

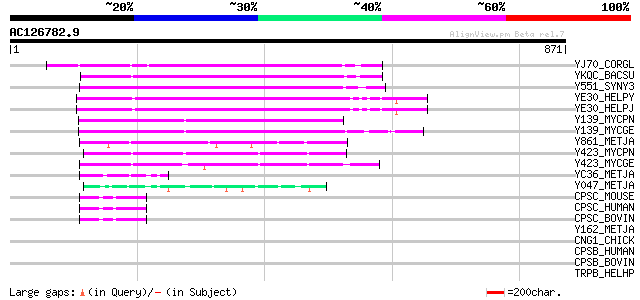
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126782.9 + phase: 1 /pseudo
(871 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YJ70_CORGL (P54122) Hypothetical UPF0036 protein Cgl1970/cg2160 313 1e-84
YKQC_BACSU (Q45493) Hypothetical UPF0036 protein ykqC 284 6e-76
Y551_SYNY3 (P54123) Hypothetical UPF0036 protein slr0551 279 2e-74
YE30_HELPY (P56185) Hypothetical UPF0036 protein HP1430 278 6e-74
YE30_HELPJ (Q9ZJI6) Hypothetical UPF0036 protein JHP1323 275 4e-73
Y139_MYCPN (P75497) Hypothetical UPF0036 protein MG139 homolog (... 242 4e-63
Y139_MYCGE (P47385) Hypothetical UPF0036 protein MG139 241 5e-63
Y861_METJA (Q58271) Hypothetical UPF0036 protein MJ0861 177 8e-44
Y423_MYCPN (P75174) Hypothetical protein MG423 homolog (C12_orf561) 93 3e-18
Y423_MYCGE (P47662) Hypothetical protein MG423 82 6e-15
YC36_METJA (Q58633) Hypothetical protein MJ1236 48 1e-04
Y047_METJA (Q60355) Hypothetical protein MJ0047 47 2e-04
CPSC_MOUSE (Q9QXK7) Cleavage and polyadenylation specificity fac... 45 8e-04
CPSC_HUMAN (Q9UKF6) Cleavage and polyadenylation specificity fac... 45 8e-04
CPSC_BOVIN (P79101) Cleavage and polyadenylation specificity fac... 45 8e-04
Y162_METJA (Q57626) Hypothetical protein MJ0162 43 0.003
CNG1_CHICK (Q90805) Cyclic nucleotide gated channel, cone photor... 42 0.007
CPSB_HUMAN (Q9P2I0) Cleavage and polyadenylation specificity fac... 39 0.046
CPSB_BOVIN (Q10568) Cleavage and polyadenylation specificity fac... 39 0.046
TRPB_HELHP (Q7VGA7) Tryptophan synthase beta chain (EC 4.2.1.20) 39 0.060
>YJ70_CORGL (P54122) Hypothetical UPF0036 protein Cgl1970/cg2160
Length = 718
Score = 313 bits (803), Expect = 1e-84
Identities = 196/534 (36%), Positives = 297/534 (54%), Gaps = 21/534 (3%)
Query: 58 SASALSASVGNDGSTSRVPQKRRRRIEGPR---KSME--DSVQRRMEQFYEGNDGPPLRV 112
S +A + N G+ +R R R G R KSM+ D QR E +G LR+
Sbjct: 100 SGNANEGANNNSGNQNRQGGNRGNRGGGRRNVVKSMQGADLTQRLPEPPKAPANG--LRI 157
Query: 113 LPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEALV 172
+GG+ EIG N + ++R +++D G++FP + GV I+PD I H+++ALV
Sbjct: 158 YALGGISEIGRNMTVFEYNNRLLIVDCGVLFPSSGEPGVDLILPDFGPIEDHLHRVDALV 217
Query: 173 ITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKNKFV 232
+THGHEDHIGA+PW++ L ++ PI AS FT+ LI + KEH P +++ N+
Sbjct: 218 VTHGHEDHIGAIPWLLK-LRNDIPILASRFTLALIAAKCKEHRQ-RPKLIEVNEQSNED- 274
Query: 233 AGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELSKEG 292
GPF I V HSIPDC GL ++ G ++HTGD K+D+TP DG+ D L EG
Sbjct: 275 RGPFNIRFWAVNHSIPDCLGLAIKTPAGLVIHTGDIKLDQTPPDGRPTDLPALSRFGDEG 334
Query: 293 VTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAADLT 352
V LM+ DSTN +PG + SE+ VA +L R + +K RVI FASN++R+ + AA +
Sbjct: 335 VDLMLCDSTNATTPGVSGSEADVAPTLKRLVGDAKQRVILASFASNVYRVQAAVDAAVAS 394
Query: 353 GRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPRAAL 412
RK+ F G S+ +E A K G T++ +D AP ++++TTG+Q EP AAL
Sbjct: 395 NRKVAFNGRSMIRNMEIAEKLGYLKAPRGTIISMDDASRMAPHKVMLITTGTQGEPMAAL 454
Query: 413 NLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHTSGH 472
+ + + D+++ S+ ++PGNE V ++N +++IG+T+V GR+ +HTSGH
Sbjct: 455 SRMARREHRQITVRDGDLIILSSSLVPGNEEAVFGVINMLAQIGATVVTGRDAKVHTSGH 514
Query: 473 AYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGIRHTAVI--KNGEMLGVSHLR 530
Y GEL + +P++ +PVHGE+ L+ ++ L STG+ V+ +NG V +
Sbjct: 515 GYSGELLFLYNAARPKNAMPVHGEWRHLRANKELAISTGVNRDNVVLAQNGV---VVDMV 571
