

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126009.4 - phase: 0
(162 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
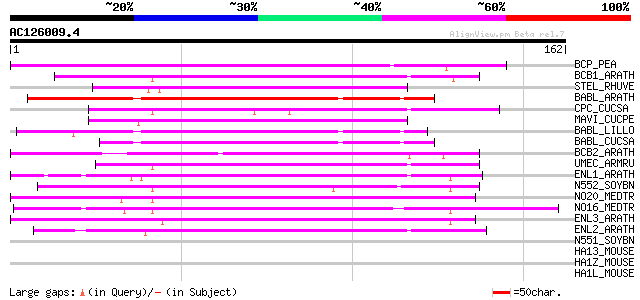
Score E
Sequences producing significant alignments: (bits) Value
BCP_PEA (Q41001) Blue copper protein precursor 99 4e-21
BCB1_ARATH (Q07488) Blue copper protein precursor (Blue copper-b... 87 2e-17
STEL_RHUVE (P00302) Stellacyanin 86 3e-17
BABL_ARATH (Q8LG89) Putative basic blue protein precursor (Plant... 86 3e-17
CPC_CUCSA (P29602) Cucumber peeling cupredoxin (CPC) (Stellacyanin) 83 3e-16
MAVI_CUCPE (P80728) Mavicyanin 82 7e-16
BABL_LILLO (P60496) Chemocyanin precursor (Basic blue protein) (... 75 8e-14
BABL_CUCSA (P00303) Basic blue protein (Cusacyanin) (Plantacyani... 74 1e-13
BCB2_ARATH (O80517) Uclacyanin II precursor (Blue copper-binding... 72 4e-13
UMEC_ARMRU (P42849) Umecyanin (UMC) 68 1e-11
ENL1_ARATH (Q9SK27) Early nodulin-like protein 1 precursor (Phyt... 64 1e-10
N552_SOYBN (Q02917) Early nodulin 55-2 precursor (N-55-2) (Nodul... 61 1e-09
NO20_MEDTR (P93329) Early nodulin 20 precursor (N-20) 61 1e-09
NO16_MEDTR (P93328) Early nodulin 16 precursor (N-16) 59 4e-09
ENL3_ARATH (Q8LC95) Early nodulin-like protein 3 precursor (Phyt... 52 6e-07
ENL2_ARATH (Q9T076) Early nodulin-like protein 2 precursor (Phyt... 46 3e-05
N551_SOYBN (Q05544) Early nodulin 55-1 (N-55-1) (Fragment) 37 0.019
HA13_MOUSE (P14426) H-2 class I histocompatibility antigen, D-K ... 33 0.21
HA1Z_MOUSE (P01896) H-2 class I histocompatibility antigen, alph... 33 0.28
HA1L_MOUSE (P01897) H-2 class I histocompatibility antigen, L-D ... 33 0.28
>BCP_PEA (Q41001) Blue copper protein precursor
Length = 189
Score = 99.0 bits (245), Expect = 4e-21
Identities = 58/147 (39%), Positives = 78/147 (52%), Gaps = 3/147 (2%)
Query: 1 MASSRVVLILSISMVLLSSVAIAATDYIVGDDKGWTVDFDYTQWAQDKVFRVGDNLVFNY 60
MA S +++ + ++ ++ AT Y VGD GW + DY+ WA DK F VGD+LVFNY
Sbjct: 1 MAFSNALVLCFLLAIINMALPSLATVYTVGDTSGWVIGGDYSTWASDKTFAVGDSLVFNY 60
Query: 61 DPSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLV 120
H V +V + ++SCT STG I LK G+ +++CGV H S MKL
Sbjct: 61 GAGAHTVDEVKESDYKSCTSGNSISTDSTGATTIPLKKAGKHYFICGVPGH-STGGMKLS 119
Query: 121 ITVLAE--GAPAPSPPPSSDAHSVVSS 145
I V A + APS PSS SS
Sbjct: 120 IKVKASSGSSAAPSATPSSSGKGSPSS 146
>BCB1_ARATH (Q07488) Blue copper protein precursor (Blue
copper-binding protein) (AtBCB) (Stellacyanin)
(Phytocyanin 1)
