

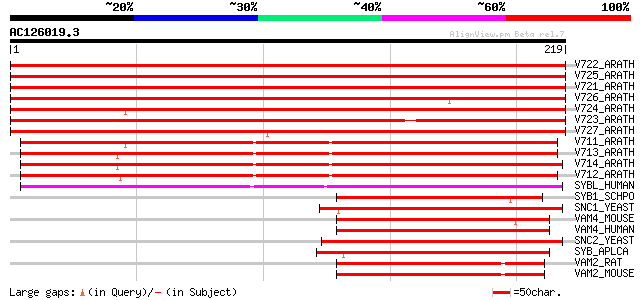
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126019.3 + phase: 0
(219 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
V722_ARATH (P47192) Vesicle-associated membrane protein 722 (AtV... 382 e-106
V725_ARATH (O48850) Vesicle-associated membrane protein 725 (AtV... 380 e-105
V721_ARATH (Q9ZTW3) Vesicle-associated membrane protein 721 (AtV... 379 e-105
V726_ARATH (Q9MAS5) Putative vesicle-associated membrane protein... 371 e-103
V724_ARATH (O23429) Vesicle-associated membrane protein 724 (AtV... 333 2e-91
V723_ARATH (Q8VY69) Vesicle-associated membrane protein 723 (AtV... 302 4e-82
V727_ARATH (Q9M376) Vesicle-associated membrane protein 727 (AtV... 281 8e-76
V711_ARATH (O49377) Vesicle-associated membrane protein 711 (AtV... 164 2e-40
V713_ARATH (Q9LFP1) Vesicle-associated membrane protein 713 (AtV... 160 2e-39
V714_ARATH (Q9FMR5) Vesicle-associated membrane protein 714 (AtV... 158 1e-38
V712_ARATH (Q9SIQ9) Vesicle-associated membrane protein 712 (AtV... 153 3e-37
SYBL_HUMAN (P51809) Synaptobrevin-like protein 1 144 1e-34
SYB1_SCHPO (Q92356) Synaptobrevin homolog 1 80 4e-15
SNC1_YEAST (P31109) Synaptobrevin homolog 1 79 6e-15
VAM4_MOUSE (O70480) Vesicle-associated membrane protein 4 (VAMP-4) 77 4e-14
VAM4_HUMAN (O75379) Vesicle-associated membrane protein 4 (VAMP-4) 76 5e-14
SNC2_YEAST (P33328) Synaptobrevin homolog 2 75 1e-13
SYB_APLCA (P35589) Synaptobrevin 72 1e-12
VAM2_RAT (P63045) Vesicle-associated membrane protein 2 (VAMP-2)... 70 4e-12
VAM2_MOUSE (P63044) Vesicle-associated membrane protein 2 (VAMP-... 70 4e-12
>V722_ARATH (P47192) Vesicle-associated membrane protein 722
(AtVAMP722) (Synaptobrevin-related protein 1)
Length = 221
Score = 382 bits (980), Expect = e-106
Identities = 182/219 (83%), Positives = 207/219 (94%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
M QQSLIYSFVARGTVIL E++DF GNFT+IA QCLQKLP+SNN+FTYNCDGHTF++LV+
Sbjct: 1 MAQQSLIYSFVARGTVILVEFTDFKGNFTSIAAQCLQKLPSSNNKFTYNCDGHTFNYLVE 60
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
NGFTYCVVAV+S GRQIP+AFLER+K+DFNKRYGGG+A TA A SLNKEFG KLKE MQY
Sbjct: 61 NGFTYCVVAVDSAGRQIPMAFLERVKEDFNKRYGGGKAATAQANSLNKEFGSKLKEHMQY 120
Query: 121 CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG 180
C++HP+E+SKLAKVKAQVSEVKGVMMENI+KV+DRGEKIE+LVDKTENLRSQAQDFR QG
Sbjct: 121 CMDHPDEISKLAKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTQG 180
Query: 181 TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
TQ+RRKMW+QNMKIKLIVLAIIIALILII+LS+C GF+C
Sbjct: 181 TQMRRKMWFQNMKIKLIVLAIIIALILIIILSICGGFNC 219