Query: 531 NRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEI 584
N R G I +G NL Y DG G L +R + G+I ++ I
Sbjct: 572 NGRAQVVGQIPVG--NL---YVDG-VTMGDIDADILADRTSLGEGGLISITAVI 619
>YKQC_BACSU (Q45493) Hypothetical UPF0036 protein ykqC
Length = 555
Score = 284 bits (727), Expect = 6e-76
Identities = 167/475 (35%), Positives = 259/475 (54%), Gaps = 14/475 (2%)
Query: 112 VLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEAL 171
V +GGLGEIG N V D +LIDAGI FP+ + LG+ +IPD T++ K KI+ L
Sbjct: 11 VFALGGLGEIGKNTYAVQFQDEIVLIDAGIKFPEDELLGIDYVIPDYTYLVKNEDKIKGL 70
Query: 172 VITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKNKF 231
ITHGHEDHIG +P+++ + N P++ + L++ +L+EHG+ ++L I +
Sbjct: 71 FITHGHEDHIGGIPYLLRQV--NIPVYGGKLAIGLLRNKLEEHGLLRQTKLNIIGEDDIV 128
Query: 232 VAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELSKE 291
+ R THSIPD G+V++ G I+HTGD+K D TP+ G+ + + E+ KE
Sbjct: 129 KFRKTAVSFFRTTHSIPDSYGIVVKTPPGNIVHTGDFKFDFTPV-GEPANLTKMAEIGKE 187
Query: 292 GVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAADL 351
GV ++SDSTN +P T SE V +S+ GR+I FASN+HR+ V AA
Sbjct: 188 GVLCLLSDSTNSENPEFTMSERRVGESIHDIFRKVDGRIIFATFASNIHRLQQVIEAAVQ 247
Query: 352 TGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPRAA 411
GRK+ G S+ + +E G +T ++ +I+ + I+ TGSQ EP AA
Sbjct: 248 NGRKVAVFGRSMESAIEIGQTLGYINCPKNTFIEHNEINRMPANKVTILCTGSQGEPMAA 307
Query: 412 LNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHTSG 471
L+ + G+ + D V++S+ IPGN V +N++ G+ ++ G ++HTSG
Sbjct: 308 LSRIANGTHRQISINPGDTVVFSSSPIPGNTISVSRTINQLYRAGAEVIHGPLNDIHTSG 367
Query: 472 HAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGI--RHTAVIKNGEMLGVSHL 529
H + E + +LR++KP+ F+P+HGEY K H L GI + ++ NGE+L L
Sbjct: 368 HGGQEEQKLMLRLIKPKFFMPIHGEYRMQKMHVKLATDCGIPEENCFIMDNGEVLA---L 424
Query: 530 RNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEI 584
+ G I G Y DG G G + L +R ++ +G+++V + I
Sbjct: 425 KGDEASVAGKIPSGS-----VYIDG-SGIGDIGNIVLRDRRILSEEGLVIVVVSI 473
>Y551_SYNY3 (P54123) Hypothetical UPF0036 protein slr0551
Length = 640
Score = 279 bits (714), Expect = 2e-74
Identities = 165/484 (34%), Positives = 270/484 (55%), Gaps = 15/484 (3%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
L++LP+GGL EIG N + D +L+DAG+ FP D GV ++PD T++R+ KI+
Sbjct: 10 LKILPLGGLHEIGKNTCVFEYDDEILLLDAGLAFPTDDMHGVNVVLPDMTYLRENREKIK 69
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKN 229
+V+THGHEDHIG + + + D I+ M L++ +L+E G+ + L+ +
Sbjct: 70 GMVVTHGHEDHIGGIAYHLKQFDIPI-IYGPRLAMALLRDKLEEAGMLERTNLQTVSPRE 128
Query: 230 KFVAGP-FEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEEL 288
G F +E IR THSI D L + G ++H+GD+KID TP+DG+ FD + + E
Sbjct: 129 MVRLGKSFVVEFIRNTHSIADSYCLAIHTPLGVVMHSGDFKIDHTPIDGEFFDLQKVAEY 188
Query: 289 SKEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAA 348
++GV ++SDSTN PG T SE+ V +L R S ++GR++ T FAS++HR+ + +
Sbjct: 189 GEKGVLCLLSDSTNAEVPGITPSEASVIPNLDRVFSQAEGRLMVTTFASSVHRVNIILSL 248
Query: 349 ADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEP 408
A RK+ VG S+ + A K G + V + + LI+TTGSQ EP
Sbjct: 249 AQKHQRKVAVVGRSMLNVIAHARKLGYIKCPDNLFVPLKAARNLPDQQQLILTTGSQGEP 308
Query: 409 RAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLH 468
AA+ S G K+ + D V++SA IPGN V+ ++R+ G+ ++ G+++ +H
Sbjct: 309 LAAMTRISNGEHPQIKIRQGDTVVFSANPIPGNTIAVVNTIDRLMMQGANVIYGKHQGIH 368
Query: 469 TSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGI--RHTAVIKNGEMLGV 526
SGHA + E + +L + +P+ F+PVHGE+ L +H + ++ GI + ++ NG+++