Length = 196
Score = 87.0 bits (214), Expect = 2e-17
Identities = 52/138 (37%), Positives = 72/138 (51%), Gaps = 15/138 (10%)
Query: 14 MVLLSSVAIAATDYIVGDDKGWTVDFD---YTQWAQDKVFRVGDNLVFNYDPSRHNVFKV 70
+++ ++V + A DY VGDD WT D YT WA K FRVGD L F++ RH+V V
Sbjct: 12 VLVFAAVVVFAEDYDVGDDTEWTRPMDPEFYTTWATGKTFRVGDELEFDFAAGRHDVAVV 71
Query: 71 NGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVITVLAEGA-- 128
+ F++C ++ I L T G ++++C V DHC Q KL ITV+A GA
Sbjct: 72 SEAAFENCEKEKPISHMTVPPVKIMLNTTGPQYFICTVGDHCRFGQ-KLSITVVAAGATG 130
Query: 129 ---------PAPSPPPSS 137
PAP PS+
Sbjct: 131 GATPGAGATPAPGSTPST 148
>STEL_RHUVE (P00302) Stellacyanin
Length = 107
Score = 86.3 bits (212), Expect = 3e-17
Identities = 45/97 (46%), Positives = 55/97 (56%), Gaps = 5/97 (5%)
Query: 25 TDYIVGDDKGWTVDF----DYT-QWAQDKVFRVGDNLVFNYDPSRHNVFKVNGTLFQSCT 79
T Y VGD GW V F DY +WA +K F +GD LVF YD HNV KV +QSC
Sbjct: 1 TVYTVGDSAGWKVPFFGDVDYDWKWASNKTFHIGDVLVFKYDRRFHNVDKVTQKNYQSCN 60
Query: 80 FPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQ 116
+ +TG B I LKT G+K+Y+CGV HC Q
Sbjct: 61 DTTPIASYNTGBBRINLKTVGQKYYICGVPKHCDLGQ 97
>BABL_ARATH (Q8LG89) Putative basic blue protein precursor
(Plantacyanin)
Length = 129
Score = 86.3 bits (212), Expect = 3e-17
Identities = 43/119 (36%), Positives = 72/119 (60%), Gaps = 4/119 (3%)
Query: 6 VVLILSISMVLLSSVAIAATDYIVGDDKGWTVDFDYTQWAQDKVFRVGDNLVFNYDPSRH 65
+V ++++S++LL + + A Y VGD WT F+ W + K FR GD LVFNY+P H
Sbjct: 15 IVTLMAVSVLLLQADYVQAATYTVGDSGIWT--FNAVGWPKGKHFRAGDVLVFNYNPRMH 72
Query: 66 NVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVITVL 124
NV KV+ + +C P + ++GKD I L ++G+ +++C +HC + MK+ +T +
Sbjct: 73 NVVKVDSGSYNNCKTPTGAKPYTSGKDRITL-SKGQNFFICNFPNHCES-DMKIAVTAV 129
>CPC_CUCSA (P29602) Cucumber peeling cupredoxin (CPC) (Stellacyanin)
Length = 137
Score = 82.8 bits (203), Expect = 3e-16
Identities = 52/125 (41%), Positives = 68/125 (53%), Gaps = 6/125 (4%)
Query: 24 ATDYIVGDDKGWTVDFD---YTQWAQDKVFRVGDNLVFNYDPSRHNVFKV-NGTLFQSCT 79
+T +IVGD+ GW+V Y+QWA K FRVGD+L FN+ + HNV ++ F +C
Sbjct: 2 STVHIVGDNTGWSVPSSPNFYSQWAAGKTFRVGDSLQFNFPANAHNVHEMETKQSFDACN 61
Query: 80 F-PPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVITVLAEGAPAPSPPPSSD 138
F N+ T I +L G ++VC V HCS Q KL I V+A A PPPSS
Sbjct: 62 FVNSDNDVERTSPVIERLDELGMHYFVCTVGTHCSNGQ-KLSINVVAANATVSMPPPSSS 120
Query: 139 AHSVV 143
S V
Sbjct: 121 PPSSV 125
>MAVI_CUCPE (P80728) Mavicyanin
Length = 108
