------NQVQMHDLIQDMGKYIVNFQ--KDPGERSRLWLAKEVEEVM 521
Query: 536 KENTGTSKIEMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVL 595
NTGT +E I+++ +S S + +A K M +L+ + + YLP++LR
Sbjct: 522 SNNTGTMAMEAIWVSSYS--STLRFSNQAVKNMKRLRVFNMGRSSTHYAIDYLPNNLRCF 579
Query: 596 -----KWKGCLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEY 650
W+ S +++ S +++ K +++ + L + +
Sbjct: 580 VCTNYPWESFPSTFELKMLVHLQLRHNSLRHLWTETK--------HLPSLRRIDLSWSKR 631
Query: 651 LTHIPDVSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLK 710
LT PD +G+ NLE ++ C NL +H+S+G +K+ L C+ L+ F + + SL+
Sbjct: 632 LTRTPDFTGMPNLEYVNLYQCSNLEEVHHSLGCCSKVIGLYLNDCKSLKRFPCVNVESLE 691
Query: 711 KLILYECECLDNFPELLCKMAHIKEIDISNTSIGELPFS 749
L L C+ L+ PE+ +M +I + + I ELP S
Sbjct: 692 YLGLRSCDSLEKLPEIYGRMKPEIQIHMQGSGIRELPSS 730
Score = 60.1 bits (144), Expect = 3e-08
Identities = 59/232 (25%), Positives = 98/232 (41%), Gaps = 39/232 (16%)
Query: 623 FSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPDVSGLSNLEKLSFTCCDNLITIHNSIG 682
+ + K + MQ ++ L +Y TH+ KL NL+ + +SI
Sbjct: 708 YGRMKPEIQIHMQG-SGIRELPSSIFQYKTHVT---------KLLLWNMKNLVALPSSIC 757
Query: 683 HLNKLEWLSAYGCRKLEHFRPLGLASLKKLILYECECLDNFPELLCKMAHIKEIDISNTS 742
L L LS GC KLE + PE + + +++ D S+T
Sbjct: 758 RLKSLVSLSVSGCSKLE----------------------SLPEEIGDLDNLRVFDASDTL 795
Query: 743 IGELPFSFQNLSELHELTVTS-----GMKFPKIV--FSNMTKLSLSFFNLSDECLPIVLK 795
I P S L++L L +FP + ++ L+LS+ NL D LP +
Sbjct: 796 ILRPPSSIIRLNKLIILMFRGFKDGVHFEFPPVAEGLHSLEYLNLSYCNLIDGGLPEEIG 855
Query: 796 WCVNMTHLDLSFSNFKILPECLRECHHLVEINVMCCESLEEIRGIPPNLKEL 847
++ LDLS +NF+ LP + + L +++ C+ L ++ +PP L EL
Sbjct: 856 SLSSLKKLDLSRNNFEHLPSSIAQLGALQSLDLKDCQRLTQLPELPPELNEL 907
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 264 bits (675), Expect = 8e-70
Identities = 240/832 (28%), Positives = 405/832 (47%), Gaps = 90/832 (10%)
Query: 16 YQVFLSFRGSD-TRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIFI 74
Y V + + +D + F +L +L +GI+ + N + A+ + R+ I
Sbjct: 668 YDVVIRYGRADISNEDFISHLRASLCRRGISVYEKFNEVD-----------ALPKCRVLI 716
Query: 75 PVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEKR 134
V + Y S+ L++I+ T+ R+V P+F+ + P +Y +R
Sbjct: 717 IVLTSTYVPSN-----LLNILEHQHTEDRVVYPIFYRLSPYDFVCNSKNY--------ER 763
Query: 135 F--QNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVKYISNKI-SRQPLHV 191
F Q++PK W+ AL + + GY + E +LI +IV+ + S +++
Sbjct: 764 FYLQDEPKK------WQAALKEITQMPGYTLTDKS-ESELIDEIVRDALKVLCSADKVNM 816
Query: 192 ATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCFL 251
+G+ +V+++ SLL S D V +GI+G G+GK+T+A +I+ ++ Q+E L
Sbjct: 817 ----IGMDMQVEEILSLLCIESLD-VRSIGIWGTVGIGKTTIAEEIFRKISVQYETCVVL 871
Query: 252 HDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSE-GIPVIKERLCRKKILLILDDVDNL 310
D+ + ++E L + +E + +S+ ++ RL RK+IL+ILDDV++
Sbjct: 872 KDLHKEVEVKGHDAVRENFLSEVLEVEPHVIRISDIKTSFLRSRLQRKRILVILDDVNDY 931
Query: 311 KQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSD 370
+ + G L++FG GSR+I+T+RN+ + I+ + V+ L+ ++L LL +
Sbjct: 932 RDVDTFLGTLNYFGPGSRIIMTSRNRRVFVLCKIDHVYEVKPLDIPKSLLLLDRGTCQIV 991
Query: 371 KVPSGYEDILNRAVAYAFGLPLVLEVVGS--NLFGKSIEDWKHTLDGYDRIPNKEIQKIL 428
P Y+ + V ++ G P VL+ + S + K ++ K T Y I I
Sbjct: 992 LSPEVYKTLSLELVKFSNGNPQVLQFLSSIDREWNKLSQEVKTTSPIY-------IPGIF 1044
Query: 429 KVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHC-ITHHLGV--LAGKSLVKI 485
+ S L++ E+ +FLDIAC F +D + D C + H+G L KSL+ I
Sbjct: 1045 EKSCCGLDDNERGIFLDIACFFNRID----KDNVAMLLDGCGFSAHVGFRGLVDKSLLTI 1100
Query: 486 STYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSKIE 545
S + N V + I+ G+E+VRQES PG+RSRLW + I HV +TGTS IE
Sbjct: 1101 SQH------NLVDMLSFIQATGREIVRQESADRPGDRSRLWNADYIRHVFINDTGTSAIE 1154
Query: 546 MIYMNLHSMESVIDKKGKAFKKMTKLKTLII-------ENGL-FSGGLKYLPSSLRVLKW 597
I++++ +++ D F+KM L+ L + ++G+ F GL+YLPS LR+L W
Sbjct: 1155 GIFLDMLNLK--FDANPNVFEKMCNLRLLKLYCSKAEEKHGVSFPQGLEYLPSKLRLLHW 1212
Query: 598 KGCLSKCLSSSI-------LNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEY 650
+ L S LN S + + +K F + +K + L Y +
Sbjct: 1213 EYYPLSSLPKSFNPENLVELNLPSSCAKK--LWKGKKARFCTTNSSLEKLKKMRLSYSDQ 1270
Query: 651 LTHIPDVSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPL-GLASL 709
LT IP +S +NLE + C++L+++ SI +L KL +L+ GC KLE+ + L SL
Sbjct: 1271 LTKIPRLSSATNLEHIDLEGCNSLLSLSQSISYLKKLVFLNLKGCSKLENIPSMVDLESL 1330
Query: 710 KKLILYECECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSGMKFPK 769
+ L L C L NFPE+ ++KE+ + T I E+P S +NL L +L + +
Sbjct: 1331 EVLNLSGCSKLGNFPEI---SPNVKELYMGGTMIQEIPSSIKNLVLLEKLDLENSRHLKN 1387
Query: 770 IVFSNMTKLSLSFFNLSD----ECLPIVLKWCVNMTHLDLSFSNFKILPECL 817
+ S L NLS E P + + LDLS ++ K LP +
Sbjct: 1388 LPTSIYKLKHLETLNLSGCISLERFPDSSRRMKCLRFLDLSRTDIKELPSSI 1439
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 236 bits (601), Expect = 3e-61
Identities = 221/870 (25%), Positives = 385/870 (43%), Gaps = 159/870 (18%)
Query: 26 DTRYGFTGNLYKALTDKGIN-TFIDKNGLQRGDEITPSLLKAIEESRIFIPVFSINYASS 84
+ RY F +L KAL KG+N FID + D ++ +E +R+ + + N S
Sbjct: 15 EVRYSFVSHLSKALQRKGVNDVFIDSD-----DSLSNESQSMVERARVSVMILPGNRTVS 69
Query: 85 SFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEKRFQNDPKNMER 144
LD+LV ++ C K K ++V+PV +GV +
Sbjct: 70 ---LDKLVKVLDCQKNKDQVVVPVLYGVRSSETE-------------------------- 100
Query: 145 LQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVKYISNKISRQPLHVATYPVGLQSRVQQ 204
W AL S +H + +L+ + V+ + K+ +G+ S++ +
Sbjct: 101 ---WLSALDSKGFSSVHHSRKECSDSQLVKETVRDVYEKLFYMER------IGIYSKLLE 151
