

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126790.7 - phase: 0 /pseudo
(625 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
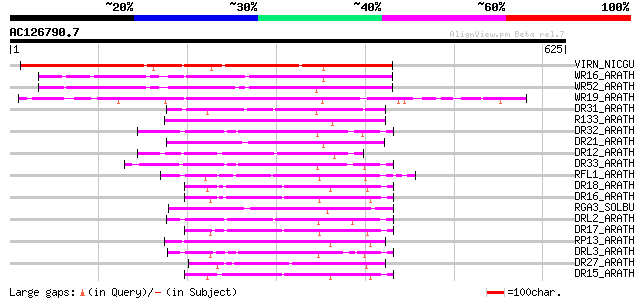
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 304 4e-82
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 175 4e-43
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 161 5e-39
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 106 2e-22
DR31_ARATH (Q9FLB4) Putative disease resistance protein At5g05400 79 4e-14
R133_ARATH (Q9STE7) Putative disease resistance RPP13-like prote... 79 5e-14
DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730 78 8e-14
DR21_ARATH (Q9LRR5) Putative disease resistance protein At3g14460 77 1e-13
DR12_ARATH (Q9LQ54) Probable disease resistance protein At1g5962... 76 3e-13
DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740 75 4e-13
RFL1_ARATH (Q8L3R3) Disease resistance protein RFL1 (RPS5-like p... 75 5e-13
DR18_ARATH (O64789) Putative disease resistance protein At1g61310 70 1e-11
DR16_ARATH (O22727) Putative disease resistance protein At1g61190 70 1e-11
RGA3_SOLBU (Q7XA40) Putative disease resistance protein RGA3 (RG... 70 2e-11
DRL2_ARATH (P60839) Probable disease resistance protein At1g12290 69 4e-11
DR17_ARATH (O64790) Putative disease resistance protein At1g61300 68 8e-11
RP13_ARATH (Q9M667) Disease resistance protein RPP13 (Resistance... 66 2e-10
DRL3_ARATH (Q9LMP6) Probable disease resistance protein At1g15890 66 2e-10
DR27_ARATH (Q9T048) Putative disease resistance protein At4g27190 66 2e-10
DR15_ARATH (Q940K0) Probable disease resistance protein At1g61180 66 2e-10
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 304 bits (779), Expect = 4e-82
Identities = 178/454 (39%), Positives = 274/454 (60%), Gaps = 43/454 (9%)
Query: 13 SSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRK 72
+SSS S +++DVF+SFRG DTR FT +LY+ L+DKGI TF DDK L G I L K
Sbjct: 2 ASSSSSSRWSYDVFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIPGELCK 61
Query: 73 SIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYG 132
+IEES+ AI++FS+NYATS +CL+ELV I+ C VIP+FY +PSHVR ++S+
Sbjct: 62 AIEESQFAIVVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQKESFA 121
Query: 133 EALAKHEVEFQNDMENMERLLKWKEALH-------------------------QFHSWIN 167
+A +HE ++++D+E ++R W+ AL+ Q S +
Sbjct: 122 KAFEEHETKYKDDVEGIQR---WRIALNEAANLKGSCDNRDKTDADCIRQIVDQISSKLC 178
Query: 168 RCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVN-- 225
+ L + +VG+++ + ++ SLL++G +GV I+GI G GG+GKTT+A A++++++
Sbjct: 179 KISLSYLQNIVGIDTHLEKIESLLEIG-INGVRIMGIWGMGGVGKTTIARAIFDTLLGRM 237
Query: 226 ----QFECRCFLYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKK 281
QF+ CFL +++EN K + LQ LLS+ + ++ +G + RL KK
Sbjct: 238 DSSYQFDGACFLKDIKEN--KRGMHSLQNALLSELLREKANYNNEEDGKHQMASRLRSKK 295
Query: 282 VLLILDDVD-KPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEES 340
VL++LDD+D K + LE L G+ WFG GSR+IITTRD++L+ + I IYE +L ES
Sbjct: 296 VLIVLDDIDNKDHYLEYLAGDLDWFGNGSRIIITTRDKHLIEKNDI--IYEVTALPDHES 353
Query: 341 LELLRKMTFKND---SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYER 397
++L ++ F + +++ + V YA GLPLALKV GS L + + +S ++ +
Sbjct: 354 IQLFKQHAFGKEVPNENFEKLSLEVVNYAKGLPLALKVWGSLLHNLRLTEWKSAIEHMKN 413
Query: 398 IPPEDIQKILKVSFDTLEEEQQSVFLDIACCFKG 431
I LK+S+D LE +QQ +FLDIAC +G
Sbjct: 414 NSYSGIIDKLKISYDGLEPKQQEMFLDIACFLRG 447