Score = 81.6 bits (200), Expect = 7e-16
Identities = 39/95 (41%), Positives = 55/95 (57%), Gaps = 2/95 (2%)
Query: 24 ATDYIVGDDKGWT--VDFDYTQWAQDKVFRVGDNLVFNYDPSRHNVFKVNGTLFQSCTFP 81
AT + VGD GWT V +DY +WA F VGD+L+FNY+ HNV +V+ F+SC
Sbjct: 1 ATVHKVGDSTGWTTLVPYDYAKWASSNKFHVGDSLLFNYNNKFHNVLQVDQEQFKSCNSS 60
Query: 82 PKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQ 116
+ ++G D I LK G +++CG+ HC Q
Sbjct: 61 SPAASYTSGADSIPLKRPGTFYFLCGIPGHCQLGQ 95
>BABL_LILLO (P60496) Chemocyanin precursor (Basic blue protein)
(Plantacyanin)
Length = 126
Score = 74.7 bits (182), Expect = 8e-14
Identities = 43/122 (35%), Positives = 63/122 (51%), Gaps = 6/122 (4%)
Query: 3 SSRVVLILSISMVLL--SSVAIAATDYIVGDDKGWTVDFDYTQWAQDKVFRVGDNLVFNY 60
S+ L+L + +V L + + Y VGD GWT F + W K FR GD LVF Y
Sbjct: 7 SAERALVLGVVLVFLVFNCEVAESVVYTVGDGGGWT--FGTSGWPAGKTFRAGDVLVFKY 64
Query: 61 DPSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLV 120
+P+ HNV V ++SCT P + +G D I L + G +++C V HC +K+
Sbjct: 65 NPAVHNVVSVPAGGYKSCTASPGSRVFKSGDDRITL-SRGTNYFICSVPGHCQG-GLKIA 122
Query: 121 IT 122
+T
Sbjct: 123 VT 124
>BABL_CUCSA (P00303) Basic blue protein (Cusacyanin) (Plantacyanin)
(CBP)
Length = 96
Score = 73.9 bits (180), Expect = 1e-13
Identities = 37/98 (37%), Positives = 56/98 (56%), Gaps = 4/98 (4%)
Query: 27 YIVGDDKGWTVDFDYTQWAQDKVFRVGDNLVFNYDPSRHNVFKVNGTLFQSCTFPPKNEA 86
Y+VG GWT F+ W + K FR GD L+FNY+P HNV VN F +C P +
Sbjct: 3 YVVGGSGGWT--FNTESWPKGKRFRAGDILLFNYNPXMHNVVVVNQGGFSTCNTPAGAKV 60
Query: 87 LSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVITVL 124
++G+D I+L +G+ +++C HC + MK+ + L
Sbjct: 61 YTSGRDQIKL-PKGQSYFICNFPGHCQS-GMKIAVNAL 96
>BCB2_ARATH (O80517) Uclacyanin II precursor (Blue copper-binding
protein II) (BCB II) (Phytocyanin 2)
Length = 202
Score = 72.4 bits (176), Expect = 4e-13
Identities = 49/140 (35%), Positives = 64/140 (45%), Gaps = 11/140 (7%)
Query: 1 MASSRVVLILSISMVLLSSVAIAATDYIVGDDKGWTVDFDYTQWAQDKVFRVGDNLVFNY 60
MA + +L + ++ +VA+ T WT DY+ WA K FRVGD L F Y
Sbjct: 9 MAVAAATALLLVLTIVPGAVAVTYTIE-------WTTGVDYSGWATGKTFRVGDILEFKY 61
Query: 61 DPSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSAR-QMKL 119
S H V V+ + C E S G I LKT G +++C HC MKL
Sbjct: 62 G-SSHTVDVVDKAGYDGCDASSSTENHSDGDTKIDLKTVGINYFICSTPGHCRTNGGMKL 120
Query: 120 VITVLA--EGAPAPSPPPSS 137
+ V+A G PA PPSS
Sbjct: 121 AVNVVAGSAGPPATPTPPSS 140
>UMEC_ARMRU (P42849) Umecyanin (UMC)
Length = 115
Score = 67.8 bits (164), Expect = 1e-11