Query: 205 VKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCFLHDVRENSAQNNLK 264
++ ++++ D + VGI+G+ G+GK+TLA+ +++ ++ +F+ CF+ D + + +
Sbjct: 152 IEKMINKQPLD-IRCVGIWGMPGIGKTTLAKAVFDQMSGEFDAHCFIEDYTKAIQEKGVY 210
Query: 265 YLQEKLLLK----TTGLEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQLHALAGGL 320
L E+ LK +G KL ++++RL K++L++LDDV + + + GG
Sbjct: 211 CLLEEQFLKENAGASGTVTKLS-------LLRDRLNNKRVLVVLDDVRSPLVVESFLGGF 263
Query: 321 DWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKVPSGYEDIL 380
DWFG S +IIT+++K + + + V+GLNE EAL+L A D ++
Sbjct: 264 DWFGPKSLIIITSKDKSVFRLCRVNQIYEVQGLNEKEALQLFSLCASIDDMAEQNLHEVS 323
Query: 381 NRAVAYAFGLPLVLEVVGSNLFGKS-IEDWKHTLDGYDRIPNKEIQKILKVSYDALEEEE 439
+ + YA G PL L + G L GK + + P +K SYD L + E
Sbjct: 324 MKVIKYANGHPLALNLYGRELMGKKRPPEMEIAFLKLKECPPAIFVDAIKSSYDTLNDRE 383
Query: 440 QSVFLDIACCFKGYQWKEFEDIL--CAHYDHCITHHLGVLAGKSLVKISTYYPSGSINDV 497
+++FLDIAC F+G +L C + H + VL KSLV IS N V
Sbjct: 384 KNIFLDIACFFQGENVDYVMQLLEGCGFFPHV---GIDVLVEKSLVTISE-------NRV 433
Query: 498 RLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGT---------------S 542
R+H+LI+D+G++++ +E+ ++ RSRLW I ++L++
Sbjct: 434 RMHNLIQDVGRQIINRET-RQTKRRSRLWEPCSIKYLLEDKEQNENEEQKTTFERAQVPE 492
Query: 543 KIEMIYMNLHSMESVIDKKGKAFKKMTKLKTLII---------ENGLFSGGLKYLPSSLR 593
+IE ++++ ++ D K AF M L+ I N G L LP+ LR
Sbjct: 493 EIEGMFLDTSNLS--FDIKHVAFDNMLNLRLFKIYSSNPEVHHVNNFLKGSLSSLPNVLR 550
Query: 594 VLKWKGCLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTH 653
+L W+ + L + + +SQ KK + + + +K + L + + L
Sbjct: 551 LLHWENYPLQFLPQNF--DPIHLVEINMPYSQLKKLW-GGTKDLEMLKTIRLCHSQQLVD 607
Query: 654 IPDVSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLG-LASLKKL 712
I D+ NLE + GC +L+ F G L L+ +
Sbjct: 608 IDDLLKAQNLEVVDL------------------------QGCTRLQSFPATGQLLHLRVV 643
Query: 713 ILYECECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQN---------LSELHELTVTS 763
L C + +FPE+ +I+ +++ T I ELP S L+E+ L+ S
Sbjct: 644 NLSGCTEIKSFPEI---PPNIETLNLQGTGIIELPLSIVKPNYRELLNLLAEIPGLSGVS 700
Query: 764 GMKFPKI-VFSNMTKLSLSFFNLSD-ECLPI----VLKWCVNMTHLDLSFSNFKILPECL 817
++ + +++ K+S S+ N CL + L+ NM +L+L
Sbjct: 701 NLEQSDLKPLTSLMKISTSYQNPGKLSCLELNDCSRLRSLPNMVNLEL------------ 748
Query: 818 RECHHLVEINVMCCESLEEIRGIPPNLKEL 847
L +++ C LE I+G P NLKEL
Sbjct: 749 -----LKALDLSGCSELETIQGFPRNLKEL 773
Score = 47.0 bits (110), Expect = 3e-04
Identities = 29/99 (29%), Positives = 54/99 (54%), Gaps = 6/99 (6%)
Query: 419 IPNKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLA 478
+ E +++L+V Y L+E +++FL IA F ++ D +++ L VLA
Sbjct: 1042 VSGNEDEEVLRVRYAGLQEIYKALFLYIAGLFNDEDVGLVAPLIANIIDMDVSYGLKVLA 1101
Query: 479 GKSLVKISTYYPSGSINDVRLHDLIKDMGKEVVRQESPK 517
+SL+++S S ++ +H L++ MGKE++ ES K
Sbjct: 1102 YRSLIRVS------SNGEIVMHYLLRQMGKEILHTESKK 1134
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein
RRS1) (Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 211 bits (538), Expect = 6e-54
Identities = 223/883 (25%), Positives = 381/883 (42%), Gaps = 124/883 (14%)
Query: 26 DTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIFIPVFSINYASSS 85
+ RY F +L +AL KGIN + + D + IE++ + + V N S
Sbjct: 18 EVRYSFVSHLSEALRRKGINNVVVDVDID--DLLFKESQAKIEKAGVSVMVLPGNCDPSE 75
Query: 86 FCLDELVHIIHCYKT-KGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEKRFQNDPKNMER 144
LD+ ++ C + K + V+ V YG++L + + D + + R
Sbjct: 76 VWLDKFAKVLECQRNNKDQAVVSVL--------------YGDSLLRDQWLSELDFRGLSR 121
Query: 145 LQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVKYISNKISRQPLHVATYPVGLQSRVQQ 204
+ +K S + L+ +IV+ + H +G+ S++ +
Sbjct: 122 IHQSRKECSDSI---------------LVEEIVRDVYET------HFYVGRIGIYSKLLE 160
Query: 205 VKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCFLHDVRENSAQNNLK 264
+++++++ G+ VGI+G+ G+GK+TLA+ +++ ++ F+ SCF+ D ++ + L
Sbjct: 161 IENMVNK-QPIGIRCVGIWGMPGIGKTTLAKAVFDQMSSAFDASCFIEDYDKSIHEKGLY 219
Query: 265 YLQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQLHALAGGLDWFG 324
L E+ LL I + +++RL K++L++LDDV N + G DW G
Sbjct: 220 CLLEEQLLPGNDATIMK------LSSLRDRLNSKRVLVVLDDVCNALVAESFLEGFDWLG 273
Query: 325 CGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALEL-LRWMAFKSDKVPSGYEDILNRA 383
GS +IIT+R+K + GI + V+GLNE EA +L L + D ++ R
Sbjct: 274 PGSLIIITSRDKQVFRLCGINQIYEVQGLNEKEARQLFLLSASIMEDMGEQNLHELSVRV 333
Query: 384 VAYAFGLPLVLEVVGSNLFG-KSIEDWKHTLDGYDRIPNKEIQKILKVSYDALEEEEQSV 442
++YA G PL + V G L G K + + + R P +I K SYD L + E+++
Sbjct: 334 ISYANGNPLAISVYGRELKGKKKLSEMETAFLKLKRRPPFKIVDAFKSSYDTLSDNEKNI 393
Query: 443 FLDIACCFKGYQWKEFEDIL--CAHYDHCITHHLGVLAGKSLVKISTYYPSGSINDVRLH 500
FLDIAC F+G +L C + H + VL K LV IS N V LH
Sbjct: 394 FLDIACFFQGENVNYVIQLLEGCGFFPHV---EIDVLVDKCLVTISE-------NRVWLH 443
Query: 501 DLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKEN---------------TGTSKIE 545
L +D+G+E++ E+ + R RLW I ++L+ N G+ +IE
Sbjct: 444 KLTQDIGREIINGET-VQIERRRRLWEPWSIKYLLEYNEHKANGEPKTTFKRAQGSEEIE 502
Query: 546 MIYMNLHSMESVIDKKGKAFKKMTKLKTLII---------ENGLFSGGLKYLPSSLRVLK 596
++++ ++ D + AFK M L+ L I +G L LP+ LR+L
Sbjct: 503 GLFLDTSNLR--FDLQPSAFKNMLNLRLLKIYCSNPEVHPVINFPTGSLHSLPNELRLLH 560
Query: 597 WKGCLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPD 656
W+ K L + + +SQ +K + + + ++ + L + ++L I D
Sbjct: 561 WENYPLKSLPQNF--DPRHLVEINMPYSQLQKLW-GGTKNLEMLRTIRLCHSQHLVDIDD 617
Query: 657 VSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLE-------HFRPLGLASL 709
+ NLE + C L + G L +L ++ GC K++ + L L
Sbjct: 618 LLKAENLEVIDLQGCTRLQNF-PAAGRLLRLRVVNLSGCIKIKSVLEIPPNIEKLHLQGT 676