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 175 bits (443), Expect = 4e-43
Identities = 116/406 (28%), Positives = 211/406 (51%), Gaps = 31/406 (7%)
Query: 33 DTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAIIIFSKNYATSS 92
+ R+ F +L KAL KG++ D + D ++ + +E +R++++I N S
Sbjct: 15 EVRYSFVSHLSKALQRKGVNDVFIDSD----DSLSNESQSMVERARVSVMILPGNRTVS- 69
Query: 93 FCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLE--DSYGEALAKHEVEFQNDMENME 150
LD+LV ++ C + K V+PV YG S L DS G + H + +D + ++
Sbjct: 70 --LDKLVKVLDCQKNKDQVVVPVLYGVRSSETEWLSALDSKGFSSVHHSRKECSDSQLVK 127
Query: 151 RLLKWKEALHQFHSWINRCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGL 210
++ +++ ++ R +G+ S++ E+ +++ D + +GI G G+
Sbjct: 128 ETVR---DVYEKLFYMER---------IGIYSKLLEIEKMINKQPLD-IRCVGIWGMPGI 174
Query: 211 GKTTLAEAVYNSIVNQFECRCFLYN-VRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEG 269
GKTTLA+AV++ + +F+ CF+ + + K L+EQ L ++ G +
Sbjct: 175 GKTTLAKAVFDQMSGEFDAHCFIEDYTKAIQEKGVYCLLEEQFLKENAGASGTVTK---- 230
Query: 270 IEIIKQRLCRKKVLLILDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKI 329
+ +++ RL K+VL++LDDV P +E +G WFG S +IIT++D+ + + +I
Sbjct: 231 LSLLRDRLNNKRVLVVLDDVRSPLVVESFLGGFDWFGPKSLIIITSKDKSVFRLCRVNQI 290
Query: 330 YEADSLNKEESLELLRKMTFKND---SSYDYILNRAVEYASGLPLALKVVGSNLFGKS-I 385
YE LN++E+L+L +D + + + ++YA+G PLAL + G L GK
Sbjct: 291 YEVQGLNEKEALQLFSLCASIDDMAEQNLHEVSMKVIKYANGHPLALNLYGRELMGKKRP 350
Query: 386 ADCESTLDKYERIPPEDIQKILKVSFDTLEEEQQSVFLDIACCFKG 431
+ E K + PP +K S+DTL + ++++FLDIAC F+G
Sbjct: 351 PEMEIAFLKLKECPPAIFVDAIKSSYDTLNDREKNIFLDIACFFQG 396
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein
RRS1) (Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 161 bits (407), Expect = 5e-39
Identities = 117/408 (28%), Positives = 213/408 (51%), Gaps = 31/408 (7%)
Query: 33 DTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAIIIFSKNYATSS 92
+ R+ F +L +AL KGI+ + D ++ D + + IE++ +++++ N S
Sbjct: 18 EVRYSFVSHLSEALRRKGINNVVVDVDID--DLLFKESQAKIEKAGVSVMVLPGNCDPSE 75
Query: 93 FCLDELVHIIHCFRE-KVTKVIPVFYGTEPSHVRKLEDSYGEALAK-HEVEFQ-NDMENM 149
LD+ ++ C R K V+ V YG + L + L++ H+ + +D +
Sbjct: 76 VWLDKFAKVLECQRNNKDQAVVSVLYGDSLLRDQWLSELDFRGLSRIHQSRKECSDSILV 135
Query: 150 ERLLKWKEALHQFHSWINRCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGG 209
E +++ +++ H ++ R +G+ S++ E+ ++++ G+ +GI G G
Sbjct: 136 EEIVR---DVYETHFYVGR---------IGIYSKLLEIENMVNKQPI-GIRCVGIWGMPG 182
Query: 210 LGKTTLAEAVYNSIVNQFECRCFLYNVRENSFKHSLK-YLQEQLLSKSIGYDTPLEHDNE 268
+GKTTLA+AV++ + + F+ CF+ + ++ + L L+EQLL G D +
Sbjct: 183 IGKTTLAKAVFDQMSSAFDASCFIEDYDKSIHEKGLYCLLEEQLLP---GNDATIMK--- 236
Query: 269 GIEIIKQRLCRKKVLLILDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITK 328
+ ++ RL K+VL++LDDV E + W G GS +IIT+RD+ + GI +
Sbjct: 237 -LSSLRDRLNSKRVLVVLDDVCNALVAESFLEGFDWLGPGSLIIITSRDKQVFRLCGINQ 295
Query: 329 IYEADSLNKEESLELL----RKMTFKNDSSYDYILNRAVEYASGLPLALKVVGSNLFG-K 383
IYE LN++E+ +L M + + + R + YA+G PLA+ V G L G K
Sbjct: 296 IYEVQGLNEKEARQLFLLSASIMEDMGEQNLHELSVRVISYANGNPLAISVYGRELKGKK 355
Query: 384 SIADCESTLDKYERIPPEDIQKILKVSFDTLEEEQQSVFLDIACCFKG 431
+++ E+ K +R PP I K S+DTL + ++++FLDIAC F+G
Sbjct: 356 KLSEMETAFLKLKRRPPFKIVDAFKSSYDTLSDNEKNIFLDIACFFQG 403
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 106 bits (265), Expect = 2e-22
Identities = 149/616 (24%), Positives = 270/616 (43%), Gaps = 95/616 (15%)
Query: 11 PTSSSSLSYDFNFDVFISFRGTD-TRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPS 69