Identities = 41/115 (35%), Positives = 52/115 (44%), Gaps = 4/115 (3%)
Query: 26 DYIVGDDKGWTVDFD---YTQWAQDKVFRVGDNLVFNYDPSRHNVFKVNGTLFQSCTFPP 82
DY VG D W D Y WA K FRVGD L F++ H+V V F +C
Sbjct: 2 DYDVGGDMEWKRPSDPKFYITWATGKTFRVGDELEFDFAAGMHDVAVVTKDAFDNCKKEN 61
Query: 83 KNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVITVLAEGAPAPSPPPSS 137
++T I L T G ++Y+C V DHC Q KL I V+ G P +
Sbjct: 62 PISHMTTPPVKIMLNTTGPQYYICTVGDHCRVGQ-KLSINVVGAGGAGGGATPGA 115
>ENL1_ARATH (Q9SK27) Early nodulin-like protein 1 precursor
(Phytocyanin-like protein)
Length = 176
Score = 64.3 bits (155), Expect = 1e-10
Identities = 45/147 (30%), Positives = 74/147 (49%), Gaps = 12/147 (8%)
Query: 1 MASSRVVLILSISMVLLSSVAIAATDYIVGDDKG-WTV----DFDYTQWAQDKVFRVGDN 55
MASS + + + S++ L S+A AA + VG G W + + +T+WAQ F+VGD
Sbjct: 1 MASSSLHVAI-FSLIFLFSLA-AANEVTVGGKSGDWKIPPSSSYSFTEWAQKARFKVGDF 58
Query: 56 LVFNYDPSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSAR 115
+VF Y+ + +V +V + SC + G+ ++L G +++ G HC
Sbjct: 59 IVFRYESGKDSVLEVTKEAYNSCNTTNPLANYTDGETKVKLDRSGPFYFISGANGHCEKG 118
Query: 116 QMKLVITVLAEG----APAPSPPPSSD 138
Q KL + V++ +PAPSP D
Sbjct: 119 Q-KLSLVVISPRHSVISPAPSPVEFED 144
>N552_SOYBN (Q02917) Early nodulin 55-2 precursor (N-55-2)
(Nodulin-315)
Length = 187
Score = 61.2 bits (147), Expect = 1e-09
Identities = 38/143 (26%), Positives = 66/143 (45%), Gaps = 15/143 (10%)
Query: 9 ILSISMVLLSSVAIAATDYIVGDDKGWTVDFD----YTQWAQDKVFRVGDNLVFNYDPSR 64
++ ++M LL S + A + G +K W + WA F++GD L+F Y+
Sbjct: 12 LVMLAMCLLISTSEAEKYVVGGSEKSWKFPLSKPDSLSHWANSHRFKIGDTLIFKYEKRT 71
Query: 65 HNVFKVNGTLFQSCTFPPKNEALSTGKDI-IQLKTEGRKWYVCGVADHCSARQMKLVITV 123
+V + N T ++ C K + G + + L G + ++ G HC +KL + V
Sbjct: 72 ESVHEGNETDYEGCNTVGKYHIVFNGGNTKVMLTKPGFRHFISGNQSHCQ-MGLKLAVLV 130
Query: 124 LAEG---------APAPSPPPSS 137
++ +P+PSPPPSS
Sbjct: 131 ISSNKTKKNLLSPSPSPSPPPSS 153
>NO20_MEDTR (P93329) Early nodulin 20 precursor (N-20)
Length = 268
Score = 60.8 bits (146), Expect = 1e-09
Identities = 38/141 (26%), Positives = 64/141 (44%), Gaps = 5/141 (3%)
Query: 1 MASSRVVLILSISMVLLSSVAIAATDYIVGD-DKGWTVDFD----YTQWAQDKVFRVGDN 55
M+SS +L++ I + + +TDY+VGD + W T+WA + F VGD
Sbjct: 1 MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT 60
Query: 56 LVFNYDPSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSAR 115
+ F Y+ +V +V + C ++ G ++ LK G ++ G HC
Sbjct: 61 ITFQYNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLG 120
