

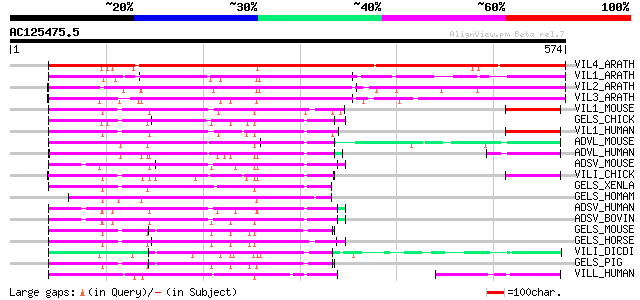
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125475.5 + phase: 0 /pseudo
(574 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VIL4_ARATH (O65570) Villin 4 647 0.0
VIL1_ARATH (O81643) Villin 1 350 6e-96
VIL2_ARATH (O81644) Villin 2 292 1e-78
VIL3_ARATH (O81645) Villin 3 290 5e-78
VIL1_MOUSE (Q62468) Villin 1 131 5e-30
GELS_CHICK (O93510) Gelsolin precursor (Actin-depolymerizing fac... 130 1e-29
VIL1_HUMAN (P09327) Villin 1 129 2e-29
ADVL_MOUSE (O88398) Advillin (p92) (Actin-binding protein DOC6) 125 2e-28
ADVL_HUMAN (O75366) Advillin (p92) 125 4e-28
ADSV_MOUSE (Q60604) Adseverin (Scinderin) (Gelsolin-like protein) 124 9e-28
VILI_CHICK (P02640) Villin 123 1e-27
GELS_XENLA (P14885) Gelsolin (Actin-depolymerizing factor) (ADF)... 122 3e-27
GELS_HOMAM (Q27319) Gelsolin, cytoplasmic (Actin-depolymerizing ... 122 3e-27
ADSV_HUMAN (Q9Y6U3) Adseverin (Scinderin) 122 3e-27
ADSV_BOVIN (Q28046) Adseverin (Scinderin) (SC) 122 3e-27
GELS_MOUSE (P13020) Gelsolin precursor, plasma (Actin-depolymeri... 121 6e-27
GELS_HORSE (Q28372) Gelsolin (Actin-depolymerizing factor) (ADF)... 120 7e-27
VILI_DICDI (P36418) Protovillin (100 kDa actin-binding protein) 120 1e-26
GELS_PIG (P20305) Gelsolin precursor, plasma (Actin-depolymerizi... 119 2e-26
VILL_HUMAN (O15195) Villin-like protein 116 1e-25
>VIL4_ARATH (O65570) Villin 4
Length = 974
Score = 647 bits (1670), Expect = 0.0
Identities = 356/584 (60%), Positives = 422/584 (71%), Gaps = 55/584 (9%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVE------- 93
QVWRVNGQ K LL A D SKFYSGDCY+FQYSYPGE++EE LIGTW GK SVE
Sbjct: 395 QVWRVNGQAKTLLQAADHSKFYSGDCYVFQYSYPGEEKEEVLIGTWFGKQSVEEERGSAV 454
Query: 94 SFTSKQ---------NGRINE------VYSIYGSYL*RQ*NNSILFHP----PKLDCF*D 134
S SK RI E + I S++ + S + ++D D
Sbjct: 455 SMASKMVESMKFVPAQARIYEGKEPIQFFVIMQSFIVFKGGISSGYKKYIAEKEVD---D 511
Query: 135 ETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQ 194
+TY E+GVALFRIQGSGPE+MQAIQV+ A+SLNSSY YIL ++S VFTW GNL+ + DQ
Sbjct: 512 DTYNENGVALFRIQGSGPENMQAIQVDPVAASLNSSYYYILHNDSSVFTWAGNLSTATDQ 571
Query: 195 ELAERMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAENDPHLFSCNFSE 254
ELAER LDLIKP+ Q R QKEG+E+EQFWELLG K EYSSQK+ +E E DPHLFSC F++
Sbjct: 572 ELAERQLDLIKPNQQSRAQKEGSESEQFWELLGGKAEYSSQKLTKEPERDPHLFSCTFTK 631
Query: 255 ------EIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLL 308
EI+NF+QDDLMTEDIFI+DCHS+IFVWVGQ+V PK ++ AL IGEKF+E+D LL
Sbjct: 632 EVLKVTEIYNFTQDDLMTEDIFIIDCHSEIFVWVGQEVVPKNKLLALTIGEKFIEKDSLL 691
Query: 309 ETISCSAPIYIVMEGSEPPFFTRFF-KWDSAKSAMLGNSYQRKLAIMKNGGTPPLVKPKR 367
E +S APIY++MEG EP FFTRFF WDS+KSAM GNS+QRKL I+KNGGTP KPKR
Sbjct: 692 EKLSPEAPIYVIMEGGEPSFFTRFFTSWDSSKSAMHGNSFQRKLKIVKNGGTPVADKPKR 751
Query: 368 RASVSYGGRSGGLPEKS-QRSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPP 426
R SYGGR+ +P+KS QRSRSMS SPDRVRVRGRSPAFNALAATFE+ N RNLSTPPP
Sbjct: 752 RTPASYGGRA-SVPDKSQQRSRSMSFSPDRVRVRGRSPAFNALAATFESQNARNLSTPPP 810
Query: 427 MIRKLYPKSKTPDLATL--APKSSAISHLTSTFEPPSAREKLIPRSLKDTSKS----NPE 480
++RKLYP+S TPD + APKSSAI+ ++ FE +E IP+ +K + K+ PE
Sbjct: 811 VVRKLYPRSVTPDSSKFAPAPKSSAIASRSALFEKIPPQEPSIPKPVKASPKTPESPAPE 870
Query: 481 TNS----------DNENSTGSREESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMP 530
+NS E S SR ESLTIQED EG ED E LP +PY+ +KT STDP+
Sbjct: 871 SNSKEQEEKKENDKEEGSMSSRIESLTIQEDAKEG-VEDEEDLPAHPYDRLKTTSTDPVS 929
Query: 531 DIDVTKREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
DIDVT+REAYLS EEF+E+ GMT+ FYKLPKWKQNK KMAVQL
Sbjct: 930 DIDVTRREAYLSSEEFKEKFGMTKEAFYKLPKWKQNKFKMAVQL 973
Score = 43.9 bits (102), Expect = 0.001
Identities = 75/357 (21%), Positives = 138/357 (38%), Gaps = 58/357 (16%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYI-FQYSYPGEDREEHLIGTWIGKNSVESFTSKQ 99
++WR+ + + KF++GD YI + + H I W+GK++ +
Sbjct: 22 EIWRIENFIPTPIPKSSIGKFFTGDSYIVLKTTALKTGALRHDIHYWLGKDTSQDEAGTA 81
Query: 100 NGRINEVYSIYGS----YL*RQ*NNSILFHPPKLDCF*DETYKEDGVA------------ 143
+ E+ + G Y Q + + F C + E GVA
Sbjct: 82 AVKTVELDAALGGRAVQYREVQGHETEKFLSYFKPCIIPQ---EGGVASGFKHVVAEEHI 138
Query: 144 --LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERML 201
LF +G + +V A SSLN YIL ++S +F + G+ ++ ++ A ++
Sbjct: 139 TRLFVCRGK--HVVHVKEVPFARSSLNHDDIYILDTKSKIFQFNGSNSSIQERAKALEVV 196
Query: 202 DLIKP---DLQCRPQ--KEG-----AETEQFWELLGVKTEYSSQKIVREAENDP------ 245
IK D C ++G A++ +FW G + R+ ND
Sbjct: 197 QYIKDTYHDGTCEVATVEDGKLMADADSGEFWGFFG-----GFAPLPRKTANDEDKTYNS 251
Query: 246 ---HLFSCNFSE----EIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIG 298
LF + E ++ L T +ILDC ++FVW+G+ R A
Sbjct: 252 DITRLFCVEKGQANPVEGDTLKREMLDTNKCYILDCGIEVFVWMGRTTSLDDRKIASKAA 311
Query: 299 EKFLEQDFLLETISCSAPIYIVMEGSEP-PFFTRFFKWDSAKSAMLGNSYQRKLAIM 354
E+ + + + + ++EG E PF ++F W + + + ++A +
Sbjct: 312 EEMIR-----SSERPKSQMIRIIEGFETVPFRSKFESWTQETNTTVSEDGRGRVAAL 363
>VIL1_ARATH (O81643) Villin 1
Length = 910
Score = 350 bits (898), Expect = 6e-96
Identities = 215/571 (37%), Positives = 307/571 (53%), Gaps = 91/571 (15%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVESF----- 95
+VWRV+G + +LL+ DQ+K ++GDCY+ QY Y ++R EHL+ WIG S++
Sbjct: 393 KVWRVDGDDVSLLSIPDQTKLFTGDCYLVQYKYTYKERTEHLLYVWIGCESIQQDRADAI 452
Query: 96 --------TSKQNGRINEVYS---------------IYGSYL*RQ*NNSILFHPPKLDCF 132
T+K + +Y ++ L R+ +L K+
Sbjct: 453 TNASAIVGTTKGESVLCHIYQGNEPSRFFPMFQSLVVFKGGLSRR-YKVLLAEKEKIG-- 509
Query: 133 *DETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSD 192
E Y E+ +LFR+ G+ P +MQAIQVN A+SLNSSY YILQ + FTW G L++
Sbjct: 510 --EEYNENKASLFRVVGTSPRNMQAIQVNLVATSLNSSYSYILQYGASAFTWIGKLSSDS 567
Query: 193 DQELAERMLDLIKPDLQCRPQ--KEGAETEQFWELLGVKTEYSSQKIVREAENDPHLFSC 250
D E+ +RML + D C+P +EG ET+ FW LLG K+EY +K +R+ +PHLF+C
Sbjct: 568 DHEVLDRMLYFL--DTSCQPTYIREGNETDTFWNLLGGKSEYPKEKEMRKQIEEPHLFTC 625
Query: 251 NFS------EEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQ 304
+ S +EI+NF QDDL TED+F+LDC S+++VW+G + K + +AL +G KFLE
Sbjct: 626 SCSSDVLKVKEIYNFVQDDLTTEDVFLLDCQSEVYVWIGSNSNIKSKEEALTLGLKFLEM 685
Query: 305 DFLLETISCSAPIYIVMEGSEPPFFTRFFKWDSAKSAMLGNSYQRKLAIMKNGGTPPLVK 364
D L E ++ P+Y+V EG EPPFFTRFF+W K+ M GNS++RKLA +K T
Sbjct: 686 DILEEGLTMRTPVYVVTEGHEPPFFTRFFEWVPEKANMHGNSFERKLASLKGKKT----S 741
Query: 365 PKRRASVSYGGRSGGLPEKSQRSRSMSVSPDRVRVRGRSP-AFNALAATFENSNVRNLST 423
KR + Y +S + +SRS+S + RG SP + L + ++ N S
Sbjct: 742 TKRSSGSQYRSQSKDNASRDLQSRSVSSNGSE---RGVSPCSSEKLLSLSSAEDMTNSSN 798
Query: 424 PPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETNS 483
P+++KL+ +S D P + S D SK P
Sbjct: 799 STPVVKKLFSESLVVD-------------------PNDGVARQESSSKSDISKQKPRVGI 839
Query: 484 DNENSTGSREESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMPDIDVTKREAYLSP 543
NS S ESL Y YE ++ DS P+ DID T+REAYL+
Sbjct: 840 ---NSDLSSLESL------------------AYSYEQLRVDSQKPVTDIDATRREAYLTE 878
Query: 544 EEFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