P SSS YD V I + D + F +L +L +GI + E+
Sbjct: 660 PRFSSSKDYD----VVIRYGRADISNEDFISHLRASLCRRGISVYEKFNEVD-------- 707
Query: 70 LRKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEP----SHVR 125
++ + R+ II+ + Y S+ L++I+ + V P+FY P + +
Sbjct: 708 ---ALPKCRVLIIVLTSTYVPSN-----LLNILEHQHTEDRVVYPIFYRLSPYDFVCNSK 759
Query: 126 KLEDSYGEALAKHEVEFQNDMENMERLLKWKEALHQFHSWINRCHLHVA-----EYLVGL 180
E Y + K ++ M ++ + I R L V ++G+
Sbjct: 760 NYERFYLQDEPKKWQAALKEITQMPGYTLTDKSESELIDEIVRDALKVLCSADKVNMIGM 819
Query: 181 ESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCFLYNVRENS 240
+ ++ E+ SLL + D V IGI GT G+GKTT+AE ++ I Q+E L ++ +
Sbjct: 820 DMQVEEILSLLCIESLD-VRSIGIWGTVGIGKTTIAEEIFRKISVQYETCVVLKDLHKEV 878
Query: 241 FKHSLKYLQEQLLSKSIGYDTPLEHDNE-GIEIIKQRLCRKKVLLILDDVDKPNQLEKLV 299
++E LS+ + + + ++ ++ RL RK++L+ILDDV+ ++ +
Sbjct: 879 EVKGHDAVRENFLSEVLEVEPHVIRISDIKTSFLRSRLQRKRILVILDDVNDYRDVDTFL 938
Query: 300 GEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFK---NDSSYD 356
G +FG GSR+I+T+R+R + I +YE L+ +SL LL + T + + Y
Sbjct: 939 GTLNYFGPGSRIIMTSRNRRVFVLCKIDHVYEVKPLDIPKSLLLLDRGTCQIVLSPEVYK 998
Query: 357 YILNRAVEYASGLPLALKVVGS--NLFGKSIADCESTLDKYERIPPEDIQKILKVSFDTL 414
+ V++++G P L+ + S + K + ++T Y I I + S L
Sbjct: 999 TLSLELVKFSNGNPQVLQFLSSIDREWNKLSQEVKTTSPIY-------IPGIFEKSCCGL 1051
Query: 415 EEEQQSVFLDIACCFKGCDWQK---------FQRHFNF-------IMAIQ*HIMLECWLM 458
++ ++ +FLDIAC F D F H F ++ I H +++
Sbjct: 1052 DDNERGIFLDIACFFNRIDKDNVAMLLDGCGFSAHVGFRGLVDKSLLTISQHNLVD---- 1107
Query: 459 NLS*RLVLQIHTTLLILLHYMT**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF*NKT- 517
++ I T ++ + + D+++L + + I +F N T
Sbjct: 1108 -----MLSFIQATGREIVRQES-----ADRPGDRSRLWNAD--------YIRHVFINDTG 1149
Query: 518 -HKIEMIYLNCSSMEPININEKAFKKMKKLKTLII------EKG--YFSKGLKYLPKSLI 568
IE I+L+ +++ + N F+KM L+ L + EK F +GL+YLP L
Sbjct: 1150 TSAIEGIFLDMLNLK-FDANPNVFEKMCNLRLLKLYCSKAEEKHGVSFPQGLEYLPSKLR 1208
Query: 569 VLKWKGF--TSEPLSF 582
+L W+ + +S P SF
Sbjct: 1209 LLHWEYYPLSSLPKSF 1224
>DR31_ARATH (Q9FLB4) Putative disease resistance protein At5g05400
Length = 874
Score = 79.0 bits (193), Expect = 4e-14
Identities = 73/256 (28%), Positives = 125/256 (48%), Gaps = 17/256 (6%)
Query: 177 LVGLESRISEV-NSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYN---SIVNQFECRCF 232
+VG E+ + NS++++G V ++GI G GG+GKTTL + N ++ N F+ +
Sbjct: 156 IVGQEAIVESTWNSMMEVG----VGLLGIYGMGGVGKTTLLSQINNKFRTVSNDFDIAIW 211
Query: 233 LYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKP 292
+ + + K + + ++L + G++ E NE IK+ L KK +L+LDD+
Sbjct: 212 VVVSKNPTVKRIQEDIGKRLDLYNEGWEQKTE--NEIASTIKRSLENKKYMLLLDDMWTK 269
Query: 293 NQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELL---RKMTF 349
L +G P GS++ T+R + G+ K E L +++ +L K T
Sbjct: 270 VDLAN-IGIPVPKRNGSKIAFTSRSNEVCGKMGVDKEIEVTCLMWDDAWDLFTRNMKETL 328
Query: 350 KNDSSYDYILNRAVEYASGLPLALKVVGSNLF-GKSIADCESTLDKYERIPPEDIQKILK 408
++ + +GLPLAL V+G + KSI + + + I DI ILK
Sbjct: 329 ESHPKIPEVAKSIARKCNGLPLALNVIGETMARKKSIEEWHDAVGVFSGI-EADILSILK 387
Query: 409 VSFDTLE-EEQQSVFL 423
S+D L+ E+ +S FL
Sbjct: 388 FSYDDLKCEKTKSCFL 403
>R133_ARATH (Q9STE7) Putative disease resistance RPP13-like protein
3
Length = 847
Score = 78.6 bits (192), Expect = 5e-14
Identities = 79/263 (30%), Positives = 117/263 (44%), Gaps = 14/263 (5%)
Query: 175 EYLVGLESRISEVN-SLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNS--IVNQFECRC 231
E +VGLE + + LL D YII I G GGLGKT LA +YNS + +F+CR