Query: 116 QMKLVITVLAEGAPAPSPPPS 136
V+ ++A +P PPPS
Sbjct: 121 LKLAVVVMVAPVLSSPPPPPS 141
>NO16_MEDTR (P93328) Early nodulin 16 precursor (N-16)
Length = 180
Score = 59.3 bits (142), Expect = 4e-09
Identities = 46/178 (25%), Positives = 82/178 (45%), Gaps = 23/178 (12%)
Query: 2 ASSRVVLILSISMVLLSSVAIAATDYIVGDD-KGWTVDFD----YTQWAQDKVFRVGDNL 56
+SS ++L++ SM LL S + +TDY++GD W V + +WA F VGD +
Sbjct: 3 SSSPILLMIIFSMWLLISHS-ESTDYLIGDSHNSWKVPLPSRRAFARWASAHEFTVGDTI 61
Query: 57 VFNYDPSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQ 116
+F YD +V +VN + C ++ G + L G ++ G HC +
Sbjct: 62 LFEYDNETESVHEVNEHDYIMCHTNGEHVEHHDGNTKVVLDKIGVYHFISGTKRHC---K 118
Query: 117 MKLVITVLAEG--------------APAPSPPPSSDAHSVVSSLFGVVMAIMVAIAVI 160
M L + V+ + P+PSP P+S + ++ G +M + V++ ++
Sbjct: 119 MGLKLAVVVQNKHDLVLPPLITMPMPPSPSPSPNSSGNKGGAAGLGFIMWLGVSLVMM 176
>ENL3_ARATH (Q8LC95) Early nodulin-like protein 3 precursor
(Phytocyanin-like protein)
Length = 186
Score = 52.0 bits (123), Expect = 6e-07
Identities = 32/150 (21%), Positives = 65/150 (43%), Gaps = 14/150 (9%)
Query: 1 MASSRVVLILSISMVLLSSVAIAATDYIVGDDKGWTVDFDYTQ----WAQDKVFRVGDNL 56
MA +V + LL+++ + + G W + ++ WA+ FRVGD L
Sbjct: 1 MAQRTLVATFFLIFFLLTNLVCSKEIIVGGKTSSWKIPSSPSESLNKWAESLRFRVGDTL 60
Query: 57 VFNYDPSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQ 116
V+ YD + +V +V + +C S G ++L+ G +++ G +C +
Sbjct: 61 VWKYDEEKDSVLQVTKDAYINCNTTNPAANYSNGDTKVKLERSGPYFFISGSKSNCVEGE 120
Query: 117 MKLVITVLAEG----------APAPSPPPS 136
++ + + G +P+P+P P+
Sbjct: 121 KLHIVVMSSRGGHTGGFFTGSSPSPAPSPA 150
>ENL2_ARATH (Q9T076) Early nodulin-like protein 2 precursor
(Phytocyanin-like protein)
Length = 349
Score = 46.2 bits (108), Expect = 3e-05
Identities = 34/134 (25%), Positives = 59/134 (43%), Gaps = 6/134 (4%)
Query: 8 LILSISMVLLSSVAIAATDYIVGDDKGWTVD--FDYTQWAQDKVFRVGDNLVFNYDPSRH 65
++LS+S + S A + VG W + +Y W+ F V D L F+Y
Sbjct: 15 ILLSLSTLFTIS---NARKFNVGGSGAWVTNPPENYESWSGKNRFLVHDTLYFSYAKGAD 71
Query: 66 NVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVITVLA 125
+V +VN + +C + + G I L G +++ G D+C Q KL + V++
Sbjct: 72 SVLEVNKADYDACNTKNPIKRVDDGDSEISLDRYGPFYFISGNEDNCKKGQ-KLNVVVIS 130
Query: 126 EGAPAPSPPPSSDA 139
P+ + P + A
Sbjct: 131 ARIPSTAQSPHAAA 144
>N551_SOYBN (Q05544) Early nodulin 55-1 (N-55-1) (Fragment)
Length = 137
Score = 37.0 bits (84), Expect = 0.019
Identities = 24/90 (26%), Positives = 40/90 (43%), Gaps = 14/90 (15%)