Query: 710 KKLILYECECLDNFPELLCKMAHIKEIDISN-----TSIGELPFSFQNLSELHELTVTSG 764
L L N EL+ + I + ++ TS+ E S Q+L +L L +
Sbjct: 677 GILALPVSTVKPNHRELVNFLTEIPGLSEASKLERLTSLLESNSSCQDLGKLICLELKDC 736
Query: 765 MKFPKIVFSNMTKLSLSFFNLSD-------ECLPIVLKWCV-------NMTHLDLSFSNF 810
+ NM L L+ +LS + P LK + L S
Sbjct: 737 SCLQSL--PNMANLDLNVLDLSGCSSLNSIQGFPRFLKQLYLGGTAIREVPQLPQSLEIL 794
Query: 811 KILPECLRECHHLVE------INVMCCESLEEIRGIPPNLKEL 847
CLR ++ +++ C LE I+G P NLKEL
Sbjct: 795 NAHGSCLRSLPNMANLEFLKVLDLSGCSELETIQGFPRNLKEL 837
Score = 45.1 bits (105), Expect = 0.001
Identities = 31/90 (34%), Positives = 51/90 (56%), Gaps = 7/90 (7%)
Query: 426 KILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKI 485
++L+VSYD L+E ++ +FL IA F + +F L A D ++ L VLA SL+ +
Sbjct: 1087 EVLRVSYDDLQEMDKVLFLYIASLFND-EDVDFVAPLIAGIDLDVSSGLKVLADVSLISV 1145
Query: 486 STYYPSGSINDVRLHDLIKDMGKEVVRQES 515
S S ++ +H L + MGKE++ +S
Sbjct: 1146 S------SNGEIVMHSLQRQMGKEILHGQS 1169
>DR42_ARATH (Q9FKZ1) Probable disease resistance protein At5g66900
Length = 809
Score = 105 bits (261), Expect = 8e-22
Identities = 151/646 (23%), Positives = 263/646 (40%), Gaps = 129/646 (19%)
Query: 196 VGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQI--------------YNFV 241
VGL + ++K L DD V + + G GK+TL ++ +N V
Sbjct: 169 VGLDWPLGELKKRL---LDDSVVTLVVSAPPGCGKTTLVSRLCDDPDIKGKFKHIFFNVV 225
Query: 242 ADQFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRKKIL 301
++ + ++ +++ N L + + GL L+ + E P IL
Sbjct: 226 SNTPNFRVIVQNLLQHNGYNALTFENDSQA--EVGLRKLLEELKENGP----------IL 273
Query: 302 LILDDVDNLKQLHALAGGLDWFGCGS------------RVIITTRNKDLLSSHGIESTHA 349
L+LDDV W G S ++++T+R +S +
Sbjct: 274 LVLDDV--------------WRGADSFLQKFQIKLPNYKILVTSR----FDFPSFDSNYR 315
Query: 350 VEGLNETEALELL-RWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIED 408
++ L + +A LL W + + P YED+L + + G P+V+EVVG +L G+S+
Sbjct: 316 LKPLEDDDARALLIHWASRPCNTSPDEYEDLLQKILKRCNGFPIVIEVVGVSLKGRSLNT 375
Query: 409 WKHTLDGYDR------IPNKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQ-------- 454
WK ++ + P + + L+ S+DAL+ + FLD+ + +
Sbjct: 376 WKGQVESWSEGEKILGKPYPTVLECLQPSFDALDPNLKECFLDMGSFLEDQKIRASVIID 435
Query: 455 -WKEFEDILCAHYDHCITHHLGVLAGKSLVKI----STYYPSGSIND--VRLHDLIKDMG 507
W E L + +L LA ++L+K+ + + G ND V HD+++++
Sbjct: 436 MWVE----LYGKGSSILYMYLEDLASQNLLKLVPLGTNEHEDGFYNDFLVTQHDILREL- 490
Query: 508 KEVVRQESPKEPGERSRL-----------W----RQEDIIHVLKENTGTSK--------I 544
+ Q KE ER RL W ++ + ++ +SK +
Sbjct: 491 --AICQSEFKENLERKRLNLEILENTFPDWCLNTINASLLSISTDDLFSSKWLEMDCPNV 548
Query: 545 EMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGG-------LKYLPSSLRVLKW 597
E + +NL S + + M KLK L I N F L LP+ R+
Sbjct: 549 EALVLNLSSSDYALP---SFISGMKKLKVLTITNHGFYPARLSNFSCLSSLPNLKRIRLE 605
Query: 598 KGCLSKC----LSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTH 653
K ++ L S L K S + F +V ++ + +DYC L
Sbjct: 606 KVSITLLDIPQLQLSSLKKLSLVMCS-FGEVFYDTEDIVVSNALSKLQEIDIDYCYDLDE 664
Query: 654 IPD-VSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPL--GLASLK 710
+P +S + +L+ LS T C+ L + +IG+L++LE L L GL++L+
Sbjct: 665 LPYWISEIVSLKTLSITNCNKLSQLPEAIGNLSRLEVLRLCSSMNLSELPEATEGLSNLR 724
Query: 711 KLILYECECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSEL 756
L + C L P+ + K+ ++K+I + S ELP S NL L
Sbjct: 725 FLDISHCLGLRKLPQEIGKLQNLKKISMRKCSGCELPESVTNLENL 770
>DR23_ARATH (O82484) Putative disease resistance protein At4g10780
Length = 892
Score = 86.3 bits (212), Expect = 4e-16
Identities = 133/518 (25%), Positives = 219/518 (41%), Gaps = 69/518 (13%)
Query: 214 DDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCFLHDVRENSAQNNLKYLQEKL--L 271
DDGV +G+YG+GG+GK+TL QI+N + D G + V S+ + +QE +
Sbjct: 170 DDGVGTMGLYGMGGVGKTTLLTQIHNTLHDTKNGVDIVIWV-VVSSDLQIHKIQEDIGEK 228
Query: 272 LKTTGLEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVII 331
L G E S+ I L +K+ +L+LDD+ L + +V+
Sbjct: 229 LGFIGKEWNKKQESQKAVDILNCLSKKRFVLLLDDIWKKVDLTKIGIPSQTRENKCKVVF 288
Query: 332 TTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKVPSG-YEDILNRAVAYA--- 387
TTR+ D+ + G+ V+ L+ +A EL + K ++ G + DIL A A
Sbjct: 289 TTRSLDVCARMGVHDPMEVQCLSTNDAWELFQ---EKVGQISLGSHPDILELAKKVAGKC 345
Query: 388 FGLPLVLEVVGSNLFGK-SIEDWKHTLDGYDRIP------NKEIQKILKVSYDALEEEE- 439
GLPL L V+G + GK ++++W H +D + I ILK SYD L ++
Sbjct: 346 RGLPLALNVIGETMAGKRAVQEWHHAVDVLTSYAAEFSGMDDHILLILKYSYDNLNDKHV 405
Query: 440 QSVFLDIACCFKGYQWKEFEDI---LCAHY-DHCITHHLGVLAG----KSLVKISTYYPS 491
+S F A + Y K++ I +C + D I V G +LV+
Sbjct: 406 RSCFQYCALYPEDYSIKKYRLIDYWICEGFIDGNIGKERAVNQGYEILGTLVRACLLSEE 465
Query: 492 GSIN-DVRLHDLIK--------DMGKEVVRQESPKEPGERSRLWRQED---IIHVLKENT 539
G +V++HD+++ D+GK R G R ++ + ED + + N
Sbjct: 466 GKNKLEVKMHDVVREMALWTLSDLGKNKERCIVQAGSGLR-KVPKVEDWGAVRRLSLMNN 524
Query: 540 GTSKI-------EMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSL 592
G +I E+ + L +S++ G+ F+ M KL L + GL S L
Sbjct: 525 GIEEISGSPECPELTTLFLQENKSLVHISGEFFRHMRKLVVLDLSENHQLDGLPEQISEL 584
Query: 593 RVLKWKGCLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLT 652
L++ L S N + C+Q + + L L+ L
Sbjct: 585 VALRY-------LDLSHTN---------------IEGLPACLQDLKTLIHLNLECMRRLG 622
Query: 653 HIPDVSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWL 690
I +S LS+L L + ++ + S+ L+ LE L
Sbjct: 623 SIAGISKLSSLRTLGLRNSNIMLDV-MSVKELHLLEHL 659
Score = 39.3 bits (90), Expect = 0.056
Identities = 34/118 (28%), Positives = 56/118 (46%), Gaps = 16/118 (13%)
Query: 733 IKEIDISNTSIGELPFSFQNLSELHELTVTSGMKFPKIV------FSNMTKLSLSFFNLS 786