Sbjct: 160 ELVVGLEDDVKILLVKLLSDNEKDKSYIISIFGMGGLGKTALARKLYNSGDVKRRFDCRA 219
Query: 232 FLYNVRENSFKHSLKYLQEQL--LSKSIGYDTPLEHDNEGIEIIKQRLCR-KKVLLILDD 288
+ Y +E + L + L +S + ++E +E+ L K ++++DD
Sbjct: 220 WTYVSQEYKTRDILIRIIRSLGIVSAEEMEKIKMFEEDEELEVYLYGLLEGKNYMVVVDD 279
Query: 289 VDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLL-SCHGITKIYEADSLNKEESLELLRKM 347
V P+ E L +GS+VIITTR R + G ++ L EES L +
Sbjct: 280 VWDPDAWESLKRALPCDHRGSKVIITTRIRAIAEGVEGTVYAHKLRFLTFEESWTLFERK 339
Query: 348 TFKNDSSYDYILNRA----VEYASGLPLALKVVGSNLFGKSIADC-ESTLDKYERIPPED 402
F N D L R V+ GLPLA+ V+ L K + E + R+
Sbjct: 340 AFSNIEKVDEDLQRTGKEMVKKCGGLPLAIVVLSGLLSRKRTNEWHEVCASLWRRLKDNS 399
Query: 403 --IQKILKVSFDTLEEEQQSVFL 423
I + +SF + E + FL
Sbjct: 400 IHISTVFDLSFKEMRHELKLCFL 422
>DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730
Length = 848
Score = 77.8 bits (190), Expect = 8e-14
Identities = 81/309 (26%), Positives = 137/309 (44%), Gaps = 40/309 (12%)
Query: 145 DMENMERLLKWKEALHQFHSWINRCHLHVAEYLVGLESRISEV-NSLLDLGCTDGVYIIG 203
++E ++ LL K I + + VGL++ + SL+D D + +G
Sbjct: 121 NLEEVKELLSKKNFEVVAQKIIPKAEKKHIQTTVGLDTMVGIAWESLID----DEIRTLG 176
Query: 204 ILGTGGLGKTTLAEAVYNSIVN-QFECRCFLYNVRENSFKHSLKYLQEQLLSKSIGYDTP 262
+ G GG+GKTTL E++ N V + E ++ V F+ L+ +Q+Q+L + + D
Sbjct: 177 LYGMGGIGKTTLLESLNNKFVELESEFDVVIWVVVSKDFQ--LEGIQDQILGR-LRPDKE 233
Query: 263 LEHDNEG--IEIIKQRLCRKKVLLILDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYL 320
E + E +I L RKK +L+LDD+ L K+ P GS+++ TTR + +
Sbjct: 234 WERETESKKASLINNNLKRKKFVLLLDDLWSEVDLIKIGVPPPSRENGSKIVFTTRSKEV 293
Query: 321 LSCHGITKIYEADSLNKEESLELLR----KMTFKNDSSYDYILNRAVEYASGLPLALKVV 376
K + D L+ +E+ EL R + ++ + GLPLAL V+
Sbjct: 294 CKHMKADKQIKVDCLSPDEAWELFRLTVGDIILRSHQDIPALARIVAAKCHGLPLALNVI 353
Query: 377 GSNLFGKSIADCESTLDKY-----------ERIP--PEDIQKILKVSFDTLEEEQQSVFL 423
G + C+ T+ ++ + P E I ILK S+D+L+
Sbjct: 354 GKAMV------CKETVQEWRHAINVLNSPGHKFPGMEERILPILKFSYDSLKNG------ 401
Query: 424 DIACCFKGC 432
+I CF C
Sbjct: 402 EIKLCFLYC 410
>DR21_ARATH (Q9LRR5) Putative disease resistance protein At3g14460
Length = 1424
Score = 77.4 bits (189), Expect = 1e-13
Identities = 73/261 (27%), Positives = 121/261 (45%), Gaps = 21/261 (8%)
Query: 177 LVG-LESRISEVNSLL--DLGCTDGVYIIGILGTGGLGKTTLAEAVYNS--IVNQFECRC 231
LVG +E +++ VN LL D +I ++G G+GKTTL E V+N + FE +
Sbjct: 168 LVGRVEDKLALVNLLLSDDEISIGKPAVISVVGMPGVGKTTLTEIVFNDYRVTEHFEVKM 227
Query: 232 FLY-NVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEI-IKQRLCRKKVLLILDDV 289
++ + N F + LQ+ S D P ++I +K+ L K+ LL+LDD
Sbjct: 228 WISAGINFNVFTVTKAVLQDITSSAVNTEDLP------SLQIQLKKTLSGKRFLLVLDDF 281
Query: 290 --DKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKM 347
+ ++ E +GS++++TTR + + KIY+ + EE EL+ +
Sbjct: 282 WSESDSEWESFQVAFTDAEEGSKIVLTTRSEIVSTVAKAEKIYQMKLMTNEECWELISRF 341
Query: 348 TFKN------DSSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPE 401
F N + + I R E GLPLA + + S+L K D + K
Sbjct: 342 AFGNISVGSINQELEGIGKRIAEQCKGLPLAARAIASHLRSKPNPDDWYAVSKNFSSYTN 401
Query: 402 DIQKILKVSFDTLEEEQQSVF 422
I +LK+S+D+L + + F
Sbjct: 402 SILPVLKLSYDSLPPQLKRCF 422
>DR12_ARATH (Q9LQ54) Probable disease resistance protein At1g59620
(CW9)
Length = 695
Score = 75.9 bits (185), Expect = 3e-13
Identities = 74/268 (27%), Positives = 136/268 (50%), Gaps = 32/268 (11%)
Query: 145 DMENMERLLKWKEALHQFHSWINRCHLHVAEYLVGLESRISE-VNSLLDLGCTDGVYIIG 203
DME + + + K+ + ++ N LVGLE + + V L+++ D ++