+EF+ER GM +SEFY LPKWKQNKLK+++ L
Sbjct: 879 KEFEERFGMAKSEFYALPKWKQNKLKISLHL 909
Score = 61.2 bits (147), Expect = 7e-09
Identities = 63/241 (26%), Positives = 108/241 (44%), Gaps = 30/241 (12%)
Query: 135 ETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQ 194
ETY+ V L R +G ++ +V SSLN +IL + S VF + G +++ ++
Sbjct: 134 ETYQ---VTLLRCKGD--HVVRVKEVPFLRSSLNHDDVFILDTASKVFLFAGCNSSTQEK 188
Query: 195 ELAERMLDLIKP---DLQCRPQ--KEG-----AETEQFWELLG--VKTEYSSQKIVREAE 242
A +++ IK D +C ++G ++ +FW G S +E
Sbjct: 189 AKAMEVVEYIKDNKHDGRCEVATIEDGKFSGDSDAGEFWSFFGGYAPIPKLSSSTTQEQT 248
Query: 243 NDP--HLFSCNFSEEIH-----NFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQAL 295
P LF + +H + +D L ++LDCHS++FVW+G+ R ++
Sbjct: 249 QTPCAELFWIDTKGNLHPTGTSSLDKDMLEKNKCYMLDCHSEVFVWMGRNTSLTERKTSI 308
Query: 296 PIGEKFLEQDFLLETISCSAPIYIVMEGSEPPFFTRFF-KWDSAK-SAMLGNSYQRKLAI 353
E+FL + E S + + ++ EG E F FF KW S + S N + K+A
Sbjct: 309 SSSEEFLRK----EGRSTTTSLVLLTEGLENARFRSFFNKWPSDRWSLAFYNEGREKVAA 364
Query: 354 M 354
+
Sbjct: 365 L 365
>VIL2_ARATH (O81644) Villin 2
Length = 976
Score = 292 bits (748), Expect = 1e-78
Identities = 191/593 (32%), Positives = 294/593 (49%), Gaps = 69/593 (11%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSY-PGEDREEHLIGTWIGKNSVESFTSKQ 99
+VW VNG+ K L D K YSGDCY+ Y+Y GE ++E+ + W GK S+ Q
Sbjct: 393 EVWYVNGKVKTPLPKEDIGKLYSGDCYLVLYTYHSGERKDEYFLSCWFGKKSIPE---DQ 449
Query: 100 NGRINEVYSIYGSYL*RQ*NNSILF--HPPKLDCF*------------------------ 133
+ I ++ S R I PP+
Sbjct: 450 DTAIRLANTMSNSLKGRPVQGRIYEGKEPPQFVALFQPMVVLKGGLSSGYKSSMGESEST 509
Query: 134 DETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDD 193
DETY + +AL ++ G+G + +A+QV + A+SLNS C++LQS + +F W+GN + +
Sbjct: 510 DETYTPESIALVQVSGTGVHNNKAVQVETVATSLNSYECFLLQSGTSMFLWHGNQSTHEQ 569
Query: 194 QELAERMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAENDPHLFSCNFS 253
ELA ++ + +KP + + KEG E+ FW LG K ++S+K E DPHLFS F+
Sbjct: 570 LELATKVAEFLKPGITLKHAKEGTESSTFWFALGGKQNFTSKKASSETIRDPHLFSFAFN 629
Query: 254 ------EEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFL 307
EEI+NF+QDDL+TEDI+ LD H+++FVWVGQ V+PK + IG+K+++
Sbjct: 630 RGKFQVEEIYNFAQDDLLTEDIYFLDTHAEVFVWVGQCVEPKEKQTVFEIGQKYIDLAGS 689
Query: 308 LETISCSAPIYIVMEGSEPPFFTRFFKWDSAKSAMLGNSYQRKLAIMKNGGTPPLVKPKR 367
LE + PIY + EG+EP FFT +F WD+ K+ + GNS+Q+K +++ GT +V+ K
Sbjct: 690 LEGLHPKVPIYKINEGNEPCFFTTYFSWDATKAIVQGNSFQKKASLL--FGTHHVVEDK- 746
Query: 368 RASVSYGGRSG-----------GLPEKSQRSRSMSVSPDRVR-----VRGRSPAFNALAA 411
S GG G S +R S DR+ R R+ A AL++
Sbjct: 747 ----SNGGNQGLRQRAEALAALNSAFNSSSNRPAYSSQDRLNESHDGPRQRAEALAALSS 802
Query: 412 TFENSNVRNLSTPPPM-IRKLYPKSKTPDLATLAP------KSSAISHLTSTFEPPSARE 464
F +S+ S PPP + + +A L+ K S + T + +
Sbjct: 803 AFNSSSSSTKSPPPPRPVGTSQASQRAAAVAALSQVLVAENKKSPDTSPTRRSTSSNPAD 862
Query: 465 KLIPRSLKDTSKSNPETN---SDNENSTGSREESLTIQEDVNEGEPEDNEGLPVYPYESV 521
+ KD +++ + E + + +E+ QE +G+ E + YE +
Sbjct: 863 DIPLTEAKDEEEASEVAGLEAKEEEEVSPAADETEAKQETEEQGDSEIQPSGATFTYEQL 922
Query: 522 KTDSTDPMPDIDVTKREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
+ S +P+ ID +REAYLS EEFQ G+ + F LP+WKQ+ LK L
Sbjct: 923 RAKSENPVTGIDFKRREAYLSEEEFQSVFGIEKEAFNNLPRWKQDLLKKKFDL 975
Score = 50.4 bits (119), Expect = 1e-05
Identities = 78/358 (21%), Positives = 138/358 (37%), Gaps = 49/358 (13%)
Query: 40 SQVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHL-IGTWIGKNSVESFTSK 98
+++WR+ E + ++ KFY GD YI + + I WIGK++ +
Sbjct: 19 TEIWRIENFEAVPVPKSEHGKFYMGDTYIVLQTTQNKGGAYLFDIHFWIGKDTSQDEAGT 78
Query: 99 QNGRINEVYSIYGSYL*R----Q*NNSILFHPPKLDCF*D---------ETYKEDGVALF 145
+ E+ ++ G + Q + S F C +T +E+
Sbjct: 79 AAVKTVELDAVLGGRAVQHREIQGHESDKFLSYFKPCIIPLEGGVASGFKTVEEEVFETR 138
Query: 146 RIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDLIK 205
G +++ QV A SSLN +IL +E ++ + G +N ++ A ++ +K
Sbjct: 139 LYTCKGKRAIRLKQVPFARSSLNHDDVFILDTEEKIYQFNGANSNIQERAKALEVVQYLK 198
Query: 206 PDLQ--------CRPQKEGAETEQ--FWELLGVKTEYSSQKIVREAENDPHLFSCNFSEE 255
K E++ FW L G I R+ ND + + +
Sbjct: 199 DKYHEGTCDVAIVDDGKLDTESDSGAFWVLFG-----GFAPIGRKVANDDDIVPESTPPK 253
Query: 256 IH------------NFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLE 303
++ + S+ L ++LDC ++I++WVG+ R A E+FL
Sbjct: 254 LYCITDGKMEPIDGDLSKSMLENTKCYLLDCGAEIYIWVGRVTQVDERKAASQSAEEFLA 313
Query: 304 QDFLLETISCSAPIYIVMEGSEP-PFFTRFFKWDSAKSAMLGNSYQR--KLAIMKNGG 358
E + + V++G E F + F W S SA GN R A++K G
Sbjct: 314 S----ENRPKATHVTRVIQGYESHSFKSNFDSWPSG-SATPGNEEGRGKVAALLKQQG 366
>VIL3_ARATH (O81645) Villin 3
Length = 966
Score = 290 bits (743), Expect = 5e-78
Identities = 189/594 (31%), Positives = 283/594 (46%), Gaps = 83/594 (13%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSY-PGEDREEHLIGTWIGKNS-------- 91
+VW ++ K +L+ K YSGDCY+ Y+Y GE +E++ + W GKNS
Sbjct: 395 EVWYIDANSKTVLSKDHVGKLYSGDCYLVLYTYHSGERKEDYFLCCWFGKNSNQEDQETA 454
Query: 92 ---VESFTSKQNGRINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*D-------------- 134
+ T+ GR + G PP+
Sbjct: 455 VRLASTMTNSLKGRPVQARIFEGK------------EPPQFVALFQHMVVLKGGLSSGYK 502
Query: 135 ----------ETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTW 184
ETY + +AL ++ G+G + +A+QV + A+SLNS C++LQS + +F W
Sbjct: 503 NSMTEKGSSGETYTPESIALIQVSGTGVHNNKALQVEAVATSLNSYDCFLLQSGTSMFLW 562
Query: 185 YGNLTNSDDQELAERMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAEND 244
GN + + QELA ++ + +KP + KEG E+ FW LG K ++S+K+ E D
Sbjct: 563 VGNHSTHEQQELAAKVAEFLKPGTTIKHAKEGTESSSFWFALGGKQNFTSKKVSSETVRD 622
Query: 245 PHLFSCNFS------EEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIG 298
PHLFS +F+ EEIHNF QDDL+TE++ +LD H+++FVWVGQ VDPK + A IG
Sbjct: 623 PHLFSFSFNRGKFQVEEIHNFDQDDLLTEEMHLLDTHAEVFVWVGQCVDPKEKQTAFEIG 682
Query: 299 EKFLEQDFLLETISCSAPIYIVMEGSEPPFFTRFFKWDSAKSAMLGNSYQRKLAIMKNGG 358
++++ LE +S P+Y + EG+EP FFT +F WDS K+ + GNSYQ+K A++ G
Sbjct: 683 QRYINLAGSLEGLSPKVPLYKITEGNEPCFFTTYFSWDSTKATVQGNSYQKKAALLL--G 740
Query: 359 TPPLVK---------PKRRASVSYGGRSGGLPEKSQRSRSMSVSPDRVRVRG-------R 402
T +V+ P++RA+ L S + SP R R G R
Sbjct: 741 THHVVEDQSSSGNQGPRQRAAA-----LAALTSAFNSSSGRTSSPSRDRSNGSQGGPRQR 795
Query: 403 SPAFNALAATFENSNVRNLSTPPPMIRKLYPKSKTPDLATLAPKSSAIS--HLTSTFEPP 460
+ A AL + F +S S+ P R + A +A S ++ S P
Sbjct: 796 AEALAALTSAFNSSP----SSKSPPRRSGLTSQASQRAAAVAALSQVLTAEKKKSPDTSP 851
Query: 461 SAREKLIPRSLKDTSKSNPETNSDNENSTGSREESLTIQEDVNEGEPEDNEGLPVYPYES 520
SA K S + + T + E E+ + + + E + YE
Sbjct: 852 SAEAKDEETSFSEVEATEEATEAKEEEEVSPAAEASAEEAKPKQDDSEVETTGVTFTYER 911
Query: 521 VKTDSTDPMPDIDVTKREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
++ S P+ ID +REAYLS EF+ GM + FYKLP WKQ+ LK L
Sbjct: 912 LQAKSEKPVTGIDFKRREAYLSEVEFKTVFGMEKESFYKLPGWKQDLLKKKFNL 965
Score = 57.4 bits (137), Expect = 1e-07
Identities = 76/349 (21%), Positives = 137/349 (38%), Gaps = 40/349 (11%)
Query: 40 SQVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHL-IGTWIGKNSVESFTSK 98