++ + + N I E+ S E ELT + +V F +M KL + +LS
Sbjct: 516 VRRLSLMNNGIEEISGS----PECPELTTLFLQENKSLVHISGEFFRHMRKLVV--LDLS 569
Query: 787 D----ECLPIVLKWCVNMTHLDLSFSNFKILPECLRECHHLVEINVMCCESLEEIRGI 840
+ + LP + V + +LDLS +N + LP CL++ L+ +N+ C L I GI
Sbjct: 570 ENHQLDGLPEQISELVALRYLDLSHTNIEGLPACLQDLKTLIHLNLECMRRLGSIAGI 627
Score = 32.7 bits (73), Expect = 5.2
Identities = 24/87 (27%), Positives = 43/87 (48%), Gaps = 5/87 (5%)
Query: 696 RKLEHFRPLGLASLKKLI---LYECECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQN 752
+ L H ++KL+ L E LD PE + ++ ++ +D+S+T+I LP Q+
Sbjct: 547 KSLVHISGEFFRHMRKLVVLDLSENHQLDGLPEQISELVALRYLDLSHTNIEGLPACLQD 606
Query: 753 LSELHELTVTSGMKFPKIVFSNMTKLS 779
L L L + + I + ++KLS
Sbjct: 607 LKTLIHLNLECMRRLGSI--AGISKLS 631
>DR19_ARATH (Q9C8T9) Putative disease resistance protein At1g63350
Length = 898
Score = 85.9 bits (211), Expect = 5e-16
Identities = 151/667 (22%), Positives = 277/667 (40%), Gaps = 108/667 (16%)
Query: 173 IGKIVKYISNKISRQPLHVATYPVGLQSRVQQVKSLLDEG----SDDGVHMVGIYGIGGL 228
+ K+ + + IS Q LQ + +++LD +DGV ++G+YG+GG+
Sbjct: 124 VEKLERRVFEVISDQASTSEVEEQQLQPTIVGQETMLDNAWNHLMEDGVGIMGLYGMGGV 183
Query: 229 GKSTLARQIYNFVADQ---FEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVS 285
GK+TL QI N + F+ ++ +E + +N L + +K+ + + K +
Sbjct: 184 GKTTLLTQINNKFSKYMCGFDSVIWVVVSKEVNVENILDEIAQKVHISGEKWDTKYKY-Q 242
Query: 286 EGIPVIKERLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIE 345
+G+ + L + + +L LDD+ L + +V+ TTR+ D+ +S G+E
Sbjct: 243 KGV-YLYNFLRKMRFVLFLDDIWEKVNLVEIGVPFPTIKNKCKVVFTTRSLDVCTSMGVE 301
Query: 346 STHAVEGLNETEALELLRWMAFKSDKVPSGYED---ILNRAVA-YAFGLPLVLEVVGSNL 401
V+ L + +A +L + K ++ G + L+R VA GLPL L VV +
Sbjct: 302 KPMEVQCLADNDAYDLFQ---KKVGQITLGSDPEIRELSRVVAKKCCGLPLALNVVSETM 358
Query: 402 FGK-SIEDWKHTLDGYDRIPNK------EIQKILKVSYDALEEEEQSVFLDIACCFKG-- 452
K ++++W+H + + K +I +LK SYD+L+ E+ + L F
Sbjct: 359 SCKRTVQEWRHAIYVLNSYAAKFSGMDDKILPLLKYSYDSLKGEDVKMCLLYCALFPEDA 418
Query: 453 -------YQWKEFEDILCAHYDHCITHHLGVLAGKSLVKISTYYPSGSI---NDVRLHDL 502
++ E+I+ + G SLV+ S + N V LHD+
Sbjct: 419 KIRKENLIEYWICEEIIDGSEGIDKAENQGYEIIGSLVRASLLMEEVELDGANIVCLHDV 478
Query: 503 IK--------DMGKE----VVRQESPKEPGERSRLWR--------QEDIIH--------- 533
++ D+GK+ +VR + W + +I H
Sbjct: 479 VREMALWIASDLGKQNEAFIVRASVGLREILKVENWNVVRRMSLMKNNIAHLDGRLDCME 538
Query: 534 ---VLKENTGTSKIEMIYMNLHSMESVIDKKGKAF-----KKMTKLKTLIIENGLFSGGL 585
+L ++T KI + N +V+D G + +++L +L N L S G+
Sbjct: 539 LTTLLLQSTHLEKISSEFFNSMPKLAVLDLSGNYYLSELPNGISELVSLQYLN-LSSTGI 597
Query: 586 KYLPSSLRVLKWKGCLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTL 645
++LP L+ LK +++ E TSQ +V + N+KVL L
Sbjct: 598 RHLPKGLQELK-----------KLIHLYLERTSQ--------LGSMVGISCLHNLKVLKL 638
Query: 646 DYCEY---LTHIPDVSGLSNLEKLSFTC------CDNLITIHNSIGHLNKLEWLSAYGCR 696
Y L + ++ L +LE L+ T D ++ H + + L+ +S R
Sbjct: 639 SGSSYAWDLDTVKELEALEHLEVLTTTIDDCTLGTDQFLSSHRLMSCIRFLK-ISNNSNR 697
Query: 697 KLEHFR---PLGLASLKKLILYECECLDNFPELLCKMAHIKEIDISN-TSIGELPFSF-- 750
R P+ + L++ + C + +C + + E+++SN + EL F
Sbjct: 698 NRNSSRISLPVTMDRLQEFTIEHCHTSEIKMGRICSFSSLIEVNLSNCRRLRELTFLMFA 757
Query: 751 QNLSELH 757
NL LH
Sbjct: 758 PNLKRLH 764
>DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730
Length = 848
Score = 81.3 bits (199), Expect = 1e-14
Identities = 73/238 (30%), Positives = 122/238 (50%), Gaps = 16/238 (6%)
Query: 214 DDGVHMVGIYGIGGLGKSTLARQIYN-FVADQFEGSCFLHDVRENSAQNNLKYLQEKLLL 272
DD + +G+YG+GG+GK+TL + N FV + E + V Q L+ +Q+++L
Sbjct: 169 DDEIRTLGLYGMGGIGKTTLLESLNNKFVELESEFDVVIWVVVSKDFQ--LEGIQDQILG 226
Query: 273 K-TTGLEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVII 331
+ E + + S+ +I L RKK +L+LDD+ + L + GS+++
Sbjct: 227 RLRPDKEWERETESKKASLINNNLKRKKFVLLLDDLWSEVDLIKIGVPPPSRENGSKIVF 286
Query: 332 TTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKVPSGYEDI--LNRAV-AYAF 388
TTR+K++ + V+ L+ EA EL R D + ++DI L R V A
Sbjct: 287 TTRSKEVCKHMKADKQIKVDCLSPDEAWELFRLTV--GDIILRSHQDIPALARIVAAKCH 344
Query: 389 GLPLVLEVVGSNLFGK-SIEDWKHTLDGYD----RIPNKE--IQKILKVSYDALEEEE 439
GLPL L V+G + K ++++W+H ++ + + P E I ILK SYD+L+ E
Sbjct: 345 GLPLALNVIGKAMVCKETVQEWRHAINVLNSPGHKFPGMEERILPILKFSYDSLKNGE 402
>DR37_ARATH (Q9LVT4) Probable disease resistance protein At5g47250
Length = 843
Score = 80.1 bits (196), Expect = 3e-14
Identities = 124/493 (25%), Positives = 207/493 (41%), Gaps = 65/493 (13%)
Query: 126 EALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGKI--VKYISNK 183
E + E+ K+ + + A+ + + SG S K+ K+ VK +S K
Sbjct: 76 EIIEENTKQLMDVASARDASSQNASAVRRRLSTSGCWFSTCNLGEKVFKKLTEVKSLSGK 135
Query: 184 ----ISRQP------LHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTL 233
++ QP + + VGL + +++ L + D M+GI+G+GG+GK+TL
Sbjct: 136 DFQEVTEQPPPPVVEVRLCQQTVGLDTTLEKTWESLRK---DENRMLGIFGMGGVGKTTL 192
Query: 234 ARQIYN---FVADQFEGSCFLHDVRENSAQNNLKYLQEKLLL-----KTTGLEIKLDHVS 285
I N V+D ++ ++ ++ + E+L + T K +S
Sbjct: 193 LTLINNKFVEVSDDYDVVIWVESSKDADVGKIQDAIGERLHICDNNWSTYSRGKKASEIS 252
Query: 286 EGIPVIKERLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIE 345
+ +K R +L+LDD+ L A+ G+ G +V+ TTR+KD+ S
Sbjct: 253 RVLRDMKPR-----FVLLLDDLWEDVSLTAI--GIPVLGKKYKVVFTTRSKDVCSVMRAN 305
Query: 346 STHAVEGLNETEALELLRWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKS 405
V+ L+E +A +L M D + + DI + VA GLPL LEV+ + KS
Sbjct: 306 EDIEVQCLSENDAWDLFD-MKVHCDGL-NEISDIAKKIVAKCCGLPLALEVIRKTMASKS 363
Query: 406 -IEDWKHTLDGYD------RIPNKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEF 458