Sbjct: 369 VSGHASQEEHKMLLALTRPKFFVPVHGEHRMLVKHSQMAQAQGIPSENIVIVNNGDVI-- 426
Query: 527 SHLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIFR 586
L R+ G + G E + D+A G E + ER ++A DG++ V+ + +
Sbjct: 427 -ELTGDRIRVAGQVPSGIELV-------DQA-GIVHESTMAERQQMAEDGLVTVAAALSK 477
Query: 587 PKNL 590
L
Sbjct: 478 TGTL 481
>YE30_HELPY (P56185) Hypothetical UPF0036 protein HP1430
Length = 689
Score = 278 bits (710), Expect = 6e-74
Identities = 180/558 (32%), Positives = 293/558 (52%), Gaps = 21/558 (3%)
Query: 105 NDGPPLRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKW 164
N +++ P+GGLGEIG N M++ I+IDAG+ FP GV +IPD +++ +
Sbjct: 136 NSKASVKITPLGGLGEIGGNMMVIETPKSAIVIDAGMSFPKEGLFGVDILIPDFSYLHQI 195
Query: 165 SHKIEALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIF-LPSRLK 223
KI ++ITH HEDHIGA P++ L P++ + ++ LI + EHG+ S K
Sbjct: 196 KDKIAGIIITHAHEDHIGATPYLFKELQF--PLYGTPLSLGLIGSKFDEHGLKKYRSYFK 253
Query: 224 IFRTKNKFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDRE 283
I + G F IE I +THSI D L ++ GTI+HTGD+KID TP+D D
Sbjct: 254 IVEKRCPISVGEFIIEWIHITHSIIDSSALAIQTKAGTIIHTGDFKIDHTPVDNLPTDLY 313
Query: 284 GLEELSKEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIG 343
L ++GV L++SDSTN G T SES +A + ++GRVI + F+SN+HR+
Sbjct: 314 RLAHYGEKGVMLLLSDSTNSHKSGTTPSESTIAPAFDTLFKEAQGRVIMSTFSSNIHRVY 373
Query: 344 SVKAAADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTG 403
RK+ +G S+ L+ A + G + ++A ++ Y ++LIVTTG
Sbjct: 374 QAIQYGIKYNRKIAVIGRSMEKNLDIARELGYIHLPYQSFIEANEVAKYPDNEILIVTTG 433
Query: 404 SQAEPRAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGR 463
SQ E +AL + + D+V+ SAK IPGNE+ V ++N + + + +
Sbjct: 434 SQGETMSALYRMATDEHRHISIKPNDLVIISAKAIPGNEASVSAVLNFLIKKEAKVAYQE 493
Query: 464 NENLHTSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGI--RHTAVIKNG 521
+N+H SGHA + E + +LR++KP+ FLPVHGEY + H+ S G+ ++ ++++G
Sbjct: 494 FDNIHVSGHAAQEEQKLMLRLIKPKFFLPVHGEYNHVARHKQTAISCGVPEKNIYLMEDG 553
Query: 522 EMLGVSHLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVS 581
+ + V ++V G I GK + + + + TS + +R +A G+ V +
Sbjct: 554 DQVEVGPAFIKKV---GTIKSGKSYVD---NQSNLSIDTS---IVQQREEVASAGVFVAT 604
Query: 582 MEIFRPKNLESLAGNTLKGKIRIT----TRCLWLDKGKLLDALYKAAHAALSSCPVKSPL 637
IF KN ++L ++ + + + L + L+ L K+++A + + P K
Sbjct: 605 --IFVNKNKQALLESSQFSSLGLVGFKDEKPLIKEIQGGLEVLLKSSNAEILNNPKKLE- 661
Query: 638 PHMERTVSEVLRKMVRKY 655
H + + L K RKY
Sbjct: 662 DHTRNFIRKALFKKFRKY 679
>YE30_HELPJ (Q9ZJI6) Hypothetical UPF0036 protein JHP1323
Length = 692
Score = 275 bits (703), Expect = 4e-73
Identities = 178/558 (31%), Positives = 292/558 (51%), Gaps = 21/558 (3%)
Query: 105 NDGPPLRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKW 164
N +++ P+GGLGEIG N M++ I+IDAG+ FP GV +IPD +++ +
Sbjct: 139 NSKASVKITPLGGLGEIGGNMMVIETPKSAIVIDAGMSFPKEGLFGVDILIPDFSYLHQI 198
Query: 165 SHKIEALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIF-LPSRLK 223
KI ++ITH HEDHIGA P++ L P++ + ++ LI + EHG+ S K
Sbjct: 199 KDKIAGIIITHAHEDHIGATPYLFKELQF--PLYGTPLSLGLIGSKFDEHGLKKYRSYFK 256
Query: 224 IFRTKNKFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDRE 283
I + G F IE I +THSI D L ++ GTI+HTGD+KID TP+D D
Sbjct: 257 IVEKRCPISVGEFIIEWIHITHSIIDSSALAIQTKAGTIIHTGDFKIDHTPVDNLPTDLY 316
Query: 284 GLEELSKEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIG 343
L ++GV L++SDSTN G T SES +A + ++GRVI + F+SN+HR+
Sbjct: 317 RLAHYGEKGVMLLLSDSTNSHKSGTTPSESTIAPAFDTLFKEAQGRVIMSTFSSNIHRVH 376