+++WR+ E + ++ KFY GD YI + + I WIGK++ +
Sbjct: 21 TEIWRIENFEPVPVPKSEHGKFYMGDTYIVLQTTQNKGGAYLFDIHFWIGKDTSQDEAGT 80
Query: 99 QNGRINEVYSIYGS----YL*RQ*NNSILFHPPKLDC-----------F*DETYKEDGVA 143
+ E+ + G Y Q + S F C F +E
Sbjct: 81 AAVKTVELDAALGGRAVQYREIQGHESDKFLSYFKPCIIPLEGGVASGFKKPEEEEFETR 140
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDL 203
L+ +G ++ QV A SSLN +IL ++ ++ + G +N ++ A ++
Sbjct: 141 LYTCKGK--RAVHLKQVPFARSSLNHDDVFILDTKEKIYQFNGANSNIQERAKALVVIQY 198
Query: 204 IKPDLQCRPQKEG----------AETEQFWELLG----VKTEYSSQKIVREAENDPHLFS 249
+K +++ +FW L G + + +S+ + P L+S
Sbjct: 199 LKDKFHEGTSDVAIVDDGKLDTESDSGEFWVLFGGFAPIARKVASEDEIIPETTPPKLYS 258
Query: 250 CNFS--EEIH-NFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDF 306
E I + S+ L ++LDC S+IF+WVG+ + R A+ E F+
Sbjct: 259 IADGQVESIDGDLSKSMLENNKCYLLDCGSEIFIWVGRVTQVEERKTAIQAAEDFVAS-- 316
Query: 307 LLETISCSAPIYIVMEGSEP-PFFTRFFKWDSAKSAMLGNSYQRKLAIM 354
E + I V++G EP F + F W S + + K+A +
Sbjct: 317 --ENRPKATRITRVIQGYEPHSFKSNFDSWPSGSATPANEEGRGKVAAL 363
Score = 36.2 bits (82), Expect = 0.24
Identities = 41/194 (21%), Positives = 74/194 (38%), Gaps = 42/194 (21%)
Query: 147 IQGSGPESMQAIQVNSAA-------SSLNSSYCYIL---------QSESVVFTWYGNLTN 190
++G G + I NS L S CY++ + + + W+G +N
Sbjct: 388 LEGGGKLEVWYIDANSKTVLSKDHVGKLYSGDCYLVLYTYHSGERKEDYFLCCWFGKNSN 447
Query: 191 SDDQELAERMLDLIKPDLQCRPQK----EGAETEQFWELL--------GVKTEYSSQKIV 238
+DQE A R+ + L+ RP + EG E QF L G+ + Y +
Sbjct: 448 QEDQETAVRLASTMTNSLKGRPVQARIFEGKEPPQFVALFQHMVVLKGGLSSGYKNSMTE 507
Query: 239 REAENDPH------LFSCNFSEEIHNFS-------QDDLMTEDIFILDCHSQIFVWVGQQ 285
+ + + + L + +HN L + D F+L + +F+WVG
Sbjct: 508 KGSSGETYTPESIALIQVS-GTGVHNNKALQVEAVATSLNSYDCFLLQSGTSMFLWVGNH 566
Query: 286 VDPKRRVQALPIGE 299
+++ A + E
Sbjct: 567 STHEQQELAAKVAE 580
>VIL1_MOUSE (Q62468) Villin 1
Length = 826
Score = 131 bits (329), Expect = 5e-30
Identities = 97/349 (27%), Positives = 169/349 (47%), Gaps = 57/349 (16%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVES------ 94
QVWR+ E + + FY GDCY+ Y+Y +++ +L+ W G + +
Sbjct: 400 QVWRIEDLELVPVESKWLGHFYGGDCYLLLYTYLIGEKQHYLLYIWQGSQASQDEIAASA 459
Query: 95 -----FTSKQNGRINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDGVA------ 143
K N ++ G ++ + + ++ + T +++ +
Sbjct: 460 YQAVLLDQKYNDEPVQIRVTMG----KEPPHLMSIFKGRMVVYQGGTSRKNNLEPVPSTR 515
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDL 203
LF+++G+ ++ +A +V + A+SLNS+ +IL++ S + W G + D++E+A+ + D
Sbjct: 516 LFQVRGTNADNTKAFEVTARATSLNSNDVFILKTPSCCYLWCGKGCSGDEREMAKMVADT 575
Query: 204 IKPDLQCRPQK----EGAETEQFWELLGVKTEYSSQKIVREAEN--DPHLFSCN------ 251
I R +K EG E FW LG K Y++ K ++E P LF C+
Sbjct: 576 IS-----RTEKQVVVEGQEPANFWMALGGKAPYANTKRLQEENQVITPRLFECSNQTGRF 630
Query: 252 FSEEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQ---DFLL 308
+ EI +F+QDDL ED+F+LD Q+F W+G+ + + + A +++L+ + L
Sbjct: 631 LATEIFDFNQDDLEEEDVFLLDVWDQVFFWIGKHANEEEKKAAATTVQEYLKTHPGNRDL 690
Query: 309 ETISCSAPIYIVMEGSEPPFFTRF------FKWDSAKS-----AMLGNS 346
ET PI +V +G EPP FT + FKW + KS A LGNS
Sbjct: 691 ET-----PIIVVKQGHEPPTFTGWFLAWDPFKWSNTKSYDDLKAELGNS 734
Score = 52.8 bits (125), Expect = 2e-06
Identities = 23/58 (39%), Positives = 37/58 (63%), Gaps = 1/58 (1%)
Query: 513 LPVYPYESVKTDSTDPMPD-IDVTKREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLK 569
LP +P E + S + +P+ +D +++E +LS E+F LGMT + F LP+WKQ +K
Sbjct: 763 LPTFPLEQLVNKSVEDLPEGVDPSRKEEHLSTEDFTRALGMTPAAFSALPRWKQQNIK 820
Score = 39.7 bits (91), Expect = 0.022
Identities = 70/350 (20%), Positives = 130/350 (37%), Gaps = 75/350 (21%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVES------ 94
Q+WR+ + + ++ F+ GDCY+ + + I WIG++S +
Sbjct: 19 QIWRIEAMQMVPVPSSTFGSFFDGDCYVVLAIHKTSSTLSYDIHYWIGQDSSQDEQGAAA 78
Query: 95 -FTSKQN--------------GRINEVYSIY---GSYL*RQ*NNSILFHPPKLDCF*DET 136
+T++ + G +E + Y G + + S + H C
Sbjct: 79 IYTTQMDDYLKGRAVQHREVQGNESETFRSYFKQGLVIRKGGVASGMKHVETNSC----- 133
Query: 137 YKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQEL 196
D L ++G ++ A +V + S N ++L ++ W G +N ++
Sbjct: 134 ---DVQRLLHVKGK--RNVLAGEVEMSWKSFNRGDVFLLDLGKLIIQWNGPESNRMERL- 187
Query: 197 AERMLDLIKPDLQCRPQKEGAET-------------EQFWELLG--------VKTEYSSQ 235
R + L K + R Q+ G T Q ++ +K S
Sbjct: 188 --RGMPLAK---EIRDQERGGRTYVGVVDGEKEGDSPQLMAIMNHVLGPRKELKAAISDS 242
Query: 236 KIVREAENDPHLFSCNFSE--------EIHNFSQDDLMTEDIFILDCHS-QIFVWVGQQV 286
+ A+ L+ + SE +QD L ED +ILD +IFVW G+
Sbjct: 243 VVEPAAKAALKLYHVSDSEGKLVVREVATRPLTQDLLKHEDCYILDQGGLKIFVWKGKNA 302
Query: 287 DPKRRVQALPIGEKFLEQDFLLETISCSAPIYIVMEGSEPPFFTRFF-KW 335
+ + R A+ F++ + S + + +G+E P F + F KW
Sbjct: 303 NAQERSGAMSQALNFIK----AKQYPPSTQVEVQNDGAESPIFQQLFQKW 348
Score = 33.5 bits (75), Expect = 1.5
Identities = 41/195 (21%), Positives = 81/195 (41%), Gaps = 41/195 (21%)
Query: 141 GVALFRIQGSGPESMQAIQVNSAA-SSLNSSYCYIL----QSESV----VFTWYGNLTNS 191
G+ ++RI E+MQ + V S+ S CY++ ++ S + W G ++
Sbjct: 17 GIQIWRI-----EAMQMVPVPSSTFGSFFDGDCYVVLAIHKTSSTLSYDIHYWIGQDSSQ 71
Query: 192 DDQELAE----RMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVRE---AEND 244
D+Q A +M D +K + +G E+E F ++ + ++R+ A
Sbjct: 72 DEQGAAAIYTTQMDDYLKGRAVQHREVQGNESETF------RSYFKQGLVIRKGGVASGM 125
Query: 245 PHLF--SCNFSEEIH------------NFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKR 290
H+ SC+ +H S D+F+LD I W G + +
Sbjct: 126 KHVETNSCDVQRLLHVKGKRNVLAGEVEMSWKSFNRGDVFLLDLGKLIIQWNGPESNRME 185
Query: 291 RVQALPIGEKFLEQD 305
R++ +P+ ++ +Q+
Sbjct: 186 RLRGMPLAKEIRDQE 200
>GELS_CHICK (O93510) Gelsolin precursor (Actin-depolymerizing
factor) (ADF) (Brevin) (Homogenin)
Length = 778
Score = 130 bits (326), Expect = 1e-29
Identities = 92/326 (28%), Positives = 158/326 (48%), Gaps = 39/326 (11%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVE------S 94
Q+WR+ G EK + +FY GD YI Y Y ++ +I TW G +S + +
Sbjct: 444 QIWRIEGSEKVPVDPATYGQFYGGDSYIILYDYRHAGKQGQIIYTWQGAHSTQDEIATSA 503
Query: 95 FTSKQ-----NGRINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDG------VA 143
F + Q G + + G + +F L + T +E G
Sbjct: 504 FLTVQLDEELGGSPVQKRVVQGK---EPPHLMSMFGGKPLIVYKGGTSREGGQTTPAQTR 560
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDL 203
LF+++ S + +A++++ AAS LNS+ ++L++ S + W G +NS + A+ +L +
Sbjct: 561 LFQVRSSTSGATRAVELDPAASQLNSNDAFVLKTPSAAYLWVGRGSNSAELSGAQELLKV 620
Query: 204 IKPDLQCRPQK--EGAETEQFWELLGVKTEYSSQKIVREAEND---PHLFSCN------- 251
L RP + EG E + FW LG K Y + +++ + D P LF+C+
Sbjct: 621 ----LGARPVQVSEGREPDNFWVALGGKAPYRTSPRLKDKKMDAYPPRLFACSNKSGRFT 676
Query: 252 FSEEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETI 311
E + +QDDL T+D+ ILD Q+FVW+G+ + + +AL ++++E D +
Sbjct: 677 IEEVPGDLTQDDLATDDVMILDTWDQVFVWIGKDAQEEEKTEALKSAKRYIETD--PASR 734
Query: 312 SCSAPIYIVMEGSEPPFFTRFF-KWD 336
P+ +V +G EPP F+ +F WD
Sbjct: 735 DKRTPVTLVKQGLEPPTFSGWFLGWD 760
Score = 68.2 bits (165), Expect = 6e-11
Identities = 58/224 (25%), Positives = 104/224 (45%), Gaps = 27/224 (12%)