Sbjct: 111 DMEGLSKRIAKKDKRNMRQTFSNNNE----SVLVGLEENVKKLVGHLVEV--EDSSQVVS 164
Query: 204 ILGTGGLGKTTLAEAVYN--SIVNQFECRCFLYNVRENSFKHSLKYLQEQLLSKSIGYDT 261
I G GG+GKTTLA V+N ++ + F ++ S + + KY+ + +L K
Sbjct: 165 ITGMGGIGKTTLARQVFNHETVKSHFAQLAWVC----VSQQFTRKYVWQTILRKVGPEYI 220
Query: 262 PLE-HDNEGIEIIKQRLCRKKVLLILDDVDKPNQLEKLVGEPGW-FGQGSRVIITTRDR- 318
LE ++E E + + L +K L++LDD+ + + + EP + G+G +V++T+R+
Sbjct: 221 KLEMTEDELQEKLFRLLGTRKALIVLDDIWREEDWDMI--EPIFPLGKGWKVLLTSRNEG 278
Query: 319 YLLSCHGITKIYEADSLNKEESLELLRKMTFKNDSSYDYILNRAVE--------YASGLP 370
L + I++ D L EES + R++ F +++ +Y ++ +E + GLP
Sbjct: 279 VALRANPNGFIFKPDCLTPEESWTIFRRIVFPGENTTEYKVDEKMEELGKQMIKHCGGLP 338
Query: 371 LALKVVGSNLFGKSIADCESTLDKYERI 398
LALKV+G L TLD+++RI
Sbjct: 339 LALKVLGGLLV------VHFTLDEWKRI 360
>DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740
Length = 862
Score = 75.5 bits (184), Expect = 4e-13
Identities = 80/323 (24%), Positives = 138/323 (41%), Gaps = 45/323 (13%)
Query: 130 SYGEALAKHEVEFQNDMENMERLLKWKEALHQFHSWINRCHLHVAEYLVGLESRISEVNS 189
+YGE ++K +E ++ LL K+ I++ + + VGL+ + S
Sbjct: 112 NYGEKVSKM-------LEEVKELLSKKDFRMVAQEIIHKVEKKLIQTTVGLDKLVEMAWS 164
Query: 190 LLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVN-QFECRCFLYNVRENSFKHSLKYL 248
L D + +G+ G GG+GKTTL E++ N V + E ++ V F+ + +
Sbjct: 165 SL---MNDEIGTLGLYGMGGVGKTTLLESLNNKFVELESEFDVVIWVVVSKDFQ--FEGI 219
Query: 249 QEQLLSKSIGYDTPLEHDNEG--IEIIKQRLCRKKVLLILDDVDKPNQLEKLVGEPGWFG 306
Q+Q+L + + D E + E +I L RKK +L+LDD+ + K+ P
Sbjct: 220 QDQILGR-LRSDKEWERETESKKASLIYNNLERKKFVLLLDDLWSEVDMTKIGVPPPTRE 278
Query: 307 QGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLR----KMTFKNDSSYDYILNRA 362
GS+++ TTR + K + L+ +E+ EL R + ++ +
Sbjct: 279 NGSKIVFTTRSTEVCKHMKADKQIKVACLSPDEAWELFRLTVGDIILRSHQDIPALARIV 338
Query: 363 VEYASGLPLALKVVGSNLFGKSIADCESTLDKYERI-------------PPEDIQKILKV 409
GLPLAL V+G + C+ T+ ++ E I ILK
Sbjct: 339 AAKCHGLPLALNVIGKAM------SCKETIQEWSHAINVLNSAGHEFPGMEERILPILKF 392
Query: 410 SFDTLEEEQQSVFLDIACCFKGC 432
S+D+L+ +I CF C
Sbjct: 393 SYDSLKNG------EIKLCFLYC 409
>RFL1_ARATH (Q8L3R3) Disease resistance protein RFL1 (RPS5-like
protein 1) (pNd13/pNd14)
Length = 885
Score = 75.1 bits (183), Expect = 5e-13
Identities = 81/303 (26%), Positives = 143/303 (46%), Gaps = 36/303 (11%)
Query: 171 LHVAEYLVGLESRISEV-NSLLDLGCTDGVYIIGILGTGGLGKTTLAEAV---YNSIVNQ 226
L + +VG +S + +V N L++ D V+I+G+ G GG+GKTTL + ++ +
Sbjct: 151 LPIQSTIVGQDSMLDKVWNCLME----DKVWIVGLYGMGGVGKTTLLTQINNKFSKLGGG 206
Query: 227 FECRCFLYNVRENSFKHSL-KYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLI 285
F+ ++ V +N+ H + K + E+L +G + ++ N+ I L RKK +L+
Sbjct: 207 FDVVIWVV-VSKNATVHKIQKSIGEKL--GLVGKNWDEKNKNQRALDIHNVLRRKKFVLL 263
Query: 286 LDDVDKPNQLEKLVGEPGWFGQ-GSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELL 344
LDD+ + +L K++G P G+ G +V TT + + G+ E L+ + +LL
Sbjct: 264 LDDIWEKVEL-KVIGVPYPSGENGCKVAFTTHSKEVCGRMGVDNPMEISCLDTGNAWDLL 322
Query: 345 RKM----TFKNDSSYDYILNRAVEYASGLPLALKVVGSNL-FGKSIADCESTLDKYERIP 399
+K T + + + E GLPLAL V+G + F ++I + +
Sbjct: 323 KKKVGENTLGSHPDIPQLARKVSEKCCGLPLALNVIGETMSFKRTIQEWRHATEVLTSAT 382
Query: 400 -----PEDIQKILKVSFDTLEEEQQSVFLDIACCFKGCDWQKFQRHFNFIMAIQ*HIMLE 454
++I ILK S+D+L E D CF C F F I+ +++E
Sbjct: 383 DFSGMEDEILPILKYSYDSLNGE------DAKSCFLYC--SLFPEDFE----IRKEMLIE 430
Query: 455 CWL 457
W+
Sbjct: 431 YWI 433
>DR18_ARATH (O64789) Putative disease resistance protein At1g61310
Length = 925
Score = 70.5 bits (171), Expect = 1e-11