Query: 60 YDPSRHNVFKVNGTLFQSCTFPPKNEAL-STGKDIIQLKTEGRKWYVCGVADHCSARQMK 118
YD +V +VN T ++ C K L + G + L G + ++ G HC +K
Sbjct: 2 YDERTESVHEVNETDYEQCNTVGKEHVLFNDGNTKVMLTKSGFRHFISGNQSHCQ-MGLK 60
Query: 119 LVITVLAE------------GAPAPSPPPS 136
L++ V++ +P+PSP PS
Sbjct: 61 LMVVVMSNNTKKKLIHSPSPSSPSPSPSPS 90
>HA13_MOUSE (P14426) H-2 class I histocompatibility antigen, D-K
alpha chain precursor (H-2D(K))
Length = 362
Score = 33.5 bits (75), Expect = 0.21
Identities = 23/91 (25%), Positives = 43/91 (46%), Gaps = 12/91 (13%)
Query: 79 TFPPKNEALSTGKDIIQLKTEG----RKWYVCGVADHCSARQMKLVITVLAEGAPAP--- 131
T+ E L+ ++++ + G +KW V ++ V EG P P
Sbjct: 240 TWQLNGEELTQDMELVETRPAGDGTFQKWASVVVP---LGKEQNYTCHVYHEGLPEPLTL 296
Query: 132 --SPPPSSDAHSVVSSLFGVVMAIMVAIAVI 160
PPPS+D++ V+ ++ GV+ A+ + AV+
Sbjct: 297 RWEPPPSTDSYMVIVAVLGVLGAVAIIGAVV 327
>HA1Z_MOUSE (P01896) H-2 class I histocompatibility antigen, alpha
chain (Clone PH-2D-2) (Fragment)
Length = 185
Score = 33.1 bits (74), Expect = 0.28
Identities = 23/91 (25%), Positives = 43/91 (46%), Gaps = 12/91 (13%)
Query: 79 TFPPKNEALSTGKDIIQLKTEG----RKWYVCGVADHCSARQMKLVITVLAEGAPAP--- 131
T+ E L+ ++++ + G +KW V ++ V EG P P
Sbjct: 50 TWQLNGEELTQDMELVETRPAGDGTFQKWASVVVP---LGKEQNYTCRVYHEGLPEPLTL 106
Query: 132 --SPPPSSDAHSVVSSLFGVVMAIMVAIAVI 160
PPPS+D++ V+ ++ GV+ A+ + AV+
Sbjct: 107 RWEPPPSTDSYMVIVAVLGVLGAMAIIGAVV 137
>HA1L_MOUSE (P01897) H-2 class I histocompatibility antigen, L-D
alpha chain precursor
Length = 362
Score = 33.1 bits (74), Expect = 0.28
Identities = 23/91 (25%), Positives = 43/91 (46%), Gaps = 12/91 (13%)
Query: 79 TFPPKNEALSTGKDIIQLKTEG----RKWYVCGVADHCSARQMKLVITVLAEGAPAP--- 131
T+ E L+ ++++ + G +KW V ++ V EG P P
Sbjct: 240 TWQLNGEELTQDMELVETRPAGDGTFQKWASVVVP---LGKEQNYTCRVYHEGLPEPLTL 296
Query: 132 --SPPPSSDAHSVVSSLFGVVMAIMVAIAVI 160
PPPS+D++ V+ ++ GV+ A+ + AV+
Sbjct: 297 RWEPPPSTDSYMVIVAVLGVLGAMAIIGAVV 327
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,053,706
Number of Sequences: 164201
Number of extensions: 725740
Number of successful extensions: 2517
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 2473
Number of HSP's gapped (non-prelim): 40
length of query: 162
length of database: 59,974,054
effective HSP length: 102
effective length of query: 60
effective length of database: 43,225,552
effective search space: 2593533120
effective search space used: 2593533120
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC126009.4