Query: 147 IQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDLIKP 206
+Q G +++A +V + S N+ C+IL S ++ W G+ +N ++ A + I+
Sbjct: 186 LQVKGRRTVRATEVPVSWESFNTGDCFILDLGSNIYQWCGSNSNRQERLKATVLAKGIRD 245
Query: 207 D-----LQCRPQKEGAETEQFWELLG------------VKTEYSSQKIVR--EAENDPHL 247
+ + +EGAE E+ ++LG KT+ +++K+ + + N
Sbjct: 246 NEKNGRAKVFVSEEGAEREEMLQVLGPKPSLPQGASDDTKTDTANRKLAKLYKVSNGAGN 305
Query: 248 FSCNFSEEIHNFSQDDLMTEDIFILD--CHSQIFVWVGQQVDPKRRVQALPIGEKFLEQD 305
+ + + + FSQ L TED FILD +IFVW G+ + R AL F+++
Sbjct: 306 MAVSLVADENPFSQAALNTEDCFILDHGTDGKIFVWKGRSANSDERKAALKTATDFIDK- 364
Query: 306 FLLETISCSAPIYIVMEGSEPPFFTRFFK-W-DSAKSAMLGNSY 347
+ ++ E E P F +FFK W D ++ LG +Y
Sbjct: 365 ---MGYPKHTQVQVLPESGETPLFKQFFKNWRDKDQTEGLGEAY 405
>VIL1_HUMAN (P09327) Villin 1
Length = 826
Score = 129 bits (324), Expect = 2e-29
Identities = 91/336 (27%), Positives = 162/336 (48%), Gaps = 48/336 (14%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVES------ 94
QVWR+ E + + FY GDCY+ Y+Y +++ +L+ W G + +
Sbjct: 400 QVWRIENLELVPVDSKWLGHFYGGDCYLLLYTYLIGEKQHYLLYVWQGSQASQDEITASA 459
Query: 95 -----FTSKQNGRINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDGVA------ 143
K NG ++ G ++ + + ++ + T + + +
Sbjct: 460 YQAVILDQKYNGEPVQIRVPMG----KEPPHLMSIFKGRMVVYQGGTSRTNNLETGPSTR 515
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDL 203
LF++QG+G + +A +V + A+ LNS+ ++L+++S + W G + D++E+A+ + D
Sbjct: 516 LFQVQGTGANNTKAFEVPARANFLNSNDVFVLKTQSCCYLWCGKGCSGDEREMAKMVADT 575
Query: 204 IKPDLQCRPQK----EGAETEQFWELLGVKTEYSSQKIVREAEN---DPHLFSCN----- 251
I R +K EG E FW LG K Y++ K ++E EN P LF C+
Sbjct: 576 IS-----RTEKQVVVEGQEPANFWMALGGKAPYANTKRLQE-ENLVITPRLFECSNKTGR 629
Query: 252 -FSEEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLET 310
+ EI +F+QDDL +D+F+LD Q+F W+G+ + + + A +++L+
Sbjct: 630 FLATEIPDFNQDDLEEDDVFLLDVWDQVFFWIGKHANEEEKKAAATTAQEYLKTH--PSG 687
Query: 311 ISCSAPIYIVMEGSEPPFFTRF------FKWDSAKS 340
PI +V +G EPP FT + FKW + KS
Sbjct: 688 RDPETPIIVVKQGHEPPTFTGWFLAWDPFKWSNTKS 723
Score = 51.2 bits (121), Expect = 7e-06
Identities = 22/58 (37%), Positives = 37/58 (62%), Gaps = 1/58 (1%)
Query: 513 LPVYPYESVKTDSTDPMPD-IDVTKREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLK 569
LP++P E + + +P+ +D +++E +LS E+F + GMT + F LP+WKQ LK
Sbjct: 763 LPIFPLEQLVNKPVEELPEGVDPSRKEEHLSIEDFTQAFGMTPAAFSALPRWKQQNLK 820
Score = 33.1 bits (74), Expect = 2.0
Identities = 14/51 (27%), Positives = 25/51 (48%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNS 91
Q+WR+ + + ++ F+ GDCYI + + I WIG++S
Sbjct: 19 QIWRIEAMQMVPVPSSTFGSFFDGDCYIILAIHKTASSLSYDIHYWIGQDS 69
>ADVL_MOUSE (O88398) Advillin (p92) (Actin-binding protein DOC6)
Length = 819
Score = 125 bits (315), Expect = 2e-28
Identities = 84/318 (26%), Positives = 158/318 (49%), Gaps = 25/318 (7%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVESFTSKQN 100
+VWR+ E + FY GDCY+ Y+Y + +++ W G+++ +
Sbjct: 398 EVWRIENLELVPVEYQWHGFFYGGDCYLVLYTYDVNGKPHYILYIWQGRHASRDELAASA 457
Query: 101 GRINEVYSIYGSY-------L*RQ*NNSILFHPPKLDCF*DETYKEDG------VALFRI 147
R EV + + ++ + + KL + T ++ V LF+I
Sbjct: 458 YRAVEVDQQFDRAPVQVRVSMGKEPRHFMAIFKGKLVIYEGGTSRKGNEEPDPPVRLFQI 517
Query: 148 QGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDLIKPD 207
G+ + +A++V+++ASSL S+ ++L++++ + WYG ++ D++ +A+ ++DL+ D
Sbjct: 518 HGNDKSNTKAVEVSASASSLISNDVFLLRTQAEHYLWYGKGSSGDERAMAKELVDLL-CD 576
Query: 208 LQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAEND--PHLFSCN------FSEEIHNF 259
EG E +FW+LLG KT Y++ K +++ D LF C+ E+ +F
Sbjct: 577 GNADTVAEGQEPPEFWDLLGGKTAYANDKRLQQETLDVQVRLFECSNKTGRFLVTEVTDF 636
Query: 260 SQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISCSAPIYI 319
+Q+DL D+ +LD Q+F+W+G + + + AL +++L PI I
Sbjct: 637 TQEDLSPGDVMLLDTWDQVFLWIGAEANATEKKGALSTAQEYLVTH--PSGRDPDTPILI 694
Query: 320 VMEGSEPPFFTRFF-KWD 336
+ +G EPP FT +F WD
Sbjct: 695 IKQGFEPPTFTGWFLAWD 712
Score = 49.3 bits (116), Expect = 3e-05
Identities = 70/330 (21%), Positives = 120/330 (36%), Gaps = 69/330 (20%)
Query: 260 SQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISCSAPIYI 319
S L++ D+F+L ++ ++W G+ R A + + + C
Sbjct: 533 SASSLISNDVFLLRTQAEHYLWYGKGSSGDERAMAKELVD-----------LLCDGNADT 581
Query: 320 VMEGSEPPFFTRFFKWDSAKSAMLGNSYQRKLAIMKNGGTPPLVKPKRRASVSYGGRSGG 379
V EG EPP F WD GG KR + +
Sbjct: 582 VAEGQEPPEF-----WDLL------------------GGKTAYANDKRLQQETLDVQVRL 618
Query: 380 LPEKSQRSRSMSVSPDRVRVRGRSPAFNALAATFE----------NSNVRN--LSTPPPM 427
++ R + SP L T++ N+ + LST
Sbjct: 619 FECSNKTGRFLVTEVTDFTQEDLSPGDVMLLDTWDQVFLWIGAEANATEKKGALSTAQEY 678
Query: 428 IRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRS--LKDTSKSNPETNSDN 485
+ +P + PD L K FEPP+ + + KS + ++
Sbjct: 679 L-VTHPSGRDPDTPILIIKQG--------FEPPTFTGWFLAWDPHIWSEGKSYEQLKNEL 729
Query: 486 ENSTG-----SREESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMP-DIDVTKREA 539
++T + ++ T+ + ++GEP+ YP E + +P D+D K+E
Sbjct: 730 GDATAIVRITADMKNATLYLNPSDGEPK------YYPVEVLLKGQNQELPEDVDPAKKEN 783
Query: 540 YLSPEEFQERLGMTRSEFYKLPKWKQNKLK 569
YLS ++F G+TR +F LP WK+ +LK
Sbjct: 784 YLSEQDFVSVFGITRGQFTALPGWKRLQLK 813
Score = 40.4 bits (93), Expect = 0.013
Identities = 75/352 (21%), Positives = 133/352 (37%), Gaps = 66/352 (18%)
Query: 43 WRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVESFTSKQNGR 102
WR+ E L+ + FY GDCYI + I WIGK+S + S
Sbjct: 19 WRIEKMELALVPLSAHGNFYEGDCYIVLSTRRVGSLLSQNIHFWIGKDSSQDEQSCAAIY 78
Query: 103 INEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DE-----TYKEDGVA-------------- 143
++ G + + + +H + D F YK+ GVA
Sbjct: 79 TTQLDDYLGGSPVQ--HREVQYH--ESDTFRGYFKQGIIYKKGGVASGMKHVETNTYDVK 134
Query: 144 -LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLD 202
L ++G ++QA +V + S N ++L V+ W G +NS ++ A
Sbjct: 135 RLLHVKGK--RNIQATEVEMSWDSFNRGDVFLLDLGMVIIQWNGPESNSGERLKAM---- 188
Query: 203 LIKPDLQCRPQKEGAE---------------TEQFWELLG----VKTEYSSQKIVREAEN 243
L+ D++ R + AE + LG +K S + + ++ ++
Sbjct: 189 LLAKDIRDRERGGRAEIGVIEGDKEAASPGLMTVLQDTLGRRSMIKPAVSDEIMDQQQKS 248
Query: 244 DPHLFSCNFSEEIHNFS----------QDDLMTEDIFILD-CHSQIFVWVGQQVDPKRRV 292
L+ + S+ S QD L +D +ILD ++I+VW G+ +
Sbjct: 249 SIMLY--HVSDTAGQLSVTEVATRPLVQDLLNHDDCYILDQSGTKIYVWKGKGATKVEKQ 306
Query: 293 QALPIGEKFLEQDFLLETISCSAPIYIVMEGSEPPFFTRFFKWDSAKSAMLG 344
A+ F++ ++ S + V +G+E F + F+ S K G
Sbjct: 307 AAMSKALDFIK----MKGYPSSTNVETVNDGAESAMFKQLFQKWSVKDQTTG 354
>ADVL_HUMAN (O75366) Advillin (p92)
Length = 819
Score = 125 bits (313), Expect = 4e-28
Identities = 86/318 (27%), Positives = 154/318 (48%), Gaps = 25/318 (7%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVESFTSKQN 100
+VWR+ E + FY GDCY+ Y+Y + H++ W +++ + +
Sbjct: 398 EVWRIENLELVPVEYQWYGFFYGGDCYLVLYTYEVNGKPHHILYIWQDRHASQDELAASA 457
Query: 101 GRINEVYSIYGSY-------L*RQ*NNSILFHPPKLDCF*DETYKEDG------VALFRI 147
+ EV + + + + + KL F T ++ V LF+I
Sbjct: 458 YQAVEVDRQFDGAAVQVRVRMGTEPRHFMAIFKGKLVIFEGGTSRKGNAEPDPPVRLFQI 517