>V725_ARATH (O48850) Vesicle-associated membrane protein 725
(AtVAMP725)
Length = 220
Score = 380 bits (975), Expect = e-105
Identities = 180/219 (82%), Positives = 206/219 (93%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
MGQQ+LIYSFVARGTVIL EY++F GNFT +A QCLQKLP+SNN+FTYNCDGHTF++LV+
Sbjct: 1 MGQQNLIYSFVARGTVILVEYTEFKGNFTAVAAQCLQKLPSSNNKFTYNCDGHTFNYLVE 60
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
NGFTYCVVAVESVGRQIP+AFLER+K+DFNKRYGGG+ATTA A SLN+EFG KLKE MQY
Sbjct: 61 NGFTYCVVAVESVGRQIPMAFLERVKEDFNKRYGGGKATTAQANSLNREFGSKLKEHMQY 120
Query: 121 CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG 180
CV+HP+E+SKLAKVKAQV+EVKGVMMENI+KV+DRGEKIE+LVDKTENLRSQAQDFR QG
Sbjct: 121 CVDHPDEISKLAKVKAQVTEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTQG 180
Query: 181 TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
T++RRKMW++NMKIKLIVL III LILII+LSVC GF C
Sbjct: 181 TKIRRKMWFENMKIKLIVLGIIITLILIIILSVCGGFKC 219
>V721_ARATH (Q9ZTW3) Vesicle-associated membrane protein 721
(AtVAMP721) (v-SNARE synaptobrevin 7B) (AtVAMP7B)
Length = 219
Score = 379 bits (973), Expect = e-105
Identities = 182/219 (83%), Positives = 205/219 (93%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
M QQSLIYSFVARGTVIL E++DF GNFT+IA QCLQKLP+SNN+FTYNCDGHTF++LV+
Sbjct: 1 MAQQSLIYSFVARGTVILVEFTDFKGNFTSIAAQCLQKLPSSNNKFTYNCDGHTFNYLVE 60
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
+GFTYCVVAV+S GRQIP++FLER+K+DFNKRYGGG+A TA A SLNKEFG KLKE MQY
Sbjct: 61 DGFTYCVVAVDSAGRQIPMSFLERVKEDFNKRYGGGKAATAQANSLNKEFGSKLKEHMQY 120
Query: 121 CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG 180
C++HP+E+SKLAKVKAQVSEVKGVMMENI+KV+DRGEKIE+LVDKTENLRSQAQDFR G
Sbjct: 121 CMDHPDEISKLAKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTTG 180
Query: 181 TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
TQ+RRKMW QNMKIKLIVLAIIIALILIIVLSVCHGF C
Sbjct: 181 TQMRRKMWLQNMKIKLIVLAIIIALILIIVLSVCHGFKC 219
>V726_ARATH (Q9MAS5) Putative vesicle-associated membrane protein
726 (AtVAMP726)
Length = 229
Score = 371 bits (952), Expect = e-103
Identities = 176/228 (77%), Positives = 205/228 (89%), Gaps = 9/228 (3%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
MGQQSLIYSFVARGTVILAEY++F GNFT++A QCLQKLP+SNN+FTYNCDGHTF++L D
Sbjct: 1 MGQQSLIYSFVARGTVILAEYTEFKGNFTSVAAQCLQKLPSSNNKFTYNCDGHTFNYLAD 60
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
NGFTYCVV +ES GRQIP+AFLER+K+DFNKRYGGG+A+TA A SLNKEFG KLKE MQY
Sbjct: 61 NGFTYCVVVIESAGRQIPMAFLERVKEDFNKRYGGGKASTAKANSLNKEFGSKLKEHMQY 120
Query: 121 CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQ-------- 172