Identities = 74/253 (29%), Positives = 117/253 (45%), Gaps = 28/253 (11%)
Query: 197 DGVYIIGILGTGGLGKTTLAEAVYN---SIVNQFECRCFLYNVRENSFKHSLKYLQEQLL 253
DGV I+G+ G GG+GKTTL + ++N I F+ ++ V + + L LQE +
Sbjct: 172 DGVGIMGLHGMGGVGKTTLFKKIHNKFAEIGGTFDIVIWIV-VSQGA---KLSKLQEDIA 227
Query: 254 SKSIGYDTPLEHDNEGIEI--IKQRLCRKKVLLILDDVDKPNQLEKL-VGEPGWFGQGSR 310
K D ++ NE + I + L K+ +L+LDD+ + LE + + P + +
Sbjct: 228 EKLHLCDDLWKNKNESDKATDIHRVLKGKRFVLMLDDIWEKVDLEAIGIPYPSEVNK-CK 286
Query: 311 VIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKNDSSYDYIL----NRAVEYA 366
V TTR R + G K + + L E++ EL + N S D ++ +
Sbjct: 287 VAFTTRSREVCGEMGDHKPMQVNCLEPEDAWELFKNKVGDNTLSSDPVIVGLAREVAQKC 346
Query: 367 SGLPLALKVVGSNLFGKS-IADCESTLDKYERIPPE------DIQKILKVSFDTLEEEQQ 419
GLPLAL V+G + K+ + + E +D R E I ILK S+D+L +E
Sbjct: 347 RGLPLALNVIGETMASKTMVQEWEYAIDVLTRSAAEFSGMENKILPILKYSYDSLGDEH- 405
Query: 420 SVFLDIACCFKGC 432
I CF C
Sbjct: 406 -----IKSCFLYC 413
>DR16_ARATH (O22727) Putative disease resistance protein At1g61190
Length = 967
Score = 70.5 bits (171), Expect = 1e-11
Identities = 73/253 (28%), Positives = 118/253 (45%), Gaps = 28/253 (11%)
Query: 197 DGVYIIGILGTGGLGKTTLAEAVYNSIV---NQFECRCFLYNVRENSFKHSLKYLQEQLL 253
DGV I+G+ G GG+GKTTL + ++N F+ ++ V + + L LQE +
Sbjct: 171 DGVGIMGLHGMGGVGKTTLFKKIHNKFAETGGTFDIVIWIV-VSQGA---KLSKLQEDIA 226
Query: 254 SKSIGYDTPLEHDNEGIEI--IKQRLCRKKVLLILDDVDKPNQLEKL-VGEPGWFGQGSR 310
K D ++ NE + I + L K+ +L+LDD+ + LE + + P + +
Sbjct: 227 EKLHLCDDLWKNKNESDKATDIHRVLKGKRFVLMLDDIWEKVDLEAIGIPYPSEVNK-CK 285
Query: 311 VIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKM----TFKNDSSYDYILNRAVEYA 366
V TTRD+ + G K + L E++ EL + T ++D + +
Sbjct: 286 VAFTTRDQKVCGQMGDHKPMQVKCLEPEDAWELFKNKVGDNTLRSDPVIVGLAREVAQKC 345
Query: 367 SGLPLALKVVGSNLFGKS-IADCESTLDKYERIPPE--DIQK----ILKVSFDTLEEEQQ 419
GLPLAL +G + K+ + + E +D R E D+Q ILK S+D+LE+E
Sbjct: 346 RGLPLALSCIGETMASKTMVQEWEHAIDVLTRSAAEFSDMQNKILPILKYSYDSLEDEH- 404
Query: 420 SVFLDIACCFKGC 432
I CF C
Sbjct: 405 -----IKSCFLYC 412
>RGA3_SOLBU (Q7XA40) Putative disease resistance protein RGA3
(RGA1-blb) (Blight resistance protein B149)
Length = 947
Score = 69.7 bits (169), Expect = 2e-11
Identities = 70/267 (26%), Positives = 123/267 (45%), Gaps = 23/267 (8%)
Query: 179 GLESRISEVNSLL--DLGCTDGVYIIGILGTGGLGKTTLAEAVYNS--IVNQFECRCFLY 234
G E E+ +L ++ ++ V ++ ILG GGLGKTTLA+ V+N I F + ++
Sbjct: 153 GREKEEDEIVKILINNVSYSEEVPVLPILGMGGLGKTTLAQMVFNDQRITEHFNLKIWVC 212
Query: 235 NVRENSFKHSLKYLQEQLLSKSIGYD--TPLEHDNEGIEIIKQRLCRKKVLLILDDV--D 290
+ K +K + E + KS+G PL+ + +++ L K+ L+LDDV +
Sbjct: 213 VSDDFDEKRLIKAIVESIEGKSLGDMDLAPLQ------KKLQELLNGKRYFLVLDDVWNE 266
Query: 291 KPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFK 350
+ + L G+ ++ITTR + S G ++Y+ +L++E+ L ++ F
Sbjct: 267 DQEKWDNLRAVLKIGASGASILITTRLEKIGSIMGTLQLYQLSNLSQEDCWLLFKQRAFC 326
Query: 351 NDSSYD----YILNRAVEYASGLPLALKVVGSNL-FGKSIADCESTLDKYERIPPEDIQK 405
+ + I V+ G+PLA K +G L F + ++ E D P+D
Sbjct: 327 HQTETSPKLMEIGKEIVKKCGGVPLAAKTLGGLLRFKREESEWEHVRDSEIWNLPQDENS 386
Query: 406 ILKVSFDTLEEEQQSVFLDIACCFKGC 432
+L L + LD+ CF C
Sbjct: 387 VL----PALRLSYHHLPLDLRQCFAYC 409
>DRL2_ARATH (P60839) Probable disease resistance protein At1g12290
Length = 884
Score = 68.9 bits (167), Expect = 4e-11
Identities = 70/272 (25%), Positives = 123/272 (44%), Gaps = 29/272 (10%)
Query: 177 LVGLESRISEV-NSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFE-CRCFLY 234