Query: 148 QGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDLIKPD 207
G+ + +A++V + ASSLNS+ ++L++++ + WYG ++ D++ +A+ + L+ D
Sbjct: 518 HGNDKSNTKAVEVPAFASSLNSNDVFLLRTQAEHYLWYGKGSSGDERAMAKELASLL-CD 576
Query: 208 LQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAEND--PHLFSCN------FSEEIHNF 259
EG E +FW+LLG KT Y++ K +++ D LF C+ EI +F
Sbjct: 577 GSENTVAEGQEPAEFWDLLGGKTPYANDKRLQQEILDVQSRLFECSNKTGQFVVTEITDF 636
Query: 260 SQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISCSAPIYI 319
+QDDL D+ +LD Q+F+W+G + + + AL +++L PI I
Sbjct: 637 TQDDLNPTDVMLLDTWDQVFLWIGAEANATEKESALATAQQYLHTH--PSGRDPDTPILI 694
Query: 320 VMEGSEPPFFTRFF-KWD 336
+ +G EPP FT +F WD
Sbjct: 695 IKQGFEPPIFTGWFLAWD 712
Score = 45.4 bits (106), Expect = 4e-04
Identities = 75/351 (21%), Positives = 135/351 (38%), Gaps = 62/351 (17%)
Query: 42 VWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVESFTSKQNG 101
VWR+ E L+ + FY GDCY+ + I WIGK+S + S
Sbjct: 18 VWRIEKMELALVPVSAHGNFYEGDCYVILSTRRVASLLSQDIHFWIGKDSSQDEQSCAAI 77
Query: 102 RINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DE-----TYKEDGVA------------- 143
++ G + + + +H + D F YK+ GVA
Sbjct: 78 YTTQLDDYLGGSPVQ--HREVQYH--ESDTFRGYFKQGIIYKQGGVASGMKHVETNTYDV 133
Query: 144 --LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERML 201
L ++G +++A +V + S N ++L V+ W G +NS ++ A
Sbjct: 134 KRLLHVKGK--RNIRATEVEMSWDSFNRGDVFLLDLGKVIIQWNGPESNSGERLKAM--- 188
Query: 202 DLIKPDLQCRPQ------------KEGAETE---QFWELLG----VKTEYSSQKIVREAE 242
L+ D++ R + KE A E + LG +K + I ++ +
Sbjct: 189 -LLAKDIRDRERGGRAEIGVIEGDKEAASPELMKVLQDTLGRRSIIKPTVPDEIIDQKQK 247
Query: 243 NDPHLFSCNFSE--------EIHNFSQDDLMTEDIFILD-CHSQIFVWVGQQVDPKRRVQ 293
+ L+ + S QD L +D +ILD ++I+VW G+ +
Sbjct: 248 STIMLYHISDSAGQLAVTEVATRPLVQDLLNHDDCYILDQSGTKIYVWKGKGATKAEKQA 307
Query: 294 ALPIGEKFLEQDFLLETISCSAPIYIVMEGSEPPFFTRFFKWDSAKSAMLG 344
A+ F++ +++ S + V +G+E F + F+ S K +G
Sbjct: 308 AMSKALGFIK----MKSYPSSTNVETVNDGAESAMFKQLFQKWSVKDQTMG 354
Score = 45.4 bits (106), Expect = 4e-04
Identities = 24/77 (31%), Positives = 42/77 (54%), Gaps = 7/77 (9%)
Query: 494 ESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMP-DIDVTKREAYLSPEEFQERLGM 552
++ T+ + N+ EP+ YP + + +P D++ K+E YLS ++F G+
Sbjct: 743 KNATLSLNSNDSEPK------YYPIAVLLKNQNQELPEDVNPAKKENYLSEQDFVSVFGI 796
Query: 553 TRSEFYKLPKWKQNKLK 569
TR +F LP WKQ ++K
Sbjct: 797 TRGQFAALPGWKQLQMK 813
>ADSV_MOUSE (Q60604) Adseverin (Scinderin) (Gelsolin-like protein)
Length = 715
Score = 124 bits (310), Expect = 9e-28
Identities = 89/326 (27%), Positives = 161/326 (49%), Gaps = 37/326 (11%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNS------VES 94
++WRV + + + +FY GDCYI Y+YP +I TW G N+ + +
Sbjct: 399 EIWRVENSGRVQIDPSSYGEFYGGDCYIILYTYP----RGQIIYTWQGANATRDELTMSA 454
Query: 95 FTSKQNGRI--NEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDGVA------LFR 146
F + Q R + + S + LF L + + T K++G A LF+
Sbjct: 455 FLTVQLDRSLGGQAVQVRVSQGKEPAHLLSLFKDKPLIIYKNGTSKKEGQAPAPPTRLFQ 514
Query: 147 IQGSGPESMQAIQVNSAASSLNSSYCYILQ-SESVVFTWYGNLTNSDDQELAERMLDLIK 205
++ + + ++V+ A+SLNS+ ++L+ + F W G + ++++ AE + D++K
Sbjct: 515 VRRNLASITRIVEVDVDANSLNSNDTFVLKLPRNNGFIWIGKGASQEEEKGAEYVADVLK 574
Query: 206 PDLQCRPQK--EGAETEQFWELLGVKTEYSSQKIV--REAENDPHLFSCN-------FSE 254
C+ + EG E E+FW LG + +Y + ++ R ++ P L+ C+ E
Sbjct: 575 ----CKASRIQEGKEPEEFWNSLGGRGDYQTSPLLETRAEDHPPRLYGCSNKTGRFIIEE 630
Query: 255 EIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISCS 314
F+QDDL +D+ +LD QIF+W+G+ + + +++ + +LE D
Sbjct: 631 VPGEFTQDDLAEDDVMLLDAWEQIFIWIGKDANEVEKKESVKSAKMYLETD--PSGRDKR 688
Query: 315 APIYIVMEGSEPPFFTRFF-KWDSAK 339
PI I+ +G EPP FT +F WDS++
Sbjct: 689 TPIVIIKQGHEPPTFTGWFLGWDSSR 714
Score = 66.2 bits (160), Expect = 2e-10
Identities = 59/221 (26%), Positives = 100/221 (44%), Gaps = 28/221 (12%)
Query: 151 GPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDLIKPD--- 207
G ++A +V + S N C+I+ + ++ W G+ N ++ A ++ I+ +
Sbjct: 144 GRRVVRATEVPLSWESFNKGDCFIIDLGTEIYQWCGSSCNKYERLKASQVAIGIRDNERK 203
Query: 208 --LQCRPQKEGAETEQFWELLGVKTEY----SSQKIVREAENDPHLFSCNFSE------- 254
Q +EG+E + ++LG K E + +V + N S+
Sbjct: 204 GRSQLIVVEEGSEPSELMKVLGRKPELPDGDNDDDVVADISNRKMAKLYMVSDASGSMKV 263
Query: 255 ----EIHNFSQDDLMTEDIFILD--CHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLL 308
E + FS L++E+ FILD QIFVW G+ +P+ R A+ E+FL++
Sbjct: 264 TLVAEENPFSMGMLLSEECFILDHGAAKQIFVWKGKNANPQERKTAMKTAEEFLQK---- 319
Query: 309 ETISCSAPIYIVMEGSEPPFFTRFFK-W-DSAKSAMLGNSY 347
S + I ++ EG E P F +FFK W D +S G Y
Sbjct: 320 MKYSTNTQIQVLPEGGETPIFKQFFKDWKDKDQSDGFGKVY 360
>VILI_CHICK (P02640) Villin
Length = 826
Score = 123 bits (309), Expect = 1e-27
Identities = 89/324 (27%), Positives = 152/324 (46%), Gaps = 35/324 (10%)
Query: 40 SQVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVES----- 94
++VWRV QE + FY GDCY+ Y+Y + +I W G+++
Sbjct: 400 AEVWRVENQELVPVEKRWLGHFYGGDCYLVLYTYYVGPKVNRIIYIWQGRHASTDELAAS 459
Query: 95 ------FTSKQNGRINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDGVA----- 143
K N +V G ++ + + K+ + + + + G
Sbjct: 460 AYQAVFLDQKYNNEPVQVRVTMG----KEPAHLMAIFKGKMVVYENGSSRAGGTEPASST 515
Query: 144 -LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLD 202
LF + G+ + +A +V A+SLNS+ ++L++ S + WYG + D++E+ + + D
Sbjct: 516 RLFHVHGTNEYNTKAFEVPVRAASLNSNDVFVLKTPSSCYLWYGKGCSGDEREMGKMVAD 575
Query: 203 LIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAEN---DPHLFSCN------FS 253
+I + EG E +FW LG KT Y++ K ++E EN P LF C+ +
Sbjct: 576 IIS-KTEKPVVAEGQEPPEFWVALGGKTSYANSKRLQE-ENPSVPPRLFECSNKTGRFLA 633
Query: 254 EEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISC 313
EI +F+QDDL D+++LD QIF W+G+ + + A +++L +
Sbjct: 634 TEIVDFTQDDLDENDVYLLDTWDQIFFWIGKGANESEKEAAAETAQEYLRSH--PGSRDL 691
Query: 314 SAPIYIVMEGSEPPFFTRFF-KWD 336
PI +V +G EPP FT +F WD
Sbjct: 692 DTPIIVVKQGFEPPTFTGWFMAWD 715
Score = 51.6 bits (122), Expect = 5e-06
Identities = 70/343 (20%), Positives = 126/343 (36%), Gaps = 61/343 (17%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVESFTSKQN 100
Q+WR+ E + FY GDCY+ + + I W+GKNS + +
Sbjct: 20 QIWRIENMEMVPVPTKSYGNFYEGDCYVLLSTRKTGSGFSYNIHYWLGKNS----SQDEQ 75
Query: 101 GRINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DET----------YKEDGVA------- 143
G + YL S+ ++ ET YK+ GVA
Sbjct: 76 GAAAIYTTQMDEYL-----GSVAVQHREVQGHESETFRAYFKQGLIYKQGGVASGMKHVE 130
Query: 144 --LFRIQG----SGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTN------- 190
+ +Q G +++ A +V + S N ++L ++ W G +N
Sbjct: 131 TNTYNVQRLLHVKGKKNVVAAEVEMSWKSFNLGDVFLLDLGQLIIQWNGPESNRAERLRA 190
Query: 191 -------SDDQELAERMLDLIKPDLQ-CRPQKEGAETEQFWELLGVKTEYSSQKIVREAE 242
D + + +++ + + P+ A T E +K +++ +
Sbjct: 191 MTLAKDIRDRERAGRAKVGVVEGENEAASPELMQALTHVLGEKKNIKAATPDEQVHQALN 250
Query: 243 NDPHLFSCNFSE--------EIHNFSQDDLMTEDIFILD-CHSQIFVWVGQQVDPKRRVQ 293
+ L+ + + I +QD L ED +ILD +IFVW G+ + + + Q