C +HPEE+SKL+KVKAQV+EVKGVMMENI+KV+DRGEKIE+LVDKTENLRSQ
Sbjct: 121 CADHPEEISKLSKVKAQVTEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQVNNNHISN 180
Query: 173 -AQDFRQQGTQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
AQDFR QGT+++RK+W++NMKIKLIV II+ALILII+LSVCHGF C
Sbjct: 181 TAQDFRTQGTKMKRKLWFENMKIKLIVFGIIVALILIIILSVCHGFKC 228
>V724_ARATH (O23429) Vesicle-associated membrane protein 724
(AtVAMP724) (SYBL1-like protein)
Length = 222
Score = 333 bits (853), Expect = 2e-91
Identities = 157/220 (71%), Positives = 193/220 (87%), Gaps = 1/220 (0%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNN-RFTYNCDGHTFSFLV 59
MGQ+S IYSFVARGT+ILAEY++FTGNF +IA QCLQKLP+S+N +FTYNCD HTF+FLV
Sbjct: 1 MGQESFIYSFVARGTMILAEYTEFTGNFPSIAAQCLQKLPSSSNSKFTYNCDHHTFNFLV 60
Query: 60 DNGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQ 119
++G+ YCVVA +S+ +QI IAFLER+K DF KRYGGG+A+TA AKSLNKEFGP +KE M
Sbjct: 61 EDGYAYCVVAKDSLSKQISIAFLERVKADFKKRYGGGKASTAIAKSLNKEFGPVMKEHMN 120
Query: 120 YCVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQ 179
Y V+H EE+ KL KVKAQVSEVK +M+ENIDK IDRGE + VL DKTENLRSQAQ++++Q
Sbjct: 121 YIVDHAEEIEKLIKVKAQVSEVKSIMLENIDKAIDRGENLTVLTDKTENLRSQAQEYKKQ 180
Query: 180 GTQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
GTQ+RRK+WYQNMKIKL+VL I++ L+LII +SVCHGF+C
Sbjct: 181 GTQVRRKLWYQNMKIKLVVLGILLLLVLIIWISVCHGFNC 220
>V723_ARATH (Q8VY69) Vesicle-associated membrane protein 723
(AtVAMP723)
Length = 217
Score = 302 bits (774), Expect = 4e-82
Identities = 148/219 (67%), Positives = 180/219 (81%), Gaps = 4/219 (1%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
M QQSL YSF+ARGTVIL E++DF GNFT++A Q L+ LP+SNN+FTYNCDGHTF+ LV+
Sbjct: 1 MAQQSLFYSFIARGTVILVEFTDFKGNFTSVAAQYLENLPSSNNKFTYNCDGHTFNDLVE 60
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
NGFTYCVVAV+S GR+IP+AFLER+K+DF KRYGG +A T A SLNKEFG LKE MQY
Sbjct: 61 NGFTYCVVAVDSAGREIPMAFLERVKEDFYKRYGGEKAATDQANSLNKEFGSNLKEHMQY 120
Query: 121 CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG 180
C++HP+E+S LAK KAQVSEVK +MMENI+KV+ RG V+ + + SQ Q F +
Sbjct: 121 CMDHPDEISNLAKAKAQVSEVKSLMMENIEKVLARG----VICEMLGSSESQPQAFYIKR 176
Query: 181 TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
TQ++RK W+QNMKIKLIVLAIIIALILII+LSVC GF+C
Sbjct: 177 TQMKRKKWFQNMKIKLIVLAIIIALILIIILSVCGGFNC 215
>V727_ARATH (Q9M376) Vesicle-associated membrane protein 727
(AtVAMP727)
Length = 240
Score = 281 bits (719), Expect = 8e-76
Identities = 133/238 (55%), Positives = 180/238 (74%), Gaps = 19/238 (7%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