Query: 344 SVKAAADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTG 403
RK+ +G S+ L+ A + G + ++A ++ Y ++LIVTTG
Sbjct: 377 QAIQYGIKYNRKIAVIGRSMEKNLDIARELGYIHLPYQSFIEANEVAKYPDNEVLIVTTG 436
Query: 404 SQAEPRAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGR 463
SQ E +AL + + D+V+ SAK IPGNE+ V ++N + + + +
Sbjct: 437 SQGETMSALYRMATDEHRHISIKPNDLVIISAKAIPGNEASVSAVLNFLIKKEAKVAYQE 496
Query: 464 NENLHTSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGI--RHTAVIKNG 521
+N+H SGHA + E + +LR++KP+ FLPVHGEY + H+ + G+ ++ ++++G
Sbjct: 497 FDNIHVSGHAAQEEQKLMLRLIKPKFFLPVHGEYNHVARHKQTAIACGVPEKNIYLMEDG 556
Query: 522 EMLGVSHLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVS 581
+ + V ++V G I GK + + + + TS + +R +A G+ +
Sbjct: 557 DQVEVGPAFIKKV---GTIKSGKSYVD---NQSNLSIDTS---IVQQREEVASAGVFAAT 607
Query: 582 MEIFRPKNLESLAGNTLKGKIRIT----TRCLWLDKGKLLDALYKAAHAALSSCPVKSPL 637
IF KN ++L ++ + + + L + L+ L K+++A + + P K
Sbjct: 608 --IFVNKNKQALLESSQFSSLGLVGFKDEKHLIKEIQGGLEVLLKSSNAEILNNPKKLE- 664
Query: 638 PHMERTVSEVLRKMVRKY 655
H + + L K RKY
Sbjct: 665 DHTRNFIRKALFKKFRKY 682
>Y139_MYCPN (P75497) Hypothetical UPF0036 protein MG139 homolog
(A65_orf569)
Length = 569
Score = 242 bits (617), Expect = 4e-63
Identities = 137/419 (32%), Positives = 233/419 (54%), Gaps = 5/419 (1%)
Query: 109 PLRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKI 168
P ++ GG+ E+G N + D I+ID GI F D LG+ IIP ++ + K+
Sbjct: 15 PTKIFAFGGIQEVGKNMYGIEYDDEIIIIDCGIKFASDDLLGIDGIIPSFEYLIENQAKV 74
Query: 169 EALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTK 228
+AL ITHGHEDHIG +P+++ +D I+A LI K++ EH +++ ++
Sbjct: 75 KALFITHGHEDHIGGVPYLLKQVDVPV-IYAPRIAASLILKKVNEHKDAKLNKVVVYDDF 133
Query: 229 NKFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEEL 288
+ F F+I+ RV HSIPD G+ ++ +G I+ +GD++ D G++ D + ++
Sbjct: 134 SNFETKHFKIDFYRVNHSIPDAFGVCVQTPNGNIVESGDFRFDFAA-GGEMLDVHKVVKI 192
Query: 289 SKEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAA 348
++ V + M ++TN PG + SE ++ ++ + I ++GRVI T FASN+ RI +
Sbjct: 193 AERNVHVFMCETTNAEIPGFSQSEKLIYRNINKIIKEARGRVILTTFASNITRINEIIEI 252
Query: 349 ADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEP 408
A RK+ +G S+ + + K G D + +V+ DI Y + +LI+ TGSQ E
Sbjct: 253 AVNNKRKVCLLGKSMDVNVNISRKIGLMDIDSNDIVEVRDIKNYPDRSILILCTGSQGED 312
Query: 409 RAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGR-NENL 467
AALN + G + L D ++ S+ IPGN + V ++N +S+ G TI N L
Sbjct: 313 SAALNTMARGKHNWVSLKSTDTIIMSSNPIPGNYAAVENLLNELSKYGVTIFENSPNMKL 372
Query: 468 HTSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGIR--HTAVIKNGEML 524
H SGHA + EL+ +L +V P++ +P+HGEY ++ +++ + GI ++ NG+++
Sbjct: 373 HASGHATQQELQLMLNLVFPRYLIPIHGEYKMMRTIKNIAQECGINGDDVGLLANGQVM 431
>Y139_MYCGE (P47385) Hypothetical UPF0036 protein MG139
Length = 569
Score = 241 bits (616), Expect = 5e-63
Identities = 159/545 (29%), Positives = 278/545 (50%), Gaps = 21/545 (3%)
Query: 109 PLRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKI 168
P ++ GG+ E+G N + D I+ID GI F D LG+ IIP + + K+
Sbjct: 15 PTKIYAFGGIQEVGKNMYGIEYDDEIIIIDCGIKFASDDLLGINGIIPSFEHLIENQSKV 74
Query: 169 EALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTK 228
+AL ITHGHEDHIG +P+++ +D I+A LI K++ EH +++ F
Sbjct: 75 KALFITHGHEDHIGGVPYLLKQVDIPV-IYAPRIAASLILKKVNEHKDAKLNKIVTFDDF 133
Query: 229 NKFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEEL 288
++F F+I+ RV HSIPD G+ ++ +G I+ +GD++ D ++ D + ++
Sbjct: 134 SEFQTKHFKIDFYRVNHSIPDAFGICVQTPNGNIVQSGDYRFDFAA-GSEMLDVHKVVKI 192