+ W+ LD + + K I ++LK+SYD L+ + FL A K Y K+
Sbjct: 364 TVIQWRRALDTLESYRSEMKGTEKGIFQVLKLSYDYLKTKNAKCFLYCALFPKAYYIKQD 423
Query: 459 EDILCAHYDHCITHHLGVLAGK--SLVKISTYYPSG----SINDVRLHDLIKDMGKEVVR 512
E + + I G K I +G S V +HD+I+DM +V
Sbjct: 424 ELVEYWIGEGFIDEKDGRERAKDRGYEIIDNLVGAGLLLESNKKVYMHDMIRDMALWIV- 482
Query: 513 QESPKEPGERSRLWRQEDIIHVLKENTGTSKI-------EMIYMNLHSMESVIDKKGKAF 565
S GER +V+K + G S++ + M+L + E F
Sbjct: 483 --SEFRDGER----------YVVKTDAGLSQLPDVTDWTTVTKMSLFNNEIKNIPDDPEF 530
Query: 566 KKMTKLKTLIIEN 578
T L TL ++N
Sbjct: 531 PDQTNLVTLFLQN 543
>R133_ARATH (Q9STE7) Putative disease resistance RPP13-like protein
3
Length = 847
Score = 79.3 bits (194), Expect = 5e-14
Identities = 107/455 (23%), Positives = 199/455 (43%), Gaps = 58/455 (12%)
Query: 196 VGLQSRVQ-QVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNF--VADQFEGSCFLH 252
VGL+ V+ + LL + D +++ I+G+GGLGK+ LAR++YN V +F+ + +
Sbjct: 163 VGLEDDVKILLVKLLSDNEKDKSYIISIFGMGGLGKTALARKLYNSGDVKRRFDCRAWTY 222
Query: 253 DVRENSAQNNL-KYLQEKLLLKTTGLE-IKLDHVSEGIPVIKERLCR-KKILLILDDVDN 309
+E ++ L + ++ ++ +E IK+ E + V L K ++++DDV +
Sbjct: 223 VSQEYKTRDILIRIIRSLGIVSAEEMEKIKMFEEDEELEVYLYGLLEGKNYMVVVDDVWD 282
Query: 310 LKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIEST---HAVEGLNETEALELLRWMA 366
+L L GS+VIITTR + + + G+E T H + L E+ L A
Sbjct: 283 PDAWESLKRALPCDHRGSKVIITTRIRAI--AEGVEGTVYAHKLRFLTFEESWTLFERKA 340
Query: 367 FKS-DKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDG-YDRIPNK-- 422
F + +KV + V GLPL + V+ L K +W + R+ +
Sbjct: 341 FSNIEKVDEDLQRTGKEMVKKCGGLPLAIVVLSGLLSRKRTNEWHEVCASLWRRLKDNSI 400
Query: 423 EIQKILKVSYDALEEEEQSVFLDIACCFKGYQWK---------------EFEDILCAHYD 467
I + +S+ + E + FL + + Y+ K E E+++
Sbjct: 401 HISTVFDLSFKEMRHELKLCFLYFSVFPEDYEIKVEKLIHLLVAEGFIQEDEEMMMEDVA 460
Query: 468 HCITHHLGVLAGKSLVKISTYYPSGSINDVRLHDLIKDM----GKEVVRQESPKEPGERS 523
C ++ L +SLVK + G + R+HDL++D+ KE+ E S
Sbjct: 461 RC---YIDELVDRSLVK-AERIERGKVMSCRIHDLLRDLAIKKAKELNFVNVYNEKQHSS 516
Query: 524 RLWRQEDIIHVLKENTGTSKIEMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSG 583
+ R+E + H++ + + + + S + +++G + T LK +
Sbjct: 517 DICRREVVHHLMNDYYLCDR--RVNKRMRSFLFIGERRGFGYVNTTNLKLKL-------- 566
Query: 584 GLKYLPSSLRVLKWKGCL--SKCLSSSILNKASEI 616
LRVL +G L SK +S+++ + E+
Sbjct: 567 --------LRVLNMEGLLFVSKNISNTLPDVIGEL 593
>DRL2_ARATH (P60839) Probable disease resistance protein At1g12290
Length = 884
Score = 79.0 bits (193), Expect = 6e-14
Identities = 159/695 (22%), Positives = 285/695 (40%), Gaps = 106/695 (15%)
Query: 172 LIGKIVKYISNK-ISRQPLHVATYPVG----LQSRVQQVKSLLDEG----SDDGVHMVGI 222
L+ IV+ + +K I + H AT VG LQ + +++L++ DDG ++G+
Sbjct: 121 LMLNIVEDLKSKGIFEEVAHPATRAVGEERPLQPTIVGQETILEKAWDHLMDDGTKIMGL 180
Query: 223 YGIGGLGKSTLARQIYNFVADQFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLD 282
YG+GG+GK+TL QI N D +G + V S + +Q+++ K + ++ +
Sbjct: 181 YGMGGVGKTTLLTQINNRFCDTDDGVEIVIWV-VVSGDLQIHKIQKEIGEKIGFIGVEWN 239
Query: 283 HVSEGIPV--IKERLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLS 340
SE I L +K+ +L+LDD+ +L + G ++ TTR + + +
Sbjct: 240 QKSENQKAVDILNFLSKKRFVLLLDDIWKRVELTEIGIPNPTSENGCKIAFTTRCQSVCA 299
Query: 341 SHGIESTHAVEGLNETEALELLRWMAFKSDKVPSGYEDI--LNRAVAYA-FGLPLVLEVV 397
S G+ V L +A +L + D S + DI + R VA A GLPL L V+
Sbjct: 300 SMGVHDPMEVRCLGADDAWDLFKKKV--GDITLSSHPDIPEIARKVAQACCGLPLALNVI 357
Query: 398 GSNL-FGKSIEDWKHTLD-------GYDRIPNKEIQKILKVSYDALEEEEQSVFLDIACC 449
G + K+ ++W +D + + + I ILK SYD LE E
Sbjct: 358 GETMACKKTTQEWDRAVDVSTTYAANFGAV-KERILPILKYSYDNLESESVKTCFLYCSL 416
Query: 450 FKGYQWKEFEDI----LCAHYDHCITHHLGVLAG-----KSLVKISTYYPSGSIND---V 497
F E E + +C + + G + +LV S G N+ V
Sbjct: 417 FPEDDLIEKERLIDYWICEGFIDGDENKKGAVGEGYEILGTLVCASLLVEGGKFNNKSYV 476
Query: 498 RLHDLIKDMG------------KEVVRQESPKEPGERSRLW----RQEDIIHVLKENTGT 541
++HD++++M +VR + + W R + + +KE G+
Sbjct: 477 KMHDVVREMALWIASDLRKHKDNCIVRAGFRLNEIPKVKDWKVVSRMSLVNNRIKEIHGS 536
Query: 542 SKI-EMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGC 600
+ ++ + L +++ G+ F+ M +L L + + GL S L L++
Sbjct: 537 PECPKLTTLFLQDNRHLVNISGEFFRSMPRLVVLDLSWNVNLSGLPDQISELVSLRYLD- 595
Query: 601 LSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPDVSGL 660
LS S + + V + K + + L L E + + VSG+
Sbjct: 596 ----LSYSSIGRLP-----------------VGLLKLKKLMHLNL---ESMLCLESVSGI 631
Query: 661 SNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLILYECECL 720
+L L NL + +I L +LE +LE+ L + + L + C
Sbjct: 632 DHLSNLKTVRLLNL-RMWLTISLLEELE--------RLENLEVLTIEIISSSALEQLLCS 682
Query: 721 DNFPELLCKMAHIKEID------ISNTSIGELPFSFQNLSELHELTVTSGMKFPKIVFSN 774
L K++ +K +D ++ SIG+L F + ++ + F N
Sbjct: 683 HRLVRCLQKVS-VKYLDEESVRILTLPSIGDLREVFIGGCGMRDIIIERNTSLTSPCFPN 741
Query: 775 MTKLSLSFFN-LSDECLPIVLKWCV---NMTHLDL 805
++K+ ++ N L D L W + N+THL++
Sbjct: 742 LSKVLITGCNGLKD------LTWLLFAPNLTHLNV 770
>DR13_ARATH (Q9XIF0) Putative disease resistance protein At1g59780
Length = 906
Score = 78.6 bits (192), Expect = 8e-14
Identities = 154/669 (23%), Positives = 276/669 (41%), Gaps = 123/669 (18%)
Query: 196 VGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNF--VADQFEGSCFLHD 253
VGL+ V+++ L +D H V I G+GGLGK+TLARQI++ V F+G ++
Sbjct: 161 VGLEKNVEKLVEELV--GNDSSHGVSITGLGGLGKTTLARQIFDHDKVKSHFDGLAWVCV 218
Query: 254 VRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQL 313
+E + ++ K + L K ++ D + + + + E KK L++ DD+ +
Sbjct: 219 SQEFTRKDVWKTILGNLSPKYKDSDLPEDDIQKKLFQLLE---TKKALIVFDDLWKREDW 275
Query: 314 HALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKVP 373