+VG E+ + + + L+D DG I+G+ G GG+GKTTL + N + + ++
Sbjct: 156 IVGQETILEKAWDHLMD----DGTKIMGLYGMGGVGKTTLLTQINNRFCDTDDGVEIVIW 211
Query: 235 NVRENSFKHSLKYLQEQLLSK--SIGYDTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKP 292
V + + +Q+++ K IG + + +N+ I L +K+ +L+LDD+ K
Sbjct: 212 VVVSGDLQ--IHKIQKEIGEKIGFIGVEWNQKSENQKAVDILNFLSKKRFVLLLDDIWKR 269
Query: 293 NQLEKLVGEPGWFGQ-GSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRK----M 347
+L + +G P + G ++ TTR + + + G+ E L +++ +L +K +
Sbjct: 270 VELTE-IGIPNPTSENGCKIAFTTRCQSVCASMGVHDPMEVRCLGADDAWDLFKKKVGDI 328
Query: 348 TFKNDSSYDYILNRAVEYASGLPLALKVVGSNLFGKS-------IADCESTLDKYERIPP 400
T + I + + GLPLAL V+G + K D +T
Sbjct: 329 TLSSHPDIPEIARKVAQACCGLPLALNVIGETMACKKTTQEWDRAVDVSTTYAANFGAVK 388
Query: 401 EDIQKILKVSFDTLEEEQQSVFLDIACCFKGC 432
E I ILK S+D LE E + CF C
Sbjct: 389 ERILPILKYSYDNLESE------SVKTCFLYC 414
>DR17_ARATH (O64790) Putative disease resistance protein At1g61300
Length = 766
Score = 67.8 bits (164), Expect = 8e-11
Identities = 73/253 (28%), Positives = 116/253 (44%), Gaps = 28/253 (11%)
Query: 197 DGVYIIGILGTGGLGKTTLAEAVYNSIV---NQFECRCFLYNVRENSFKHSLKYLQEQLL 253
D V I+G+ G GG+GKTTL + ++N ++F+ ++ V K L LQE +
Sbjct: 59 DRVGIMGLHGMGGVGKTTLFKKIHNKFAKMSSRFD--IVIWIVVSKGAK--LSKLQEDIA 114
Query: 254 SKSIGYDTPLEHDNEGIEI--IKQRLCRKKVLLILDDVDKPNQLEKL-VGEPGWFGQGSR 310
K D ++ NE + I + L K+ +L+LDD+ + LE + V P + +
Sbjct: 115 EKLHLCDDLWKNKNESDKATDIHRVLKGKRFVLMLDDIWEKVDLEAIGVPYPSEVNK-CK 173
Query: 311 VIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKM----TFKNDSSYDYILNRAVEYA 366
V TTRD+ + G K + L E++ EL + T ++D + +
Sbjct: 174 VAFTTRDQKVCGEMGDHKPMQVKCLEPEDAWELFKNKVGDNTLRSDPVIVELAREVAQKC 233
Query: 367 SGLPLALKVVGSNLFGKS-IADCESTLDKYERIPPE------DIQKILKVSFDTLEEEQQ 419
GLPLAL V+G + K+ + + E +D R E I ILK S+D+L +E
Sbjct: 234 RGLPLALSVIGETMASKTMVQEWEHAIDVLTRSAAEFSNMGNKILPILKYSYDSLGDEH- 292
Query: 420 SVFLDIACCFKGC 432
I CF C
Sbjct: 293 -----IKSCFLYC 300
>RP13_ARATH (Q9M667) Disease resistance protein RPP13 (Resistance to
Peronospora parasitica protein 13)
Length = 835
Score = 66.2 bits (160), Expect = 2e-10
Identities = 75/263 (28%), Positives = 116/263 (43%), Gaps = 15/263 (5%)
Query: 175 EYLVGLESRISEV-NSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNS--IVNQFECRC 231
E +VGLE + LLD +II I G GGLGKT LA +YNS + +FE R
Sbjct: 161 EVVVGLEDDAKILLEKLLDYE-EKNRFIISIFGMGGLGKTALARKLYNSRDVKERFEYRA 219
Query: 232 FLYNVRENSFKHSLKYLQEQL-LSKSIGYDTPLEHDNEGIEIIKQRLCR-KKVLLILDDV 289
+ Y +E L + L ++ + + E +E+ L KK L+++DD+
Sbjct: 220 WTYVSQEYKTGDILMRIIRSLGMTSGEELEKIRKFAEEELEVYLYGLLEGKKYLVVVDDI 279
Query: 290 DKPNQLEKLVGEPGWFGQGSRVIITTRDRYLL-SCHGITKIYEADSLNKEESLELLRKMT 348
+ + L +GSRVIITTR + + G ++ L EES EL +
Sbjct: 280 WEREAWDSLKRALPCNHEGSRVIITTRIKAVAEGVDGRFYAHKLRFLTFEESWELFEQRA 339
Query: 349 FKNDSSYDYIL----NRAVEYASGLPLALKVVGSNLFGKSIADCESTLDK-YERIPPEDI 403
F+N D L V+ GLPL + V+ L K+ ++ + + R+ + I
Sbjct: 340 FRNIQRKDEDLLKTGKEMVQKCRGLPLCIVVLAGLLSRKTPSEWNDVCNSLWRRLKDDSI 399
Query: 404 QK---ILKVSFDTLEEEQQSVFL 423
+ +SF L E + FL
Sbjct: 400 HVAPIVFDLSFKELRHESKLCFL 422
>DRL3_ARATH (Q9LMP6) Probable disease resistance protein At1g15890
Length = 921
Score = 66.2 bits (160), Expect = 2e-10
Identities = 73/278 (26%), Positives = 128/278 (45%), Gaps = 44/278 (15%)
Query: 178 VGLESRISEV-NSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIV---NQFECRCFL 233
VGL++ + NSL+ D +G+ G GG+GKTTL ++ N + N F+ +