Query: 289 SKEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAA 348
++ V + MS+STN PG + SE ++ ++ + + ++GRVI T FASN+ RI +
Sbjct: 193 AERNVHVFMSESTNAEVPGFSQSEKLIYRNIQKILKEARGRVILTTFASNITRINEIIEI 252
Query: 349 ADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEP 408
A RK+ +G S+ + + K G D + +V+ DI Y +++LI+ TGSQ E
Sbjct: 253 ALNNKRKICLLGKSMDVNVNISRKIGLMAIDSNDIVEVRDIKNYPDRNILILCTGSQGEE 312
Query: 409 RAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNE-NL 467
AALN + G + L D ++ S+ IPGN + V ++N +S+ G I ++ L
Sbjct: 313 AAALNTMARGKHNWVSLKSTDTIIMSSNPIPGNYAAVENLLNELSKFGVAIYENSSQLKL 372
Query: 468 HTSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGIR--HTAVIKNGEMLG 525
H SGHA + EL+ +L ++ P++ +P+HGE+ ++ +++ GI+ A++ NG+++
Sbjct: 373 HASGHATQQELQLMLNLMFPKYLIPIHGEFKMMRTIKNIANECGIKSEDVALLSNGQVMY 432
Query: 526 VSHLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIF 585
+ + SN I+ ++ S D A + +R ++ DG+ V +
Sbjct: 433 L--IDEELYYSNEIINADPIYIESHNSSPDLA------RIIKQRQILSRDGMFAVIVVFD 484
Query: 586 RPKNLESLAGNTLKGKIRITTRCLW-LDKGKLLDALYKAAHAALSSCPVKSPLPHMERTV 644
+ N+ + TL IT C + LD L+ + + L S E+
Sbjct: 485 KNNNIIGIP--TL-----ITRGCFFALDSNPLMTKIAHSVKRTLESVIQSKKFNSHEQLT 537
Query: 645 SEVLR 649
E+ R
Sbjct: 538 KELKR 542
>Y861_METJA (Q58271) Hypothetical UPF0036 protein MJ0861
Length = 448
Score = 177 bits (450), Expect = 8e-44
Identities = 133/453 (29%), Positives = 224/453 (49%), Gaps = 41/453 (9%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPD---YDDLGVQK----------IIP 156
L ++ IGG E+G N V I++D GI ++D + K IIP
Sbjct: 3 LEIIAIGGYEEVGRNMTAVNVDGEIIILDMGIRLDRVLIHEDTDISKLHSLELIEKGIIP 62
Query: 157 DTTFIRKWSHKIEALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKR-LKEHG 215
+ T ++ +++A+V++HGH DHIGA+P + A N PI + +T+EL+K+ L E
Sbjct: 63 NDTVMKNIEGEVKAIVLSHGHLDHIGAVPKL--AHRYNAPIIGTPYTIELVKREILSEKK 120
Query: 216 IFLPSRLKIFRTKNKFVAGP-FEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETP 274
+ + L + P +E IR+THSIPD VL G+I++ D+K D P
Sbjct: 121 FDVRNPLIVLNAGESIDLTPNITLEFIRITHSIPDSVLPVLHTPYGSIVYGNDFKFDNFP 180
Query: 275 LDGKVFDREGLEELSKEGVTLMMSDSTNVLSPGRTTSESVVADSLLRH----ISASKGRV 330
+ G+ D ++++ K GV +S++T + G+T E ++A LL++ K +
Sbjct: 181 VVGERPDYRAIKKVGKNGVLCFISETTRINHEGKTPPE-IIASGLLKNDLLAADNDKHGI 239
Query: 331 ITTQFASNLHRIGSVKAAADLTGRKLVFVGMSLRTYLEAAWKDGKAPF--------DPST 382
I T F+S++ RI S+ A+ GR V +G S+ + A G F DPS+
Sbjct: 240 IVTTFSSHIARIKSITDIAEKMGRTPVLLGRSMMRFCGIAQDIGLVKFPEDLRIYGDPSS 299
Query: 383 LVKA-EDIDAYAPKDLLIVTTGSQAEPRAALNLASFGSSHAFKLTKEDIVLYSAKVIPG- 440
+ A ++I + LI+ TG Q E A L+ + + +K K D V++SA IP
Sbjct: 300 IEMALKNIVKEGKEKYLIIATGHQGEEGAVLSRMATNKT-PYKFEKYDCVVFSADPIPNP 358
Query: 441 -NESRVMEMMNRISEIGSTIVMGRNENLHTSGHAYRGELEEVLRIVKPQHFLPVHGEYLF 499
N ++ + +R+ +G I G H SGHA + + ++LR + P+H +P HG++
Sbjct: 359 MNAAQRYMLESRLKLLGVRIFKG----AHVSGHAAKEDHRDMLRWLNPEHIIPSHGDFNL 414
Query: 500 LKEHESLGKSTGIR---HTAVIKNGEMLGVSHL 529
E+ L + G R +++NG+ L +
Sbjct: 415 TAEYTKLAEEEGYRLGEDVHLLRNGQCLSFERI 447
>Y423_MYCPN (P75174) Hypothetical protein MG423 homolog (C12_orf561)
Length = 561
Score = 92.8 bits (229), Expect = 3e-18
Identities = 94/422 (22%), Positives = 186/422 (43%), Gaps = 17/422 (4%)
Query: 116 GGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEALVITH 175
GG E G NC ++ ++ + + G + P LGV+KIIPD ++I++ +I+ + I +
Sbjct: 10 GGQDERGKNCFVLEINNDVFIFNVGSLTPTTAVLGVKKIIPDFSWIQENQARIKGIFIGN 69