M Q+ LIYSFVA+GTV+LAE++ ++GNF+TIA+QCLQKLP +++++TY+CDGHTF+FLVD
Sbjct: 1 MSQKGLIYSFVAKGTVVLAEHTPYSGNFSTIAVQCLQKLPTNSSKYTYSCDGHTFNFLVD 60
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATT-------------------A 101
NGF + VVA ES GR +P FLER+K+DF KRY +
Sbjct: 61 NGFVFLVVADESTGRSVPFVFLERVKEDFKKRYEASIKNDERHPLADEDEDDDLFGDRFS 120
Query: 102 TAKSLNKEFGPKLKEQMQYCVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEV 161
A +L++EFGP LKE MQYC+ HPEE+SKL+K+KAQ++EVKG+MM+NI+KV+DRGEKIE+
Sbjct: 121 VAYNLDREFGPILKEHMQYCMSHPEEMSKLSKLKAQITEVKGIMMDNIEKVLDRGEKIEL 180
Query: 162 LVDKTENLRSQAQDFRQQGTQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
LVDKTENL+ QA F++QG QLRRKMW Q++++KL+V + + ILI+ + C GF C
Sbjct: 181 LVDKTENLQFQADSFQRQGRQLRRKMWLQSLQMKLMVAGAVFSFILIVWVVACGGFKC 238
>V711_ARATH (O49377) Vesicle-associated membrane protein 711
(AtVAMP711) (v-SNARE synaptobrevin 7C) (AtVAMP7C)
Length = 219
Score = 164 bits (414), Expect = 2e-40
Identities = 82/213 (38%), Positives = 139/213 (64%), Gaps = 3/213 (1%)
Query: 5 SLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNN-RFTYNCDGHTFSFLVDNGF 63
+++Y+ VARGTV+L+E++ + N +TIA Q L+K+P N+ +Y+ D + F +G
Sbjct: 2 AILYALVARGTVVLSEFTATSTNASTIAKQILEKVPGDNDSNVSYSQDRYVFHVKRTDGL 61
Query: 64 TYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCVE 123
T +A E+ GR+IP AFLE I F + YG TA A ++N+EF L +Q+ Y
Sbjct: 62 TVLCMAEETAGRRIPFAFLEDIHQRFVRTYGRA-VHTALAYAMNEEFSRVLSQQIDYYSN 120
Query: 124 HPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQL 183
P ++ ++K ++++V+GVM+ENIDKV+DRGE++E+LVDKT N++ FR+Q +
Sbjct: 121 DPN-ADRINRIKGEMNQVRGVMIENIDKVLDRGERLELLVDKTANMQGNTFRFRKQARRF 179
Query: 184 RRKMWYQNMKIKLIVLAIIIALILIIVLSVCHG 216
R +W++N K+ ++++ +++ +I I V +CHG
Sbjct: 180 RSNVWWRNCKLTVLLILLLLVIIYIAVAFLCHG 212
>V713_ARATH (Q9LFP1) Vesicle-associated membrane protein 713
(AtVAMP713)
Length = 221
Score = 160 bits (406), Expect = 2e-39
Identities = 83/214 (38%), Positives = 139/214 (64%), Gaps = 4/214 (1%)
Query: 5 SLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPA--SNNRFTYNCDGHTFSFLVDNG 62
++I++ VARGTV+L+E+S + N ++I+ Q L+KLP S++ +Y+ D + F +G
Sbjct: 2 AIIFALVARGTVVLSEFSATSTNASSISKQILEKLPGNDSDSHMSYSQDRYIFHVKRTDG 61
Query: 63 FTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCV 122
T +A E+ GR IP AFL+ I F K YG +A A S+N EF L +QM++
Sbjct: 62 LTVLCMADETAGRNIPFAFLDDIHQRFVKTYGRA-IHSAQAYSMNDEFSRVLSQQMEFYS 120
Query: 123 EHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQ 182
P +++++K ++S+V+ VM+ENIDKV+DRGE++E+LVDKTEN++ FR+Q +
Sbjct: 121 NDPN-ADRMSRIKGEMSQVRNVMIENIDKVLDRGERLELLVDKTENMQGNTFRFRKQARR 179
Query: 183 LRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHG 216