Sbjct: 251 SALKLYHVSDASGNLVIQEVAIRPLTQDMLQHEDCYILDQAGLKIFVWKGKNANKEEKQQ 310
Query: 294 ALPIGEKFLEQDFLLETISCSAPIYIVMEGSEPPFFTRFF-KW 335
A+ F++ L S + +GSE F + F KW
Sbjct: 311 AMSRALGFIKAKNYL----ASTSVETENDGSESAVFRQLFQKW 349
Score = 46.6 bits (109), Expect = 2e-04
Identities = 22/58 (37%), Positives = 35/58 (59%), Gaps = 1/58 (1%)
Query: 513 LPVYPYESVKTDSTDPMP-DIDVTKREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLK 569
L +P + + + + +P +D +++E +LS E+F+ GMTRS F LP WKQ LK
Sbjct: 763 LETFPLDVLVNTAAEDLPRGVDPSRKENHLSDEDFKAVFGMTRSAFANLPLWKQQNLK 820
>GELS_XENLA (P14885) Gelsolin (Actin-depolymerizing factor) (ADF)
(Brevin) (Fragment)
Length = 417
Score = 122 bits (306), Expect = 3e-27
Identities = 86/319 (26%), Positives = 147/319 (45%), Gaps = 33/319 (10%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVES------ 94
Q+WR+ EK + + +FY GD YI Y Y ++ +I TW G +S +
Sbjct: 84 QIWRIENCEKVPVLESHYGQFYGGDSYIILYHYKSGGKQGQIIYTWQGDDSTKDEITASA 143
Query: 95 -----FTSKQNGRINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDG------VA 143
+ G +V + G + LF + + T +E G V
Sbjct: 144 ILSAQLDEELGGGPVQVRVVQGK---EPAHLISLFGGKPMIIYKGGTSREGGQTKDANVR 200
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDL 203
LF+++ S +A++V++ AS+LNS+ ++L + S + W G + + ++ A+ +L +
Sbjct: 201 LFQVRTSSSGFSRAVEVDNTASNLNSNDAFVLTTPSASYLWVGQGSTNVEKNGAKELLKI 260
Query: 204 IKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAEN--DPHLFSCN-------FSE 254
+ P EG ET+ FW LG K +Y + +++ N P LF+C+ E
Sbjct: 261 LGVSASEIP--EGQETDDFWGALGGKADYRTSARLKDKLNAHPPRLFACSNKTGRFIIEE 318
Query: 255 EIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISCS 314
SQDDL T+D+ +LD Q++VWVG + + +A+ K++E D
Sbjct: 319 VPGEISQDDLATDDVMLLDTWDQVYVWVGNEAQEDEKKEAIASAYKYIESD--PANRDKR 376
Query: 315 APIYIVMEGSEPPFFTRFF 333
P+ I +G EPP F +F
Sbjct: 377 TPVAITKQGFEPPTFIGWF 395
>GELS_HOMAM (Q27319) Gelsolin, cytoplasmic (Actin-depolymerizing
factor) (ADF)
Length = 754
Score = 122 bits (305), Expect = 3e-27
Identities = 78/299 (26%), Positives = 140/299 (46%), Gaps = 27/299 (9%)
Query: 61 FYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVES-----------FTSKQNGRINEVYSI 109
F+ GD Y+ +Y Y E +++ W G S + ++ G+ +V +
Sbjct: 427 FFGGDSYVLKYIYEVNGNERYILYFWQGCASSQDEKASSAIHTVRLDNELCGKAVQVRVV 486
Query: 110 YG----SYL*RQ*NNSILFHPPKLDCF*D----ETYKEDGVALFRIQGSGPESMQAIQVN 161
G +L ++F K F + +TY DG LFR++G+ +AIQ
Sbjct: 487 QGYEPAHFLRIFKGRMVIFLGGKASGFKNVHDHDTYDVDGTRLFRVRGTCDFDTRAIQQT 546
Query: 162 SAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDLIKPDLQCRPQKEGAETEQ 221
A SLNS ++L++ + W G + +++ + E++++L+ P EG E +
Sbjct: 547 EVAGSLNSDDVFVLETPGKTYLWIGKGASEEEKAMGEKVVELVSPGRDMVTVAEGEEDDD 606
Query: 222 FWELLGVKTEYSSQKIVREAENDPHLFSCNFS-------EEIHNFSQDDLMTEDIFILDC 274
FW LG K +Y + + + P LF C S E+ +F+Q+DL +D+ +LD
Sbjct: 607 FWGGLGGKGDYQTARDLDRPLLYPRLFHCTISPAGCLRVNEMSDFAQEDLNEDDVMVLDS 666
Query: 275 HSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISCSAPIYIVMEGSEPPFFTRFF 333
+++VWVGQ D + + +A + E +++ D T+ + I + +G EP FT F
Sbjct: 667 GDEVYVWVGQGSDDQEKEKAFTMAENYIKTDPTERTLDATV-ILRINQGEEPAAFTSIF 724
Score = 37.4 bits (85), Expect = 0.11
Identities = 44/218 (20%), Positives = 85/218 (38%), Gaps = 34/218 (15%)
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDL 203
LF ++G +++ QV S+N C+IL S V+ + G + D+ A + +
Sbjct: 134 LFHVKGR--RNIRIRQVEVGVGSMNKGDCFILDCGSQVYAYMGPSSRKMDRLKAIQAANP 191
Query: 204 IKPDLQCRPQK--------EGAETEQFWELLG-------------------VKTEYSSQK 236
++ D K G+E + LG ++E +
Sbjct: 192 VRADDHAGKAKVIVIDETASGSEAGESSPGLGGGSPDDVADEDTGVDDSAFERSEVNVVT 251
Query: 237 IVREAENDPHLFSCNFSEEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALP 296
+ E+ + N E Q L + D F+LD ++VW+G K +V+++
Sbjct: 252 LHHIFEDGDGVIQTNMIGE-KPLLQSMLDSGDCFLLDTGVGVYVWIGSGSSKKEKVKSME 310
Query: 297 IGEKFLEQDFLLETISCSAPIYIVMEGSEPPFFTRFFK 334
+ ++E+ + + V+E +EP F +FK
Sbjct: 311 LAAGYMEK----KGYPTYTNVQRVVEKAEPAVFKAYFK 344
>ADSV_HUMAN (Q9Y6U3) Adseverin (Scinderin)
Length = 715
Score = 122 bits (305), Expect = 3e-27
Identities = 93/326 (28%), Positives = 158/326 (47%), Gaps = 37/326 (11%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVE------S 94
++WRV + + +FY GDCYI Y+YP +I TW G N+ +
Sbjct: 399 EIWRVENNGRIQVDQNSYGEFYGGDCYIILYTYP----RGQIIYTWQGANATRDELTTSA 454
Query: 95 FTSKQNGRI--NEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDGVA------LFR 146
F + Q R + I S + LF L + + T K+ G A LF+
Sbjct: 455 FLTVQLDRSLGGQAVQIRVSQGKEPVHLLSLFKDKPLIIYKNGTSKKGGQAPAPPTRLFQ 514
Query: 147 IQGSGPESMQAIQVNSAASSLNSSYCYILQ-SESVVFTWYGNLTNSDDQELAERMLDLIK 205
++ + + ++V+ A+SLNS+ ++L+ ++ + W G + ++++ AE + ++K
Sbjct: 515 VRRNLASITRIVEVDVDANSLNSNDVFVLKLPQNSGYIWVGKGASQEEEKGAEYVASVLK 574
Query: 206 PDLQCRPQK--EGAETEQFWELLGVKTEYSSQKIVREAEND--PHLFSCNFS------EE 255
C+ + EG E E+FW LG K +Y + ++ D P L+ C+ EE
Sbjct: 575 ----CKTLRIQEGEEPEEFWNSLGGKKDYQTSPLLETQAEDHPPRLYGCSNKTGRFVIEE 630
Query: 256 IHN-FSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISCS 314
I F+QDDL +D+ +LD QIF+W+G+ + + ++L + +LE D
Sbjct: 631 IPGEFTQDDLAEDDVMLLDAWEQIFIWIGKDANEVEKKESLKSAKMYLETD--PSGRDKR 688
Query: 315 APIYIVMEGSEPPFFTRFF-KWDSAK 339
PI I+ +G EPP FT +F WDS+K
Sbjct: 689 TPIVIIKQGHEPPTFTGWFLGWDSSK 714
Score = 68.2 bits (165), Expect = 6e-11
Identities = 78/345 (22%), Positives = 139/345 (39%), Gaps = 42/345 (12%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVES------ 94
QVWR+ E + + FY GD Y+ ++ + + W+GK +
Sbjct: 20 QVWRIEKLELVPVPQSAHGDFYVGDAYLVLHTAKTSRGFTYRLHFWLGKECSQDESTAAA 79
Query: 95 -FTSKQNGRIN-------EVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDGVALFR 146
FT + + + E+ + + + + + D A
Sbjct: 80 IFTVQMDDYLGGKPVQNRELQGYESNDFVSYFKGGLKYKAGGVASGLNHVLTNDLTAKRL 139
Query: 147 IQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDLIK- 205
+ G ++A +V + S N C+I+ + ++ W G+ N ++ A ++ I+
Sbjct: 140 LHVKGRRVVRATEVPLSWDSFNKGDCFIIDLGTEIYQWCGSSCNKYERLKANQVATGIRY 199
Query: 206 PDLQCRPQ----KEGAETEQFWELLGVKTEY----SSQKIVREAENDPHLFSCNFSE--- 254
+ + R + +EG+E + ++LG K E I+ + N S+
Sbjct: 200 NERKGRSELIVVEEGSEPSELIKVLGEKPELPDGGDDDDIIADISNRKMAKLYMVSDASG 259
Query: 255 --------EIHNFSQDDLMTEDIFILD--CHSQIFVWVGQQVDPKRRVQALPIGEKFLEQ 304
E + FS L++E+ FILD QIFVW G+ +P+ R A+ E+FL+Q
Sbjct: 260 SMRVTVVAEENPFSMAMLLSEECFILDHGAAKQIFVWKGKDANPQERKAAMKTAEEFLQQ 319
Query: 305 DFLLETISCSAPIYIVMEGSEPPFFTRFFK-W-DSAKSAMLGNSY 347
S + I ++ EG E P F +FFK W D +S G Y
Sbjct: 320 ----MNYSKNTQIQVLPEGGETPIFKQFFKDWRDKDQSDGFGKVY 360
>ADSV_BOVIN (Q28046) Adseverin (Scinderin) (SC)
Length = 715
Score = 122 bits (305), Expect = 3e-27
Identities = 91/326 (27%), Positives = 157/326 (47%), Gaps = 37/326 (11%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVE------S 94
Q+WRV + + +FY GDCYI Y+YP +I TW G N+ +
Sbjct: 399 QIWRVENNGRVEIDRNSYGEFYGGDCYIILYTYP----RGQIIYTWQGANATRDELTTSA 454
Query: 95 FTSKQNGRI--NEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDGVA------LFR 146
F + Q R + I S + LF L + + T K++G A LF+