+ +A G +V++T+RN D + H + T E L E +LL+ +AF K
Sbjct: 276 YRIAPMFPERKAGWKVLLTSRN-DAIHPHCV--TFKPELLTHDECWKLLQRIAFSKQKTI 332
Query: 374 SGY---EDILNRA---VAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKI 427
+GY ++++ A + LPL ++++G L D KHTL + I I I
Sbjct: 333 TGYIIDKEMVKMAKEMTKHCKRLPLAVKLLGGLL------DAKHTLRQWKLISENIISHI 386
Query: 428 LKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITH-------------HL 474
+ + E + SV ++ F+G L + HC+ + L
Sbjct: 387 VVGGTSSNENDSSSVNHVLSLSFEG---------LPGYLKHCLLYLASYPEDHEIEIERL 437
Query: 475 GVLAGKSLVKISTYYPSGSINDVR---LHDLIKDMGKEVVRQESPKEPGERSRLW----- 526
+ + Y +I DV + +L+K + +++ E+ +L
Sbjct: 438 SYVWAAEGITYPGNYEGATIRDVADLYIEELVK-RNMVISERDALTSRFEKCQLHDLMRE 496
Query: 527 ------RQEDIIHVLKENTGTSKIEMIYMNLHSMESVIDK---KGKAFKKMTKLKTLI-- 575
++E+ + ++ + T +S + + + V + G+ K +KL++L+
Sbjct: 497 ICLLKAKEENFLQIVTDPTSSSSVHSLASSRSRRLVVYNTSIFSGENDMKNSKLRSLLFI 556
Query: 576 -IENGLFSGGLKYLP-SSLRVLKWKGCLSK--CLSSSILNKASEITSQ*FCFSQQKKSFL 631
+ FS G ++ LRVL G K L SSI
Sbjct: 557 PVGYSRFSMGSNFIELPLLRVLDLDGAKFKGGKLPSSI---------------------- 594
Query: 632 VCMQKFQNMKVLTLDYCEYLTHIPDVSGLSNLEKLSF----TCCDNLITIHNSIGHLNKL 687
K ++K L+L Y +T++P S L NL+ L + LI + N + +L
Sbjct: 595 ---GKLIHLKYLSL-YQASVTYLP--SSLRNLKSLLYLNLRINSGQLINVPNVFKEMLEL 648
Query: 688 EWLSAYGCRKLEHFRPLGLASLKKL-ILYECECLDNFPELLCKMAHIKEIDISNTSIG-- 744
+LS R L L +L KL L D+ L +M ++ + I + G
Sbjct: 649 RYLSLPWER--SSLTKLELGNLLKLETLINFSTKDSSVTDLHRMTKLRTLQILISGEGLH 706
Query: 745 --ELPFSFQNLSELHELTVTSG-----MKFPKIVF-----------SNMTKLSLSFFNLS 786
L + L L +LTVT K PK+++ S++T +SL + L
Sbjct: 707 METLSSALSMLGHLEDLTVTPSENSVQFKHPKLIYRPMLPDVQHFPSHLTTISLVYCFLE 766
Query: 787 DECLPIVLK 795
++ +P + K
Sbjct: 767 EDPMPTLEK 775
>RFL1_ARATH (Q8L3R3) Disease resistance protein RFL1 (RPS5-like
protein 1) (pNd13/pNd14)
Length = 885
Score = 77.0 bits (188), Expect = 2e-13
Identities = 77/271 (28%), Positives = 132/271 (48%), Gaps = 20/271 (7%)
Query: 187 QPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQI---YNFVAD 243
+ L + + VG S + +V + L E D V +VG+YG+GG+GK+TL QI ++ +
Sbjct: 149 EELPIQSTIVGQDSMLDKVWNCLME---DKVWIVGLYGMGGVGKTTLLTQINNKFSKLGG 205
Query: 244 QFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRKKILLI 303
F+ ++ + + K + EKL L + K + ++ I L RKK +L+
Sbjct: 206 GFDVVIWVVVSKNATVHKIQKSIGEKLGLVGKNWDEK--NKNQRALDIHNVLRRKKFVLL 263
Query: 304 LDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLR 363
LDD+ +L + G +V TT +K++ G+++ + L+ A +LL+
Sbjct: 264 LDDIWEKVELKVIGVPYPSGENGCKVAFTTHSKEVCGRMGVDNPMEISCLDTGNAWDLLK 323
Query: 364 WMAFKSDKVPSGYEDI--LNRAVA-YAFGLPLVLEVVGSNL-FGKSIEDWKHTLDGYDRI 419
+ + DI L R V+ GLPL L V+G + F ++I++W+H +
Sbjct: 324 KKV--GENTLGSHPDIPQLARKVSEKCCGLPLALNVIGETMSFKRTIQEWRHATEVLTSA 381
Query: 420 PN-----KEIQKILKVSYDALE-EEEQSVFL 444
+ EI ILK SYD+L E+ +S FL
Sbjct: 382 TDFSGMEDEILPILKYSYDSLNGEDAKSCFL 412
>DR31_ARATH (Q9FLB4) Putative disease resistance protein At5g05400
Length = 874
Score = 76.3 bits (186), Expect = 4e-13
Identities = 83/323 (25%), Positives = 155/323 (47%), Gaps = 36/323 (11%)
Query: 205 VKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYN---FVADQFEGSCFLHDVRENSAQN 261
V+S + + GV ++GIYG+GG+GK+TL QI N V++ F+ + ++ + + +
Sbjct: 163 VESTWNSMMEVGVGLLGIYGMGGVGKTTLLSQINNKFRTVSNDFDIAIWVVVSKNPTVKR 222
Query: 262 NLKYLQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQLHALAGGLD 321
+ + ++L L G E K + +E IK L KK +L+LDD+ L + +
Sbjct: 223 IQEDIGKRLDLYNEGWEQKTE--NEIASTIKRSLENKKYMLLLDDMWTKVDLANIGIPVP 280
Query: 322 WFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALEL----LRWMAFKSDKVPSGYE 377
GS++ T+R+ ++ G++ V L +A +L ++ K+P +
Sbjct: 281 KRN-GSKIAFTSRSNEVCGKMGVDKEIEVTCLMWDDAWDLFTRNMKETLESHPKIPEVAK 339
Query: 378 DILNRAVAYAFGLPLVLEVVGSNLF-GKSIEDWKHTLDGYDRIPNKEIQKILKVSYDALE 436
I + GLPL L V+G + KSIE+W + + I +I ILK SYD L+
Sbjct: 340 SIARK----CNGLPLALNVIGETMARKKSIEEWHDAVGVFSGI-EADILSILKFSYDDLK 394
Query: 437 -EEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKISTYYPSGSI- 494
E+ +S FL A + Y+ + +D++ ++ + G++ G + Y G++
Sbjct: 395 CEKTKSCFLFSALFPEDYEIGK-DDLI----EYWVGQ--GIILGSKGINYKGYTIIGTLT 447
Query: 495 -----------NDVRLHDLIKDM 506
V++HD++++M
Sbjct: 448 RAYLLKESETKEKVKMHDVVREM 470
>DR28_ARATH (O81825) Putative disease resistance protein At4g27220
Length = 919
Score = 76.3 bits (186), Expect = 4e-13
Identities = 96/360 (26%), Positives = 158/360 (43%), Gaps = 42/360 (11%)
Query: 176 IVKYIS-NKISRQPLHVATYPV--GLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKST 232
++K IS NK SR+ + P ++ ++ + L D V +G++G+GG+GK+T
Sbjct: 90 LIKKISVNKSSREIVERVLGPSFHPQKTALEMLDKLKDCLKKKNVQKIGVWGMGGVGKTT 149
Query: 233 LARQIYNFVADQFEGSCFLHDVRENSAQN-NLKYLQEKLLLKTTGLEIKLDHVSE-GIPV 290
L R + N + F + +++ +LK +Q + K G + +++ G+ +
Sbjct: 150 LVRTLNNDLLKYAATQQFALVIWVTVSKDFDLKRVQMD-IAKRLGKRFTREQMNQLGLTI 208
Query: 291 IKERLCRKKILLILDDV------DNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGI 344
+ + K LLILDDV D L AL D S+V++T+R ++
Sbjct: 209 CERLIDLKNFLLILDDVWHPIDLDQLGIPLALERSKD-----SKVVLTSRRLEVCQQMMT 263
Query: 345 ESTHAVEGLNETEALELL---RWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNL 401
V L E EA EL SD V +D+ + GLPL + +G L
Sbjct: 264 NENIKVACLQEKEAWELFCHNVGEVANSDNVKPIAKDVSHECC----GLPLAIITIGRTL 319
Query: 402 FGK-SIEDWKHTLDGYDRI-----PNKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQW 455
GK +E WKHTL+ R ++I LK+SYD L++ +S FL A + Y
Sbjct: 320 RGKPQVEVWKHTLNLLKRSAPSIDTEEKIFGTLKLSYDFLQDNMKSCFLFCALFPEDYSI 379