R MW++N+K+ + ++ ++ ++ I + VCHG
Sbjct: 180 YRTIMWWRNVKLTIALILVLALVVYIAMAFVCHG 213
>V714_ARATH (Q9FMR5) Vesicle-associated membrane protein 714
(AtVAMP714)
Length = 221
Score = 158 bits (399), Expect = 1e-38
Identities = 82/216 (37%), Positives = 139/216 (63%), Gaps = 4/216 (1%)
Query: 5 SLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPA--SNNRFTYNCDGHTFSFLVDNG 62
+++Y+ VARGTV+LAE+S TGN + + L+KL S+ R ++ D + F L +G
Sbjct: 2 AIVYAVVARGTVVLAEFSAVTGNTGAVVRRILEKLSPEISDERLCFSQDRYIFHILRSDG 61
Query: 63 FTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCV 122
T+ +A ++ GR++P ++LE I F K YG A A A ++N EF L +QM++
Sbjct: 62 LTFLCMANDTFGRRVPFSYLEEIHMRFMKNYGKV-AHNAPAYAMNDEFSRVLHQQMEFFS 120
Query: 123 EHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQ 182
+P V L +V+ +VSE++ VM+ENI+K+++RG++IE+LVDKT ++ + FR+Q +
Sbjct: 121 SNPS-VDTLNRVRGEVSEIRSVMVENIEKIMERGDRIELLVDKTATMQDSSFHFRKQSKR 179
Query: 183 LRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFS 218
LRR +W +N K+ +++ +I+ L+ II+ S C G +
Sbjct: 180 LRRALWMKNAKLLVLLTCLIVFLLYIIIASFCGGIT 215
>V712_ARATH (Q9SIQ9) Vesicle-associated membrane protein 712
(AtVAMP712)
Length = 219
Score = 153 bits (387), Expect = 3e-37
Identities = 79/213 (37%), Positives = 134/213 (62%), Gaps = 3/213 (1%)
Query: 5 SLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPAS-NNRFTYNCDGHTFSFLVDNGF 63
S++Y+ VARGTV+LAE S + N +TIA Q L+K+P + ++ +Y+ D + F +G
Sbjct: 2 SILYALVARGTVVLAELSTTSTNASTIAKQILEKIPGNGDSHVSYSQDRYVFHVKRTDGL 61
Query: 64 TYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCVE 123
T +A E GR+IP +FLE I F + YG +A A ++N EF L +Q++Y
Sbjct: 62 TVLCMADEDAGRRIPFSFLEDIHQRFVRTYGRA-IHSAQAYAMNDEFSRVLNQQIEYYSN 120
Query: 124 HPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQL 183
P ++++K ++++V+ VM+ENID ++DRGE++E+LVDKT N++ FR+Q +
Sbjct: 121 DPN-ADTISRIKGEMNQVRDVMIENIDNILDRGERLELLVDKTANMQGNTFRFRKQTRRF 179
Query: 184 RRKMWYQNMKIKLIVLAIIIALILIIVLSVCHG 216
+W++N K+ L+++ +++ +I I V CHG
Sbjct: 180 NNTVWWRNCKLTLLLILVLLVIIYIGVAFACHG 212
>SYBL_HUMAN (P51809) Synaptobrevin-like protein 1
Length = 220
Score = 144 bits (364), Expect = 1e-34
Identities = 77/214 (35%), Positives = 125/214 (57%), Gaps = 2/214 (0%)
Query: 5 SLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVDNGFT 64
+++++ VARGT ILA+++ GNF + Q L K+P+ NN+ TY+ + F ++ +
Sbjct: 2 AILFAVVARGTTILAKHAWCGGNFLEVTEQILAKIPSENNKLTYSHGNYLFHYICQDRIV 61
Query: 65 YCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCVEH 124
Y + + R FL IK F YG RA TA ++N EF L Q+++ E+
Sbjct: 62 YLCITDDDFERSRAFNFLNEIKKRFQTTYGS-RAQTALPYAMNSEFSSVLAAQLKHHSEN 120