Sbjct: 455 FLTVQLDRSLGGQAVQIRVSQGKEPAHLLSLFKDKPLIIYKNGTSKKEGQAPAPPIRLFQ 514
Query: 147 IQGSGPESMQAIQVNSAASSLNSSYCYILQ-SESVVFTWYGNLTNSDDQELAERMLDLIK 205
++ + + ++V+ A+SLNS+ ++L+ ++ + W G + ++++ AE + ++K
Sbjct: 515 VRRNLASITRIMEVDVDANSLNSNDVFVLKLRQNNGYIWIGKGSTQEEEKGAEYVASVLK 574
Query: 206 PDLQCRPQ--KEGAETEQFWELLGVKTEYSSQKIVREAEND--PHLFSCN-------FSE 254
C+ +EG E E+FW LG K +Y + ++ D P L+ C+ E
Sbjct: 575 ----CKTSTIQEGKEPEEFWNSLGGKKDYQTSPLLESQAEDHPPRLYGCSNKTGRFIIEE 630
Query: 255 EIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISCS 314
F+QDDL +D+ +LD QIF+W+G+ + + ++L + +LE D
Sbjct: 631 VPGEFTQDDLAEDDVMLLDAWEQIFIWIGKDANEVEKSESLKSAKIYLETD--PSGRDKR 688
Query: 315 APIYIVMEGSEPPFFTRFF-KWDSAK 339
PI I+ +G EPP FT +F WDS++
Sbjct: 689 TPIVIIKQGHEPPTFTGWFLGWDSSR 714
Score = 71.6 bits (174), Expect = 5e-12
Identities = 75/345 (21%), Positives = 140/345 (39%), Gaps = 42/345 (12%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVES------ 94
QVWR+ E + + FY GD Y+ ++ + + W+GK +
Sbjct: 20 QVWRIEKLELVPVPESAYGNFYVGDAYLVLHTTQASRGFTYRLHFWLGKECTQDESTAAA 79
Query: 95 -FTSKQNGRIN-------EVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDGVALFR 146
FT + + + E+ + + + + + D A
Sbjct: 80 IFTVQMDDYLGGKPVQNRELQGYESTDFVGYFKGGLKYKAGGVASGLNHVLTNDLTAQRL 139
Query: 147 IQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDLIKP 206
+ G ++A +V + S N C+I+ + ++ W G+ N ++ A ++ I+
Sbjct: 140 LHVKGRRVVRATEVPLSWDSFNKGDCFIIDLGTEIYQWCGSSCNKYERLKASQVAIGIRD 199
Query: 207 D-----LQCRPQKEGAETEQFWELLG-------------VKTEYSSQKIVR--EAENDPH 246
+ Q +EG+E + ++LG +K + +++K+ + +
Sbjct: 200 NERKGRAQLIVVEEGSEPSELTKVLGEKPKLRDGEDDDDIKADITNRKMAKLYMVSDASG 259
Query: 247 LFSCNFSEEIHNFSQDDLMTEDIFILD--CHSQIFVWVGQQVDPKRRVQALPIGEKFLEQ 304
+ E + FS L++E+ FILD QIFVW G+ +P+ R A+ E+FL+Q
Sbjct: 260 SMKVSLVAEENPFSMAMLLSEECFILDHGAAKQIFVWKGKDANPQERKAAMKTAEEFLQQ 319
Query: 305 DFLLETISCSAPIYIVMEGSEPPFFTRFFK-W-DSAKSAMLGNSY 347
S + I ++ EG E P F +FFK W D +S G Y
Sbjct: 320 ----MNYSTNTQIQVLPEGGETPIFKQFFKDWRDRDQSDGFGKVY 360
>GELS_MOUSE (P13020) Gelsolin precursor, plasma
(Actin-depolymerizing factor) (ADF) (Brevin)
Length = 780
Score = 121 bits (303), Expect = 6e-27
Identities = 84/324 (25%), Positives = 150/324 (45%), Gaps = 35/324 (10%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVES------ 94
Q+WR+ G K + +FY GD YI Y+Y R+ +I W G S +
Sbjct: 446 QIWRIEGSNKVPVDPATYGQFYGGDSYIILYNYRHGGRQGQIIYNWQGAQSTQDEVAASA 505
Query: 95 -----FTSKQNGRINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDG------VA 143
+ G + + G + LF + + T ++ G +
Sbjct: 506 ILTAQLDEELGGTPVQSRVVQGK---EPAHLMSLFGGKPMIIYKGGTSRDGGQTAPASIR 562
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDL 203
LF+++ S + +A++V + +LNS+ ++L++ S + W G + ++ A+ +L +
Sbjct: 563 LFQVRASSSGATRAVEVMPKSGALNSNDAFVLKTPSAAYLWVGAGASEAEKTGAQELLKV 622
Query: 204 IKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAENDPH---LFSCN-------FS 253
++ Q +EG+E + FWE LG KT Y + +++ + D H LF+C+
Sbjct: 623 LRS--QHVQVEEGSEPDAFWEALGGKTAYRTSPRLKDKKMDAHPPRLFACSNRIGRFVIE 680
Query: 254 EEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISC 313
E Q+DL T+D+ +LD Q+FVWVG+ + + +AL ++++E D
Sbjct: 681 EVPGELMQEDLATDDVMLLDTWDQVFVWVGKDSQEEEKTEALTSAKRYIETD--PANRDR 738
Query: 314 SAPIYIVMEGSEPPFFTRFF-KWD 336
PI +V +G EPP F +F WD
Sbjct: 739 RTPITVVRQGFEPPSFVGWFLGWD 762
Score = 62.4 bits (150), Expect = 3e-09
Identities = 52/212 (24%), Positives = 98/212 (45%), Gaps = 27/212 (12%)
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDL 203
LF+++G ++A +V + S N+ C+IL + ++ W G+ +N ++ A ++
Sbjct: 187 LFQVKGR--RVVRATEVPVSWDSFNNGDCFILDLGNNIYQWCGSGSNKFERLKATQVSKG 244
Query: 204 IKPD-----LQCRPQKEGAETEQFWELLG------------VKTEYSSQKIVR--EAEND 244
I+ + Q +EG E E ++LG K + +++++ + + N
Sbjct: 245 IRDNERSGRAQVHVSEEGGEPEAMLQVLGPKPALPEGTEDTAKEDAANRRLAKLYKVSNG 304
Query: 245 PHLFSCNFSEEIHNFSQDDLMTEDIFILD--CHSQIFVWVGQQVDPKRRVQALPIGEKFL 302
S + + + F+Q L +ED FILD +IFVW G+Q + + R AL F+
Sbjct: 305 AGSMSVSLVADENPFAQGALRSEDCFILDHGRDGKIFVWKGKQANMEERKAALKTASDFI 364
Query: 303 EQDFLLETISCSAPIYIVMEGSEPPFFTRFFK 334
+ + ++ EG E P F +FFK
Sbjct: 365 SK----MQYPRQTQVSVLPEGGETPLFKQFFK 392
>GELS_HORSE (Q28372) Gelsolin (Actin-depolymerizing factor) (ADF)
(Brevin)
Length = 730
Score = 120 bits (302), Expect = 7e-27
Identities = 85/326 (26%), Positives = 149/326 (45%), Gaps = 35/326 (10%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVES------ 94
Q+WRV G K + +FY GD YI Y+Y R+ +I W G S +
Sbjct: 396 QIWRVEGSNKVPVDPATYGQFYGGDSYIILYNYRHGSRQGQIIYNWQGAQSTQDEVAASA 455
Query: 95 -----FTSKQNGRINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDG------VA 143
+ G + + G + LF + + T +E G
Sbjct: 456 ILTAQLDEELGGTPVQSRVVQGK---EPAHLMSLFGGKPMIVYKGGTSREGGQTAPASTR 512
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDL 203
LF+++ S + +A+++ A +LNS+ ++L++ S + W G + ++ A+ +L +
Sbjct: 513 LFQVRASSSGATRAVEIIPKAGALNSNDAFVLKTPSAAYLWVGAGASEAEKTGAQELLRV 572
Query: 204 IKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAENDPH---LFSCN-------FS 253
++ Q EG+E + FWE LG K Y + +++ + D H LF+C+
Sbjct: 573 LRA--QPVQVAEGSEPDSFWEALGGKATYRTSPRLKDKKMDAHPPRLFACSNKIGRFVIE 630
Query: 254 EEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISC 313
E F Q+DL T+D+ +LD Q+FVWVG+ + + +AL +++++ D
Sbjct: 631 EVPGEFMQEDLATDDVMLLDTWDQVFVWVGKDSQDEEKTEALTSAKRYIDTDPAHR--DR 688
Query: 314 SAPIYIVMEGSEPPFFTRFF-KWDSA 338
PI +V +G EPP F +F WD +
Sbjct: 689 RTPITVVKQGFEPPSFVGWFLGWDDS 714
Score = 66.6 bits (161), Expect = 2e-10
Identities = 60/225 (26%), Positives = 108/225 (47%), Gaps = 29/225 (12%)
Query: 147 IQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDLIKP 206
+Q G ++A +V + S N+ C+IL + ++ W G+ +N ++ A ++ I+
Sbjct: 138 LQVKGRRVVRATEVPVSWESFNNGDCFILDLGNNIYQWCGSKSNRFERLKATQVSKGIRD 197
Query: 207 D-----LQCRPQKEGAETEQFWELLG------------VKTEYSSQKIVR--EAENDPHL 247
+ Q +EGAE E ++LG VK + +++K+ + + N
Sbjct: 198 NERSGRAQVSVFEEGAEPEAMLQVLGPKPTLPEATEDTVKEDAANRKLAKLYKVSNGAGP 257
Query: 248 FSCNFSEEIHNFSQDDLMTEDIFILD--CHSQIFVWVGQQVDPKRRVQALPIGEKFL-EQ 304
+ + + F+Q L +ED FILD +IFVW G+Q + + R AL F+ +
Sbjct: 258 MVVSLVADENPFAQGALRSEDCFILDHGKDGKIFVWKGKQANMEERKAALKTASDFISKM 317
Query: 305 DFLLETISCSAPIYIVMEGSEPPFFTRFFK-W-DSAKSAMLGNSY 347
D+ +T + ++ EG E P F +FFK W D ++ LG +Y
Sbjct: 318 DYPKQT-----QVSVLPEGGETPLFRQFFKNWRDPDQTEGLGLAY 357
>VILI_DICDI (P36418) Protovillin (100 kDa actin-binding protein)
Length = 959
Score = 120 bits (301), Expect = 1e-26
Identities = 135/568 (23%), Positives = 227/568 (39%), Gaps = 123/568 (21%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYS-YPGEDREEHLIGTWIGKNS------VE 93
++W V + K ++ ++ FY+ CY+ ++ + + ++ W G+ S
Sbjct: 472 KIWHVRNRNKFEISQSEFGLFYNQSCYLVLFTLFAADGSNNSILYYWQGRFSSSEDKGAA 531
Query: 94 SFTSKQNGRINEVYSIYGSYL*RQ*NNSILFH------------PPKLDCF*DETYKEDG 141
+ +K G+ I+ + + N L H P E
Sbjct: 532 ALLAKDVGKELHRSCIHVRTVQNKEPNHFLEHFQGRMVVFKGSRPNATTEVSLENLSSSL 591