Sbjct: 226 VGLDAMVGRAWNSLMK----DERRTLGLYGMGGVGKTTLLASINNKFLEGMNGFDL--VI 279
Query: 234 YNVRENSFKHSLKYLQEQLLSKSIGYDTPLEH--DNEGIEIIKQRLCRKKVLLILDDVDK 291
+ V ++ + +QEQ+L + +G + + E I L KK +L+LDD+
Sbjct: 280 WVVVSKDLQN--EGIQEQILGR-LGLHRGWKQVTEKEKASYICNILNVKKFVLLLDDLWS 336
Query: 292 PNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRK----M 347
LEK+ P GS+++ TTR + + + + D L +E+ EL +K +
Sbjct: 337 EVDLEKIGVPPLTRENGSKIVFTTRSKDVCRDMEVDGEMKVDCLPPDEAWELFQKKVGPI 396
Query: 348 TFKNDSSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERI--------- 398
++ + + E GLPLAL V+ GK++A E T+ +++ +
Sbjct: 397 PLQSHEDIPTLARKVAEKCCGLPLALSVI-----GKAMASRE-TVQEWQHVIHVLNSSSH 450
Query: 399 ----PPEDIQKILKVSFDTLEEEQQSVFLDIACCFKGC 432
E I +LK S+D L++E+ + CF C
Sbjct: 451 EFPSMEEKILPVLKFSYDDLKDEK------VKLCFLYC 482
>DR27_ARATH (Q9T048) Putative disease resistance protein At4g27190
Length = 985
Score = 66.2 bits (160), Expect = 2e-10
Identities = 69/238 (28%), Positives = 113/238 (46%), Gaps = 21/238 (8%)
Query: 202 IGILGTGGLGKTTLAEAVYNSIVNQFECRCF---LYNVRENSFKHSLKYLQEQLLSKSIG 258
IG+ G GG+GKTTL + N + + + F ++ + F + +Q+Q +++ +
Sbjct: 167 IGVWGMGGVGKTTLVRTLNNKLREEGATQPFGLVIFVIVSKEF--DPREVQKQ-IAERLD 223
Query: 259 YDTPLEHDNEGI--EIIKQRLCRKKVLLILDDVDKPNQLEKLVGEPGWFGQGSRVIITTR 316
DT +E E + I + +K LLILDDV KP L+ L +GS+VI+T+
Sbjct: 224 IDTQMEESEEKLARRIYVGLMKERKFLLILDDVWKPIDLDLLGIPRTEENKGSKVILTS- 282
Query: 317 DRYLLSCHGITKIYE--ADSLNKEESLELLRKMT--FKNDSSYDYILNRAVEYASGLPLA 372
R+L C + + D L +E++ EL K I + GLPLA
Sbjct: 283 -RFLEVCRSMKTDLDVRVDCLLEEDAWELFCKNAGDVVRSDHVRKIAKAVSQECGGLPLA 341
Query: 373 LKVVGSNLFG-KSIADCESTLDKYERIPP------EDIQKILKVSFDTLEEEQQSVFL 423
+ VG+ + G K++ L K + P E I + LK+S+D LE++ + FL
Sbjct: 342 IITVGTAMRGKKNVKLWNHVLSKLSKSVPWIKSIEEKIFQPLKLSYDFLEDKAKFCFL 399
>DR15_ARATH (Q940K0) Probable disease resistance protein At1g61180
Length = 889
Score = 66.2 bits (160), Expect = 2e-10
Identities = 72/253 (28%), Positives = 116/253 (45%), Gaps = 28/253 (11%)
Query: 197 DGVYIIGILGTGGLGKTTLAEAVYN---SIVNQFECRCFLYNVRENSFKHSLKYLQEQLL 253
DGV I+G+ G GG+GKTTL + ++N I F+ ++ S + LQE +
Sbjct: 170 DGVGIMGLHGMGGVGKTTLFKKIHNKFAEIGGTFDIVIWIV----VSKGVMISKLQEDIA 225
Query: 254 SKSIGYDTPLEHDNEGIEI--IKQRLCRKKVLLILDDVDKPNQLEKL-VGEPGWFGQGSR 310
K D ++ NE + I + L K+ +L+LDD+ + LE + + P + +
Sbjct: 226 EKLHLCDDLWKNKNESDKATDIHRVLKGKRFVLMLDDIWEKVDLEAIGIPYPSEVNK-CK 284
Query: 311 VIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKNDSSYDYIL----NRAVEYA 366
V TTR R + G K + + L E++ EL + N S D ++ +
Sbjct: 285 VAFTTRSREVCGEMGDHKPMQVNCLEPEDAWELFKNKVGDNTLSSDPVIVELAREVAQKC 344
Query: 367 SGLPLALKVVGSNLFGKS-IADCESTLDKYERIPPE--DIQK----ILKVSFDTLEEEQQ 419
GLPLAL V+G + K+ + + E + + E D+Q ILK S+D+L +E
Sbjct: 345 RGLPLALNVIGETMSSKTMVQEWEHAIHVFNTSAAEFSDMQNKILPILKYSYDSLGDEH- 403
Query: 420 SVFLDIACCFKGC 432
I CF C
Sbjct: 404 -----IKSCFLYC 411
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.331 0.145 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,036,152
Number of Sequences: 164201
Number of extensions: 2957878
Number of successful extensions: 12217
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 12059
Number of HSP's gapped (non-prelim): 120
length of query: 625
length of database: 59,974,054
effective HSP length: 116
effective length of query: 509
effective length of database: 40,926,738
effective search space: 20831709642
effective search space used: 20831709642
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC126790.7