Query: 456 KEFEDILC----------AHYDHCITHHLGVLAGKSLVKISTYYPSGSINDVRLHDLIKD 505
K E I+ HY+ + GV + L S + V++HD+++D
Sbjct: 380 KVSELIMYWVAEGLLDGQHHYEDMMNE--GVTLVERLKDSCLLEDGDSCDTVKMHDVVRD 437
Score = 36.2 bits (82), Expect = 0.47
Identities = 22/74 (29%), Positives = 39/74 (51%), Gaps = 1/74 (1%)
Query: 706 LASLKKLILYECECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSGM 765
L SL+ L+L C+ L N P L + ++ +D+ ++I ELP + LS L + V++
Sbjct: 540 LHSLRSLVLRNCKKLRNLPSLE-SLVKLQFLDLHESAIRELPRGLEALSSLRYICVSNTY 598
Query: 766 KFPKIVFSNMTKLS 779
+ I + +LS
Sbjct: 599 QLQSIPAGTILQLS 612
>DRL5_ARATH (Q9C8K0) Probable disease resistance protein At1g51480
Length = 854
Score = 74.7 bits (182), Expect = 1e-12
Identities = 145/603 (24%), Positives = 252/603 (41%), Gaps = 106/603 (17%)
Query: 183 KISRQPLHVATYPVGLQSRVQQV-KSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYN-F 240
K+ + +H VGL + V+ KSL++ D + + ++G+GG+GK+TL I N F
Sbjct: 145 KVEEKNIHTT---VGLYAMVEMAWKSLMN----DEIRTLCLHGMGGVGKTTLLACINNKF 197
Query: 241 VADQFEGSCFLHDVRENSAQNNLKYLQEKLLLK-TTGLEIKLDHVSEGIPVIKERLCRKK 299
V + E + V Q L+ +Q+++L + E + + ++ +I L RKK
Sbjct: 198 VELESEFDVVIWVVVSKDFQ--LEGIQDQILGRLRLDKEWERETENKKASLINNNLKRKK 255
Query: 300 ILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEAL 359
+L+LDD+ + L+ + G++++ T R+K++ + V L+ EA
Sbjct: 256 FVLLLDDLWSEVDLNKIGVPPPTRENGAKIVFTKRSKEVSKYMKADMQIKVSCLSPDEAW 315
Query: 360 ELLRWMAFKSDKVPSGYEDI--LNRAV-AYAFGLPLVLEVVGSNLFGK-SIEDWKHTLDG 415
EL R D + S +EDI L R V A GLPL L V+G + K +I++W H ++
Sbjct: 316 ELFRITV--DDVILSSHEDIPALARIVAAKCHGLPLALIVIGEAMACKETIQEWHHAINV 373
Query: 416 YD-----RIPNKE--IQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDH 468
+ + P E I +LK SYD+L+ E I CF + C+ +
Sbjct: 374 LNSPAGHKFPGMEERILLVLKFSYDSLKNGE------IKLCF----------LYCSLFPE 417
Query: 469 CITHHLGVLAGKSLVKISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQ 528
L I + G IN R D + G +++ G R
Sbjct: 418 DFEIEKEKL-------IEYWICEGYINPNRYEDGGTNQGYDII--------GLLVR---- 458
Query: 529 EDIIHVLKENTGTSKIEMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYL 588
H+L E T+K++M Y+ + GK + +T+ +++G ++ +
Sbjct: 459 ---AHLLIECELTTKVKMHYVIREMALWINSDFGK------QQETICVKSG---AHVRMI 506
Query: 589 PSSLRVLKWKGCLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYC 648
P+ + W+ L S+ + K S C K N+ L L Y
Sbjct: 507 PND---INWEIVRQVSLISTQIEKIS------------------CSSKCSNLSTLLLPYN 545
Query: 649 EYLT-HIPDVSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEW--LSAYGCRKLEHFRPLG 705
+ + + + L L + +LI + I +L L++ LS+ G + L P G
Sbjct: 546 KLVNISVGFFLFMPKLVVLDLSTNMSLIELPEEISNLCSLQYLNLSSTGIKSL----PGG 601
Query: 706 LASLKKLILYECECLDNFPELLCKMAHIKEIDI-----SNTSIGE-LPFSFQNLSELHEL 759
+ L+KLI E L+ A + + + SN + + L Q++ L L
Sbjct: 602 MKKLRKLIYLNLEFSYKLESLVGISATLPNLQVLKLFYSNVCVDDILMEELQHMDHLKIL 661
Query: 760 TVT 762
TVT
Sbjct: 662 TVT 664
>DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740
Length = 862
Score = 74.3 bits (181), Expect = 2e-12
Identities = 93/355 (26%), Positives = 164/355 (46%), Gaps = 37/355 (10%)
Query: 176 IVKYISNKISRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLAR 235
+ + I +K+ ++ + VGL V+ S L +D + +G+YG+GG+GK+TL
Sbjct: 136 VAQEIIHKVEKKLIQTT---VGLDKLVEMAWSSL---MNDEIGTLGLYGMGGVGKTTLLE 189
Query: 236 QIYN-FVADQFEGSCFLHDVRENSAQNNLKYLQEKLLLK-TTGLEIKLDHVSEGIPVIKE 293
+ N FV + E + V Q + +Q+++L + + E + + S+ +I
Sbjct: 190 SLNNKFVELESEFDVVIWVVVSKDFQ--FEGIQDQILGRLRSDKEWERETESKKASLIYN 247
Query: 294 RLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGL 353
L RKK +L+LDD+ + + + GS+++ TTR+ ++ + V L
Sbjct: 248 NLERKKFVLLLDDLWSEVDMTKIGVPPPTRENGSKIVFTTRSTEVCKHMKADKQIKVACL 307
Query: 354 NETEALELLRWMAFKSDKVPSGYEDI--LNRAV-AYAFGLPLVLEVVGSNLFGK-SIEDW 409
+ EA EL R D + ++DI L R V A GLPL L V+G + K +I++W
Sbjct: 308 SPDEAWELFRLTV--GDIILRSHQDIPALARIVAAKCHGLPLALNVIGKAMSCKETIQEW 365
Query: 410 KHTLDGYD----RIPNKE--IQKILKVSYDALEEEEQSVFLDIACCF------KGYQWKE 457
H ++ + P E I ILK SYD+L+ E + F +W E
Sbjct: 366 SHAINVLNSAGHEFPGMEERILPILKFSYDSLKNGEIKLCFLYCSLFPEDSEIPKEKWIE 425
Query: 458 FEDILCA------HYDHCITHHLGVLAGKSLVKISTYYPSGSINDVRLHDLIKDM 506
+ +C Y+ T+H + G LV+ ++V++HD+I++M
Sbjct: 426 Y--WICEGFINPNRYEDGGTNHGYDIIG-LLVRAHLLIECELTDNVKMHDVIREM 477
>RPS5_ARATH (O64973) Disease resistance protein RPS5 (Resistance to
Pseudomonas syringae protein 5) (pNd3/pNd10)
Length = 889
Score = 73.9 bits (180), Expect = 2e-12
Identities = 68/238 (28%), Positives = 116/238 (48%), Gaps = 17/238 (7%)
Query: 214 DDGVHMVGIYGIGGLGKSTLARQI---YNFVADQFEGSCFLHDVRENSAQNNLKYLQEKL 270
+DG ++G+YG+GG+GK+TL +I ++ + D+F+ ++ R ++ + + + EK+
Sbjct: 173 EDGSGILGLYGMGGVGKTTLLTKINNKFSKIDDRFDVVIWVVVSRSSTVRKIQRDIAEKV 232
Query: 271 LLKTTGLEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVI 330
L G+E + ++ I L R+K +L+LDD+ L A+ G +V
Sbjct: 233 GL--GGMEWSEKNDNQIAVDIHNVLRRRKFVLLLDDIWEKVNLKAVGVPYPSKDNGCKVA 290
Query: 331 ITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKVPSGYEDI--LNRAVA-YA 387
TTR++D+ G++ V L E+ +L + K+ + DI L R VA
Sbjct: 291 FTTRSRDVCGRMGVDDPMEVSCLQPEESWDLFQMKVGKN--TLGSHPDIPGLARKVARKC 348
Query: 388 FGLPLVLEVVGSNLFGK-SIEDWKHTLDGYDRIP------NKEIQKILKVSYDALEEE 438
GLPL L V+G + K ++ +W H +D EI +LK SYD L E
Sbjct: 349 RGLPLALNVIGEAMACKRTVHEWCHAIDVLTSSAIDFSGMEDEILHVLKYSYDNLNGE 406
>DRL3_ARATH (Q9LMP6) Probable disease resistance protein At1g15890
Length = 921
Score = 73.6 bits (179), Expect = 3e-12
Identities = 96/340 (28%), Positives = 160/340 (46%), Gaps = 33/340 (9%)
Query: 190 HVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEG-S 248