Query: 125 PEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQLR 184
+ + K+ + +AQV E+KG+M+ NID V RGE++E+L+DKTENL + F+ L
Sbjct: 121 -KGLDKVMETQAQVDELKGIMVRNIDLVAQRGERLELLIDKTENLVDSSVTFKTTSRNLA 179
Query: 185 RKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFS 218
R M +N+K+ +I++ + I I IIV +C GF+
Sbjct: 180 RAMCMKNLKLTIIIIIVSIVFIYIIVSPLCGGFT 213
>SYB1_SCHPO (Q92356) Synaptobrevin homolog 1
Length = 121
Score = 80.1 bits (196), Expect = 4e-15
Identities = 37/82 (45%), Positives = 61/82 (74%), Gaps = 1/82 (1%)
Query: 130 KLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQLRRKMWY 189
K A ++ Q+ + G+M ENI KV +RGE+++ L DKT+NL AQ FR+ ++R+KMW+
Sbjct: 31 KTAAIQQQIDDTVGIMRENISKVSERGERLDSLQDKTDNLAVSAQGFRRGANRVRKKMWW 90
Query: 190 QNMKIKL-IVLAIIIALILIIV 210
++M+++L I++ III L++IIV
Sbjct: 91 KDMRMRLCIIIGIIILLVVIIV 112
>SNC1_YEAST (P31109) Synaptobrevin homolog 1
Length = 117
Score = 79.3 bits (194), Expect = 6e-15
Identities = 34/99 (34%), Positives = 68/99 (68%), Gaps = 3/99 (3%)
Query: 123 EHPEEV---SKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQ 179
E P+ V S+ A+++A++ + G+M +NI+KV +RGE++ + DK +NL AQ F++
Sbjct: 18 ERPQNVQSKSRTAELQAEIDDTVGIMRDNINKVAERGERLTSIEDKADNLAVSAQGFKRG 77
Query: 180 GTQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFS 218
++R+ MWY+++K+K+ + +II L+++I++ + FS
Sbjct: 78 ANRVRKAMWYKDLKMKMCLALVIIILLVVIIVPIAVHFS 116
>VAM4_MOUSE (O70480) Vesicle-associated membrane protein 4 (VAMP-4)
Length = 141
Score = 76.6 bits (187), Expect = 4e-14
Identities = 42/85 (49%), Positives = 58/85 (67%), Gaps = 1/85 (1%)
Query: 130 KLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQLRRKMWY 189
K+ V+ QV EV VM ENI KVI+RGE+++ L DK+E+L A F + QLRR+MW+
Sbjct: 52 KIKHVQNQVDEVIDVMQENITKVIERGERLDELQDKSESLSDNATAFSNRSKQLRRQMWW 111
Query: 190 QNMKIKLIV-LAIIIALILIIVLSV 213
+ KIK I+ LA I L++II+L V
Sbjct: 112 RGCKIKAIMALAAAILLLMIIILIV 136
>VAM4_HUMAN (O75379) Vesicle-associated membrane protein 4 (VAMP-4)
Length = 141
Score = 76.3 bits (186), Expect = 5e-14
Identities = 37/84 (44%), Positives = 56/84 (66%)
Query: 130 KLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQLRRKMWY 189
K+ V+ QV EV VM ENI KVI+RGE+++ L DK+E+L A F + QLRR+MW+
Sbjct: 52 KIKHVQNQVDEVIDVMQENITKVIERGERLDELQDKSESLSDNATAFSNRSKQLRRQMWW 111
Query: 190 QNMKIKLIVLAIIIALILIIVLSV 213
+ KIK I+ + L+L+I++ +
Sbjct: 112 RGCKIKAIMALVAAILLLVIIILI 135
>SNC2_YEAST (P33328) Synaptobrevin homolog 2
Length = 115
Score = 75.1 bits (183), Expect = 1e-13
Identities = 28/95 (29%), Positives = 65/95 (67%)
Query: 124 HPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQL 183