Query: 142 VALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQS--ESVVFTWYGNLTNSDDQELAER 199
L+ ++G+ P ++ +IQV A SSL+S+ +IL + ++ + W G SD++E A +
Sbjct: 592 QGLYHVRGTEPINIHSIQVEKAISSLDSNDSFILVNFKNTISYIWVGKY--SDEKEAALQ 649
Query: 200 MLDLIKPDLQCRPQKEGAETEQFWELLGVKT------EYSSQKIVREAENDPHLFSCNFS 253
+ + + EG ET +FWE L + +Y +Q E E LF C+ +
Sbjct: 650 ISSNVFTGYNFQLIDEGDETSEFWESLETNSSLSLLKDYYTQLRTVEQEKKTRLFQCSNN 709
Query: 254 E------EIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFL 307
EIH+FSQDDL ++D+ ILD QIFVWVG++ ++ A E LE
Sbjct: 710 SGVFKVFEIHDFSQDDLDSDDVMILDNQKQIFVWVGKESSDTEKLMA---NETALEYIMN 766
Query: 308 LETISCSAPIYIVMEGSEPPFFT-RFFKWDSAKSAMLGNSYQRKLAIM----KNGGTPPL 362
T PI+ + +G EP FT F W K+ +SY+ KL+ + +G P+
Sbjct: 767 APTHRRDDPIFTIQDGFEPHEFTFNFHAWQVNKTQQ--DSYKSKLSAILGSNNSGPASPI 824
Query: 363 VKPKRRASVSYGGRSGGLPEKSQRSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLS 422
+ P + +++ P AAT + ++
Sbjct: 825 MLP---------------------TSGVTLKP-------------TTAATPKPITTPTVT 850
Query: 423 TPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETN 482
TP P+ TP +ATL + A+ +LK T+ + P
Sbjct: 851 TPKPI--------TTPTVATLKTVTPAV-------------------TLKPTTVTTPSKV 883
Query: 483 SDNENSTGSREESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMPDIDVTKREAYLS 542
+ N++ ++T YP SV T+ DID + YLS
Sbjct: 884 ATTTNTSTPSPTTIT----------------TFYPL-SVLKQKTNLPNDIDKSCLHLYLS 926
Query: 543 PEEFQERLGMTRSEFYKLPKWKQNKLKM 570
EEF MT+ F K P WK +L++
Sbjct: 927 DEEFLSTFKMTKEIFQKTPAWKTKQLRV 954
Score = 47.8 bits (112), Expect = 8e-05
Identities = 50/216 (23%), Positives = 87/216 (40%), Gaps = 31/216 (14%)
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGN----LTNSDDQELAER 199
LF ++G +++ QV+ ++ SLNS ++L E ++ W G+ L +L R
Sbjct: 195 LFHLKGR--RNIRVKQVDISSKSLNSGDVFVLDCEDFIYQWNGSESSRLEKGKGLDLTIR 252
Query: 200 MLDLIKPDLQCRPQKEG---AETEQFWELLG------VKTEYSSQKIVREAENDPHLFSC 250
+ D + E + +FW+ LG K E E ++ +
Sbjct: 253 LRDEKSAKAKIIVMDENDTDKDHPEFWKRLGGCKDDVQKAEQGGDDFAYEKKSVEQIKLY 312
Query: 251 ---NFSEEIHN---------FSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIG 298
N + E+H +S L E +ILDC ++++VW+G+ +R A+
Sbjct: 313 QVENLNYEVHLHLIDPIGDVYSTTQLNAEFCYILDCETELYVWLGKASANDQRTVAMANA 372
Query: 299 EKFLEQDFLLETISCSAPIYIVMEGSEPPFFTRFFK 334
L +D PI + +GSE F FK
Sbjct: 373 MDLLHED----NRPSWTPIIKMTQGSENTLFKDKFK 404
>GELS_PIG (P20305) Gelsolin precursor, plasma (Actin-depolymerizing
factor) (ADF) (Brevin) (Fragment)
Length = 772
Score = 119 bits (299), Expect = 2e-26
Identities = 86/324 (26%), Positives = 149/324 (45%), Gaps = 35/324 (10%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVES------ 94
Q+WR+ G K + +FY GD YI Y+Y R+ +I W G S +
Sbjct: 438 QIWRIEGSNKVPVDPATYGQFYGGDSYIILYNYRHGGRQGQIIYNWQGAQSTQDEVAASA 497
Query: 95 -----FTSKQNGRINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*DETYKEDG------VA 143
+ G + + G + LF + + T +E G
Sbjct: 498 ILTAQLDEELGGTPVQSRVVQGK---EPAHLMSLFGGKPMIIYRGGTSREGGQTAPASTR 554
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDL 203
LF+++ S + +A++V A +LNS+ ++L++ S + W G + ++ A+ +L +
Sbjct: 555 LFQVRASSSGATRAVEVIPKAGALNSNDAFVLKTPSAAYLWVGTGASEAEKTGAQELLRV 614
Query: 204 IKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAENDPH---LFSCNFS------E 254
++ Q EG+E + FWE LG K Y + +++ + D H LF+C+ E
Sbjct: 615 LRA--QPVQVAEGSEPDSFWEALGGKAAYRTSPRLKDKKMDAHPPRLFACSNKIGRFVVE 672
Query: 255 EIHN-FSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISC 313
E+ Q+DL T+D+ +LD Q+FVWVG+ + + +AL ++++E D
Sbjct: 673 EVPGELMQEDLATDDVMLLDTWDQVFVWVGKDSQEEEKTEALTSAKRYIETD--PANRDR 730
Query: 314 SAPIYIVMEGSEPPFFTRFF-KWD 336
PI +V +G EPP F +F WD
Sbjct: 731 RTPINVVKQGFEPPSFVGWFLGWD 754
Score = 58.5 bits (140), Expect = 4e-08
Identities = 51/212 (24%), Positives = 96/212 (45%), Gaps = 27/212 (12%)
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDL 203
LF+++G ++A +V + S N C+IL + ++ W G+ +N ++ A ++
Sbjct: 179 LFQVKGR--RVVRATEVPVSWESFNRGDCFILDLGNDIYQWCGSNSNRYERLKATQVSKG 236
Query: 204 IKPD-----LQCRPQKEGAETEQFWELLG------------VKTEYSSQKIVR--EAEND 244
I+ + +E AE ++LG VK + +++K+ + + N
Sbjct: 237 IRDNERSGRAHVHVSEEDAEPAGMLQVLGPKPTLPEGTEDTVKEDAANRKLAKLYKVSNG 296
Query: 245 PHLFSCNFSEEIHNFSQDDLMTEDIFILDC--HSQIFVWVGQQVDPKRRVQALPIGEKFL 302
+ + + + F+Q L +ED FILD +IFVW G+Q + + R AL F+
Sbjct: 297 AGTMTVSLVADENPFAQGALKSEDCFILDHGKDGKIFVWKGKQANTEERKAALKTASDFI 356
Query: 303 EQDFLLETISCSAPIYIVMEGSEPPFFTRFFK 334
+ + ++ EG E P F +FFK
Sbjct: 357 SK----MNYPKQTQVSVLPEGGETPLFKQFFK 384
>VILL_HUMAN (O15195) Villin-like protein
Length = 686
Score = 116 bits (291), Expect = 1e-25
Identities = 88/322 (27%), Positives = 150/322 (46%), Gaps = 29/322 (9%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVESFTSKQN 100
+VW + + + + +G+CY+ Y+Y R ++++ W G + N
Sbjct: 224 EVWCIQDLHRQPVDPKRHGQLCAGNCYLVLYTYQRLGRVQYILYLWQGHQATADEIEALN 283
Query: 101 GRINEVYSIYGSYL*RQ*NNSILFHPP--------KLDCF*DET------YKEDGVALFR 146
E+ +YG L ++ + ++ PP +L F + LF+
Sbjct: 284 SNAEELDVMYGGVLVQE-HVTMGSEPPHFLAIFQGQLVIFQERAGHHGKGQSASTTRLFQ 342
Query: 147 IQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDLIKP 206
+QG+ + + ++V + ASSLNSS ++L + SV + W+G N D +E+A ++ +I
Sbjct: 343 VQGTDSHNTRTMEVPARASSLNSSDIFLLVTASVCYLWFGKGCNGDQREMARVVVTVISR 402
Query: 207 DLQCRPQKEGAETEQFWELLGVKTEYSSQKIVRE--AENDPHLFSCN------FSEEIHN 258
+ EG E FWE LG + Y S K + E P LF C+ E+
Sbjct: 403 KNE-ETVLEGQEPPHFWEALGGRAPYPSNKRLPEEVPSFQPRLFECSSHMGCLVLAEVGF 461
Query: 259 FSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISCSAPIY 318
FSQ+DL DI +LD +IF+W+G+ + +A+ G+++L+ S + PI
Sbjct: 462 FSQEDLDKYDIMLLDTWQEIFLWLGEAASEWK--EAVAWGQEYLKTH--PAGRSPATPIV 517
Query: 319 IVMEGSEPP-FFTRFFKWDSAK 339
+V +G EPP F FF WD K
Sbjct: 518 LVKQGHEPPTFIGWFFTWDPYK 539
Score = 44.7 bits (104), Expect = 7e-04
Identities = 33/131 (25%), Positives = 64/131 (48%), Gaps = 10/131 (7%)
Query: 441 ATLAPKSSAISHLTSTFEPPSAREKLIP-RSLKDTSKSNPETNSDNENSTGSREESLTIQ 499
+T++ ++ +++L + P + R + ++LK + S+ + S GSR S
Sbjct: 558 STISEITAEVNNLRLSRWPGNGRAGAVALQALKGSQDSSENDLVRSPKSAGSRTSS---- 613
Query: 500 EDVNEGEPEDNEGLPVYPYESVKTDSTDPMPD-IDVTKREAYLSPEEFQERLGMTRSEFY 558
V+ N GL E + + + +P+ +D +RE YLS +FQ+ G ++ EFY
Sbjct: 614 -SVSSTSATINGGLR---REQLMHQAVEDLPEGVDPARREFYLSDSDFQDIFGKSKEEFY 669
Query: 559 KLPKWKQNKLK 569
+ W+Q + K
Sbjct: 670 SMATWRQRQEK 680
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 69,070,788
Number of Sequences: 164201
Number of extensions: 2984638
Number of successful extensions: 9139
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 8786
Number of HSP's gapped (non-prelim): 262
length of query: 574
length of database: 59,974,054
effective HSP length: 116
effective length of query: 458
effective length of database: 40,926,738
effective search space: 18744446004
effective search space used: 18744446004
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC125475.5