Query: 176 GHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLP-SRLKIFRTK--NKFV 232
++IG+L ++ + PI+ S+ +IK ++ E+ + +P L+I K
Sbjct: 70 PVTENIGSLEFLFHTV-GFFPIYTSTIGAVVIKTKIHENKLNIPHDELEIHELKPLETVK 128
Query: 233 AGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELSKE- 291
G I P +V+ SIP G L DG I++ D+ + D + L ++ +
Sbjct: 129 IGHHNITPFKVSSSIPSSFGFALHTDDGYIVYVDDFIVLN---DKNIAFENQLNQIIPQV 185
Query: 292 -GVTLMMSDSTNVL--SPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAA 348
TL++ ++ + G TT + + L R I+++KGRV + SN + + ++
Sbjct: 186 ANKTLLLITGVGLVGRNTGFTTPKHKSLEQLNRIIASAKGRVFAACYDSNAYSVMTLAQI 245
Query: 349 ADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEP 408
A + R V S A + + E+I+ + ++V T +
Sbjct: 246 ARMQNRPFVIYSHSFVHLFNAIVRQKLFNNTHLNTISIEEIN--NSTNAIVVLTAPPDKL 303
Query: 409 RAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLH 468
A L + K D ++ I G E ++++ ++ + E L
Sbjct: 304 YAKLFKIGTNEDERVRYRKTDSFIFMIPRIAGYEELEAQILDDVARNEVSYYNLGREIL- 362
Query: 469 TSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGIRHTAVI--KNGEMLGV 526
S +A +++ ++ +KP++ +P G Y + K G+ + V+ NGE+L +
Sbjct: 363 -SINASDEDMKFLVTSLKPKYIIPTSGLYRDFINFTMVMKQAGVEQSQVLIPFNGEVLAI 421
Query: 527 SH 528
+H
Sbjct: 422 NH 423
>Y423_MYCGE (P47662) Hypothetical protein MG423
Length = 561
Score = 82.0 bits (201), Expect = 6e-15
Identities = 96/486 (19%), Positives = 199/486 (40%), Gaps = 37/486 (7%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
++ +GG E G NC ++ + + + G + P LGV+KIIPD ++I++ +++
Sbjct: 4 IKFFALGGQDERGKNCYVLEIDNDVFIFNVGSLTPTTAVLGVKKIIPDFSWIQENQARVK 63
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLP-SRLKIFRTK 228
+ I + +++G+L ++ + PI+ SS +IK ++ E+ + + +L+I K
Sbjct: 64 GIFIGNAITENLGSLEFLFHTV-GFFPIYTSSIGASIIKSKINENKLNIARDKLEIHELK 122
Query: 229 --NKFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLE 286
I P +V+ S+P G L +G I+ D+ + + D+
Sbjct: 123 PLETIEISNHSITPFKVSSSLPSSFGFALNTDNGYIVFIDDFIV--------LNDKNIAF 174
Query: 287 ELSKEGVTLMMSDSTNVL---------SPGRTTSESVVADSLLRHISASKGRVITTQFAS 337
E + +SD+T +L + G TT + + L R I+ +KGR+ + S
Sbjct: 175 ENQLNQIIPKLSDNTLLLITGVGLVGRNSGFTTPKHKSLEQLNRIITPAKGRIFVACYDS 234
Query: 338 NLHRIGSVKAAADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDL 397
N + + ++ A + R + S + + E+I+ +
Sbjct: 235 NAYSVMTLAQIARMQNRPFIIYSQSFVHLFNTIVRQKLFNNTHLNTISIEEIN--NSTNS 292
Query: 398 LIVTTGSQAEPRAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGS 457
++V T + A L + K D ++ + G E ++++ I+
Sbjct: 293 IVVLTSPPDKLYAKLFKIGMNEDERIRYRKSDTFIFMTPKVAGYEEIEAQILDDIARNEV 352
Query: 458 TIVMGRNENLHTSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGIRHT-- 515
+ E L S A +++ ++ +KP++ +P G Y + K G
Sbjct: 353 SYYNLGREIL--SIQASDEDMKFLVSSLKPKYIIPTGGLYRDFINFTMVLKQAGAEQNQI 410
Query: 516 AVIKNGEMLGVSHLRNRRVLSNGFISLGKENLQLKYSDGDKA-FGTSGELFLDERMRIAL 574
++ NGE+L + N + K L+L D A G + ER +++
Sbjct: 411 LILFNGEVL---------TIENKKLDSKKNELKLNPKCVDSAGLQEIGASIMFERDQMSE 461
Query: 575 DGIIVV 580
G++++
Sbjct: 462 SGVVII 467
>YC36_METJA (Q58633) Hypothetical protein MJ1236
Length = 634
Score = 48.1 bits (113), Expect = 1e-04
Identities = 39/141 (27%), Positives = 67/141 (46%), Gaps = 17/141 (12%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
+RV +GG E+G +C+ V D +LID GI D K P ++
Sbjct: 180 IRVSFLGGAREVGRSCLYVQTPDTRVLIDCGINVACED-----KAFPHFDAPEFSIEDLD 234
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKN 229
A+++TH H DH G +P + + P++ + T +L+ K++ L+I + +