+P +K A ++ ++ + G+M +NI+KV +RGE++ + DK +NL AQ F++ ++
Sbjct: 21 NPNSQNKTAALRQEIDDTVGIMRDNINKVAERGERLTSIEDKADNLAISAQGFKRGANRV 80
Query: 184 RRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFS 218
R++MW++++K+++ + ++I L+++I++ + FS
Sbjct: 81 RKQMWWKDLKMRMCLFLVVIILLVVIIVPIVVHFS 115
>SYB_APLCA (P35589) Synaptobrevin
Length = 180
Score = 71.6 bits (174), Expect = 1e-12
Identities = 38/93 (40%), Positives = 61/93 (64%), Gaps = 1/93 (1%)
Query: 122 VEHPEEVSK-LAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG 180
++ P E SK L + +AQV EV +M N++KV+DR +KI L D+ E L++ A F
Sbjct: 12 MQPPREQSKRLQQTQAQVDEVVDIMRVNVEKVLDRDQKISQLDDRAEALQAGASQFEASA 71
Query: 181 TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSV 213
+L+RK W++N K+ LI+ AII +++II++ V
Sbjct: 72 GKLKRKYWWKNCKMMLILGAIIGVIVIIIIVWV 104
>VAM2_RAT (P63045) Vesicle-associated membrane protein 2 (VAMP-2)
(Synaptobrevin 2)
Length = 115
Score = 70.1 bits (170), Expect = 4e-12
Identities = 32/82 (39%), Positives = 57/82 (69%), Gaps = 1/82 (1%)
Query: 130 KLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQLRRKMWY 189
+L + +AQV EV +M N+DKV++R +K+ L D+ + L++ A F +L+RK W+
Sbjct: 30 RLQQTQAQVDEVVDIMRVNVDKVLERDQKLSELDDRADALQAGASQFETSAAKLKRKYWW 89
Query: 190 QNMKIKLIVLAIIIALILIIVL 211
+N+K+ +I+L +I A+ILII++
Sbjct: 90 KNLKM-MIILGVICAIILIIII 110
>VAM2_MOUSE (P63044) Vesicle-associated membrane protein 2 (VAMP-2)
(Synaptobrevin 2)
Length = 115
Score = 70.1 bits (170), Expect = 4e-12
Identities = 32/82 (39%), Positives = 57/82 (69%), Gaps = 1/82 (1%)
Query: 130 KLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQLRRKMWY 189
+L + +AQV EV +M N+DKV++R +K+ L D+ + L++ A F +L+RK W+
Sbjct: 30 RLQQTQAQVDEVVDIMRVNVDKVLERDQKLSELDDRADALQAGASQFETSAAKLKRKYWW 89
Query: 190 QNMKIKLIVLAIIIALILIIVL 211
+N+K+ +I+L +I A+ILII++
Sbjct: 90 KNLKM-MIILGVICAIILIIII 110
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.137 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,515,645
Number of Sequences: 164201
Number of extensions: 927022
Number of successful extensions: 3858
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 61
Number of HSP's successfully gapped in prelim test: 47
Number of HSP's that attempted gapping in prelim test: 3757
Number of HSP's gapped (non-prelim): 120
length of query: 219
length of database: 59,974,054
effective HSP length: 106
effective length of query: 113
effective length of database: 42,568,748
effective search space: 4810268524
effective search space used: 4810268524
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC126019.3