H+ T VGL + V + + L + D +G+YG+GG+GK+TL I N + G
Sbjct: 221 HIQT-TVGLDAMVGRAWNSLMK---DERRTLGLYGMGGVGKTTLLASINNKFLEGMNGFD 276
Query: 249 CFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVS--EGIPVIKERLCRKKILLILDD 306
+ V QN + +QE++L + GL V+ E I L KK +L+LDD
Sbjct: 277 LVIWVVVSKDLQN--EGIQEQILGR-LGLHRGWKQVTEKEKASYICNILNVKKFVLLLDD 333
Query: 307 VDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMA 366
+ + L + GS+++ TTR+KD+ ++ V+ L EA EL +
Sbjct: 334 LWSEVDLEKIGVPPLTRENGSKIVFTTRSKDVCRDMEVDGEMKVDCLPPDEAWELFQ--- 390
Query: 367 FKSDKVP-SGYEDI--LNRAVA-YAFGLPLVLEVVGSNLFGK-SIEDWKHTL----DGYD 417
K +P +EDI L R VA GLPL L V+G + + ++++W+H +
Sbjct: 391 KKVGPIPLQSHEDIPTLARKVAEKCCGLPLALSVIGKAMASRETVQEWQHVIHVLNSSSH 450
Query: 418 RIPNKE--IQKILKVSYDALEEEEQSV-FLDIACCFKGYQWKEFEDI---LCAHY----- 466
P+ E I +LK SYD L++E+ + FL + + Y+ ++ E I +C +
Sbjct: 451 EFPSMEEKILPVLKFSYDDLKDEKVKLCFLYCSLFPEDYEVRKEELIEYWMCEGFIDGNE 510
Query: 467 DHCITHHLGVLAGKSLVKISTYYPSGSINDVRLHDLIKDM 506
D ++ G SLV+ V++HD+I++M
Sbjct: 511 DEDGANNKGHDIIGSLVRAHLLMDGELTTKVKMHDVIREM 550
Score = 38.5 bits (88), Expect = 0.095
Identities = 53/205 (25%), Positives = 88/205 (42%), Gaps = 22/205 (10%)
Query: 651 LTHIPDVSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEH-----FRPLG 705
L HIP +L ++S C N I +S + L L KL H FR
Sbjct: 573 LCHIPKDINWESLRRMSLMC--NQIANISSSSNSPNLSTLLLQN-NKLVHISCDFFR--F 627
Query: 706 LASLKKLILYECECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSGM 765
+ +L L L L + PE + K+ ++ I++S T I LP SF+ L +L L +
Sbjct: 628 MPALVVLDLSRNSSLSSLPEAISKLGSLQYINLSTTGIKWLPVSFKELKKLIHLNLEFTD 687
Query: 766 KFPKIVFSNMTKLSLSFFNLSDECLPIVLKWCVNMTHLDLSFSNFKILPECLRECHHLVE 825
+ IV ++ S NL VLK + +D S +L E L+ ++
Sbjct: 688 ELESIV-----GIATSLPNLQ------VLKLFSSRVCIDGSLMEELLLLEHLKVLTATIK 736
Query: 826 INVMCCESLEEIRGIPPNLKELCAR 850
+ + ES++ + + +++ LC R
Sbjct: 737 -DALILESIQGVDRLVSSIQALCLR 760
>DR27_ARATH (Q9T048) Putative disease resistance protein At4g27190
Length = 985
Score = 73.6 bits (179), Expect = 3e-12
Identities = 185/794 (23%), Positives = 314/794 (39%), Gaps = 134/794 (16%)
Query: 190 HVATYPVGLQSRVQQVKSLLDEG-SDDGVHMVGIYGIGGLGKSTLARQIYNFVADQ---- 244
HV V Q+ + + + +G + + +G++G+GG+GK+TL R + N + ++
Sbjct: 136 HVPGVSVVHQTMASNMLAKIRDGLTSEKAQKIGVWGMGGVGKTTLVRTLNNKLREEGATQ 195
Query: 245 -FEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEGIPV--IKERLCRKKIL 301
F F+ +E + K + E+L + T +E + ++ I V +KER K L
Sbjct: 196 PFGLVIFVIVSKEFDPREVQKQIAERLDIDTQ-MEESEEKLARRIYVGLMKER----KFL 250
Query: 302 LILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALEL 361
LILDDV L L GS+VI+T+R ++ S + V+ L E +A EL
Sbjct: 251 LILDDVWKPIDLDLLGIPRTEENKGSKVILTSRFLEVCRSMKTDLDVRVDCLLEEDAWEL 310
Query: 362 LRWMAFKSDKVPSGYEDILNRAVAY-AFGLPLVLEVVGSNLFG-KSIEDWKHTLDGYDR- 418
A D V S + + +AV+ GLPL + VG+ + G K+++ W H L +
Sbjct: 311 FCKNA--GDVVRSDHVRKIAKAVSQECGGLPLAIITVGTAMRGKKNVKLWNHVLSKLSKS 368
Query: 419 IP-----NKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQ---------W--KEFEDIL 462
+P ++I + LK+SYD LE++ + FL A + Y W + F + L
Sbjct: 369 VPWIKSIEEKIFQPLKLSYDFLEDKAKFCFLLCALFPEDYSIEVTEVVRYWMAEGFMEEL 428
Query: 463 CAHYDHCITHHLGVLAGKSLVKISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGER 522
+ D + + G+ +SL + V++HD+++D ++ S +
Sbjct: 429 GSQED---SMNEGITTVESLKDYCLLEDGDRRDTVKMHDVVRDFAIWIM---SSSQDDSH 482
Query: 523 SRLWRQEDIIHVLKENTGTSKIEMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFS 582
S + + + ++ S + MN + +ES+ D + K + L L+ N L
Sbjct: 483 SLVMSGTGLQDIRQDKLAPSLRRVSLMN-NKLESLPDLVEEFCVKTSVL--LLQGNFLLK 539
Query: 583 ----GGLKYLPSSLRVLKWKGCLSKCLSSSILNKASEITSQ*F--CFSQQKKSFL----- 631
G L+ P +LR+L G K S L + + S CF K L
Sbjct: 540 EVPIGFLQAFP-TLRILNLSGTRIKSFPSCSLLRLFSLHSLFLRDCFKLVKLPSLETLAK 598
Query: 632 -----VC----------MQKFQNMKVLTLDYCEYLTHIPD--VSGLSNLEKLSFTCCDNL 674
+C +++ + + L L +L IP VS LS+LE L T
Sbjct: 599 LELLDLCGTHILEFPRGLEELKRFRHLDLSRTLHLESIPARVVSRLSSLETLDMTSSHYR 658
Query: 675 ITIH----------NSIGHLNKLEWLS--AYGCRKLEHFRPLGLASLKKLILYECECLDN 722
++ IG L +L+ LS + L + R + LKK L
Sbjct: 659 WSVQGETQKGQATVEEIGCLQRLQVLSIRLHSSPFLLNKRNTWIKRLKKFQLVVGSRYIL 718
Query: 723 FPELLCKMAHIKEIDISNTSIGEL-------------------------PFSFQNLSELH 757
+ I +++S SIG L F+NL L
Sbjct: 719 RTRHDKRRLTISHLNVSQVSIGWLLAYTTSLALNHCQGIEAMMKKLVSDNKGFKNLKSLT 778
Query: 758 -ELTVTSGMKFPKIVFSNMTKLSLSFFNLSDECLPIVLK------WCVNMTHLDLSFSNF 810
E + + + ++V +N +K S +L + L+ + THL L
Sbjct: 779 IENVIINTNSWVEMVSTNTSKQSSDILDLLPNLEELHLRRVDLETFSELQTHLGLKLETL 838
Query: 811 KILPECLRECHHLVEINVMCCESLEEIRG-----IPPNLKELCARYCKSLSSSSRRMLMS 865
KI I + C L + PNL+E+ YC SL + +L
Sbjct: 839 KI-------------IEITMCRKLRTLLDKRNFLTIPNLEEIEISYCDSLQNLHEALLYH 885
Query: 866 QPSITCIFILPKGN 879
QP + + +L N
Sbjct: 886 QPFVPNLRVLKLRN 899
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 123,539,082
Number of Sequences: 164201
Number of extensions: 5441880
Number of successful extensions: 15567
Number of sequences better than 10.0: 168
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 113
Number of HSP's that attempted gapping in prelim test: 14993
Number of HSP's gapped (non-prelim): 451
length of query: 1040
length of database: 59,974,054
effective HSP length: 120
effective length of query: 920
effective length of database: 40,269,934
effective search space: 37048339280
effective search space used: 37048339280
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 71 (32.0 bits)
Medicago: description of AC126790.9