Sbjct: 235 AVIVTHAHLDHCGFIPGLF-RYGYDGPVYCTRPTRDLMTLLQKDY-------LEIAKKEG 286
Query: 230 KFVAGPFEIEPIR--VTHSIP 248
K V P+ + I+ V H+IP
Sbjct: 287 KEV--PYTSKDIKTCVKHTIP 305
>Y047_METJA (Q60355) Hypothetical protein MJ0047
Length = 428
Score = 47.0 bits (110), Expect = 2e-04
Identities = 95/433 (21%), Positives = 170/433 (38%), Gaps = 92/433 (21%)
Query: 116 GGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEALVITH 175
G E+G +C+ + IL+D G+ LG + P + ++ + I+H
Sbjct: 7 GAALEVGRSCIEIKTDKSKILLDCGVK------LGKEIEYP---ILDNSIRDVDKVFISH 57
Query: 176 GHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKNKFVAGP 235
H DH GALP V+ + P+ + + +LIK LK+ +KI T+NK + P
Sbjct: 58 AHLDHSGALP-VLFHRKMDVPVITTELSKKLIKVLLKD-------MVKIAETENKKI--P 107
Query: 236 FEIEPIR--VTHSIP-----------------------DCCGLVLRCSDG-TILHTGDWK 269
+ ++ + H+IP ++L + TIL+TGD K
Sbjct: 108 YNNHDVKEAIRHTIPLNYNDKKYYKDFSYELFSAGHIPGSASILLNYQNNKTILYTGDVK 167
Query: 270 IDETPLDGKVFDREGLE-ELSKEGVTLMMSDST--NVLSPGRTTSESVVADSLLRHISAS 326
+ +T L +G + +K+ + +++ +ST N + P R E + ++ I
Sbjct: 168 LRDTRL------TKGADLSYTKDDIDILIIESTYGNSIHPDRKAVELSFIEK-IKEILFR 220
Query: 327 KGRVITTQFASN-------LHRIGSVKAAADLTGRKLVFVGMSLR--------TYLEAAW 371
G + FA + + ++ A L G + + L + LE A
Sbjct: 221 GGVALIPVFAVDRAQEILLILNDYNIDAPIYLDGMAVEVTKLMLNYKHMLNESSQLEKAL 280
Query: 372 KDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPRAALNLASFGSSHAFKLTKEDIV 431
K+ K +KA I+ + ++VTT + L + K ++
Sbjct: 281 KNVKIIEKSEDRIKA--IENLSKNGGIVVTTAGMLDGGPILYYLKLFMHN----PKNALL 334
Query: 432 LYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNE---NLHT-----SGHAYRGELEEVLR 483
L +V N ++E I +G++E NL S HA EL E+++
Sbjct: 335 LTGYQVRDSNGRHLIET--------GKIFIGKDEIKPNLEVCMYNFSCHAGMDELHEIIK 386
Query: 484 IVKPQHFLPVHGE 496
V P+ + HGE
Sbjct: 387 KVNPELLIIQHGE 399
>CPSC_MOUSE (Q9QXK7) Cleavage and polyadenylation specificity
factor, 73 kDa subunit (CPSF 73 kDa subunit)
Length = 684
Score = 45.1 bits (105), Expect = 8e-04
Identities = 29/105 (27%), Positives = 50/105 (47%), Gaps = 6/105 (5%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
L + P+G E+G +C+++ R I++D GI + L +P I +I+
Sbjct: 12 LLIRPLGAGQEVGRSCIILEFKGRKIMLDCGI----HPGLEGMDALPYIDLIDP--AEID 65
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEH 214
L+I+H H DH GALPW + F + T + + L ++
Sbjct: 66 LLLISHFHLDHCGALPWFLQKTSFKGRTFMTHATKAIYRWLLSDY 110
>CPSC_HUMAN (Q9UKF6) Cleavage and polyadenylation specificity
factor, 73 kDa subunit (CPSF 73 kDa subunit)
Length = 684
Score = 45.1 bits (105), Expect = 8e-04
Identities = 29/105 (27%), Positives = 50/105 (47%), Gaps = 6/105 (5%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
L + P+G E+G +C+++ R I++D GI + L +P I +I+
Sbjct: 12 LLIRPLGAGQEVGRSCIILEFKGRKIMLDCGI----HPGLEGMDALPYIDLIDP--AEID 65
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEH 214
L+I+H H DH GALPW + F + T + + L ++
Sbjct: 66 LLLISHFHLDHCGALPWFLQKTSFKGRTFMTHATKAIYRWLLSDY 110
>CPSC_BOVIN (P79101) Cleavage and polyadenylation specificity
factor, 73 kDa subunit (CPSF 73 kDa subunit)
Length = 684
Score = 45.1 bits (105), Expect = 8e-04
Identities = 29/105 (27%), Positives = 50/105 (47%), Gaps = 6/105 (5%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
L + P+G E+G +C+++ R I++D GI + L +P I +I+
Sbjct: 12 LLIRPLGAGQEVGRSCIILEFKGRKIMLDCGI----HPGLEGMDALPYIDLIDP--AEID 65
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEH 214
L+I+H H DH GALPW + F + T + + L ++
Sbjct: 66 LLLISHFHLDHCGALPWFLQKTSFKGRTFMTHATKAIYRWLLSDY 110
>Y162_METJA (Q57626) Hypothetical protein MJ0162
Length = 421
Score = 43.1 bits (100), Expect = 0.003
Identities = 24/72 (33%), Positives = 38/72 (52%), Gaps = 14/72 (19%)
Query: 116 GGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHK-IEALVIT 174
GG +IGM+C+ V +L+D G+ PDT I K K ++A++++
Sbjct: 8 GGCQQIGMSCVEVETQKGRVLLDCGMS-------------PDTGEIPKVDDKAVDAVIVS 54
Query: 175 HGHEDHIGALPW 186
H H DH GA+P+
Sbjct: 55 HAHLDHCGAIPF 66
>CNG1_CHICK (Q90805) Cyclic nucleotide gated channel, cone
photoreceptor, alpha subunit (CNG channel 1) (CNG-1)
Length = 735
Score = 42.0 bits (97), Expect = 0.007
Identities = 20/54 (37%), Positives = 29/54 (53%)
Query: 762 ANNGYVSRKEHKSNTKQDDSEDIDEAKSEEMSDSEPESSKSEKKNKWKTEEVKK 815
+NN +KE K K++ E+ E K EE D + + K +KK+ K EE KK
Sbjct: 145 SNNTNEDKKEEKKEVKEEKKEEKKEEKKEEKKDDKKDDKKDDKKDDKKKEEQKK 198
>CPSB_HUMAN (Q9P2I0) Cleavage and polyadenylation specificity
factor, 100 kDa subunit (CPSF 100 kDa subunit)
Length = 782
Score = 39.3 bits (90), Expect = 0.046
Identities = 25/91 (27%), Positives = 47/91 (51%), Gaps = 8/91 (8%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
+++ + G+ E C L+ + L+D G +D+ II +RK H+I+
Sbjct: 5 IKLTTLSGVQEESALCYLLQVDEFRFLLDCG-----WDEHFSMDIIDS---LRKHVHQID 56
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFAS 200
A++++H H+GALP+ + L N I+A+
Sbjct: 57 AVLLSHPDPLHLGALPYAVGKLGLNCAIYAT 87
>CPSB_BOVIN (Q10568) Cleavage and polyadenylation specificity
factor, 100 kDa subunit (CPSF 100 kDa subunit)
Length = 782
Score = 39.3 bits (90), Expect = 0.046
Identities = 25/91 (27%), Positives = 47/91 (51%), Gaps = 8/91 (8%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
+++ + G+ E C L+ + L+D G +D+ II +RK H+I+
Sbjct: 5 IKLTTLSGVQEESALCYLLQVDEFRFLLDCG-----WDEHFSMDIIDS---LRKHVHQID 56
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFAS 200
A++++H H+GALP+ + L N I+A+
Sbjct: 57 AVLLSHPDPLHLGALPYAVGKLGLNCAIYAT 87
>TRPB_HELHP (Q7VGA7) Tryptophan synthase beta chain (EC 4.2.1.20)
Length = 420
Score = 38.9 bits (89), Expect = 0.060
Identities = 46/202 (22%), Positives = 82/202 (39%), Gaps = 17/202 (8%)
Query: 473 AYRGELEEVLR--IVKPQHFLPVHGEYLFLKEHESLGKSTGIRHTAVIKNGEMLG----V 526
AY+ EL+ + + + +P + H LK L K G+ +T K LG
Sbjct: 52 AYKKELKSLFKHFVGRPTPLIYAHNASKILKNDIYL-KFEGLANTGAHKINNALGQVLLA 110
Query: 527 SHLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIFR 586
H++ +RV++ G L + G E+F+ E + IA V +ME+F
Sbjct: 111 KHMKKKRVIAE----TGAGQHGLATAAACARLGLECEIFMGE-IDIARQRPNVFNMELFG 165
Query: 587 PK-NLESLAGNTLKGKIRITTRCLWLDKGKLLDALYKAAHAALSSCPVKSPLPHMERTVS 645
K + S TLK + R + K D + +AL P + ++ +S
Sbjct: 166 AKVHSVSSGSKTLKDAVNEALR----EWSKRSDDSFYVLGSALGPYPYPDIVRDLQSVIS 221
Query: 646 EVLRKMVRKYSGKRPEVIAIAI 667
+ L+K + Y P+++ +
Sbjct: 222 KELKKQTKAYFSSLPDILVACV 243
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 98,592,234
Number of Sequences: 164201
Number of extensions: 4262535
Number of successful extensions: 16159
Number of sequences better than 10.0: 177
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 132
Number of HSP's that attempted gapping in prelim test: 15624
Number of HSP's gapped (non-prelim): 504
length of query: 871
length of database: 59,974,054
effective HSP length: 119
effective length of query: 752
effective length of database: 40,434,135
effective search space: 30406469520
effective search space used: 30406469520
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Medicago: description of AC126782.9


