

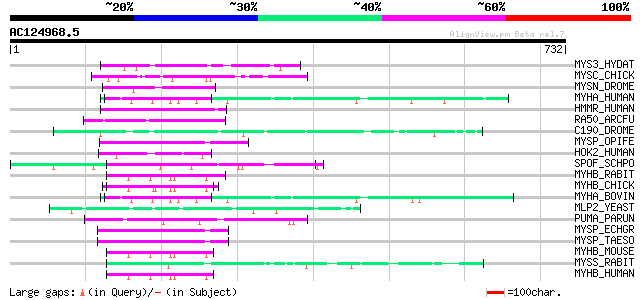
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124968.5 - phase: 0
(732 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
MYS3_HYDAT (P39922) Myosin heavy chain, clone 203 (Fragment) 54 1e-06
MYSC_CHICK (P29616) Myosin heavy chain, cardiac muscle isoform (... 53 3e-06
MYSN_DROME (Q99323) Myosin heavy chain, non-muscle (Zipper prote... 52 7e-06
MYHA_HUMAN (P35580) Myosin heavy chain, nonmuscle type B (Cellul... 52 7e-06
HMMR_HUMAN (O75330) Hyaluronan mediated motility receptor (Intra... 52 7e-06
RA50_ARCFU (O29230) DNA double-strand break repair rad50 ATPase 50 2e-05
C190_DROME (Q9VJE5) Restin homolog (Cytoplasmic linker protein 1... 49 4e-05
MYSP_OPIFE (Q9BMQ6) Paramyosin (Fragment) 49 5e-05
HOK2_HUMAN (Q96ED9) Hook homolog 2 (h-hook2) (hHK2) 49 6e-05
SPOF_SCHPO (Q10411) Sporulation-specific protein 15 48 8e-05
MYHB_RABIT (P35748) Myosin heavy chain, smooth muscle isoform (S... 48 8e-05
MYHB_CHICK (P10587) Myosin heavy chain, gizzard smooth muscle 47 1e-04
MYHA_BOVIN (Q27991) Myosin heavy chain, nonmuscle type B (Cellul... 47 2e-04
MLP2_YEAST (P40457) MLP2 protein (Myosin-like protein 2) 47 2e-04
PUMA_PARUN (O61308) 227 kDa spindle- and centromere-associated p... 46 3e-04
MYSP_ECHGR (P35417) Paramyosin 46 3e-04
MYSP_TAESO (P35418) Paramyosin (Antigen B) (AgB) 46 4e-04
MYHB_MOUSE (O08638) Myosin heavy chain, smooth muscle isoform (S... 46 4e-04
MYSS_RABIT (P02562) Myosin heavy chain, skeletal muscle (Fragments) 45 5e-04
MYHB_HUMAN (P35749) Myosin heavy chain, smooth muscle isoform (S... 45 5e-04
>MYS3_HYDAT (P39922) Myosin heavy chain, clone 203 (Fragment)
Length = 539
Score = 54.3 bits (129), Expect = 1e-06
Identities = 60/276 (21%), Positives = 118/276 (42%), Gaps = 37/276 (13%)
Query: 120 RQSSTDLFTELDHMRSLLQESKEREAKLNA---ELVECRKNQSEVDELV----KKVALLE 172
R+ DL E D L +E K+ E+ L +L E E +LV K ++ LE
Sbjct: 235 REIEQDLKKEKDSKMKLEKEKKKVESDLKDNRDKLSETETRLKETQDLVTKREKSISDLE 294
Query: 173 EEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSS 232
K GL Q+ L +R+ ++ + LE E+ R+L ++ +Q++ L R+
Sbjct: 295 NAKEGLESQISQL------QRKIQELLAKIEELEEELENERKLRQKSELQRKELESRIEE 348
Query: 233 MESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNS 292
++ QL + ++ + + K EA R L K++E L + + S
Sbjct: 349 LQDQLETAGGATSAQVEVGKKREAECNR-----LRKEIEALNIANDAAI----------S 393
Query: 293 CLRTELKNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSD 352
++ + T + + + + ++ + + ++ + NS + +KK K T+SD
Sbjct: 394 AIKAKTNATIAEIQEENEAMKKAKAKLEKEKSALNNELNETKNSLDQIKKQK----TNSD 449
Query: 353 QLS-----QVECTNNSIIDKNWIESISEGSNRRRHS 383
+ S Q+ N+ + + + S SE N + +S
Sbjct: 450 KNSRMLEEQINELNSKLAQVDELHSQSESKNSKVNS 485
Score = 40.8 bits (94), Expect = 0.013
Identities = 53/256 (20%), Positives = 106/256 (40%), Gaps = 20/256 (7%)
Query: 124 TDLFTELDHMRSLLQESKE-REAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQL 182
T+ EL S L+ K+ R+ +++ + RK + ++ + ++EE +EQL
Sbjct: 154 TEKTEELQSNISRLETEKQNRDKQIDTLNEDIRKQDETISKMNAEKKHVDEELKDRTEQL 213
Query: 183 VALSRSC-GLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSD 241
A C L + + + S + +E ++ + + +L +K+ + L +LS ++
Sbjct: 214 QAAEDKCNNLNKTKNKLESSIREIEQDLKKEKDSKMKLEKEKKKVESDLKDNRDKLSETE 273
Query: 242 N--SSESDIVAKFKAEASLLRLTNEDLSKQVEGLQ------TSRLNEVEE----LAYLRW 289
D+V K + S L E L Q+ LQ +++ E+EE LR
Sbjct: 274 TRLKETQDLVTKREKSISDLENAKEGLESQISQLQRKIQELLAKIEELEEELENERKLRQ 333
Query: 290 VNSCLRTELKNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPIT 349
+ R EL++ L D+L + S+ + +C L K+ + I
Sbjct: 334 KSELQRKELESRIEEL-QDQLETAGGATSAQVEVGKKREAECN-----RLRKEIEALNIA 387
Query: 350 SSDQLSQVECTNNSII 365
+ +S ++ N+ I
Sbjct: 388 NDAAISAIKAKTNATI 403
>MYSC_CHICK (P29616) Myosin heavy chain, cardiac muscle isoform
(Fragment)
Length = 1102
Score = 52.8 bits (125), Expect = 3e-06
Identities = 78/315 (24%), Positives = 138/315 (43%), Gaps = 52/315 (16%)
Query: 108 SQVHPHAFPTHRRQSSTDLFT-------ELDHMRSLLQESK---EREAKLNAELVECRKN 157
SQ A R ST+LF LDH+ +L +E+K E + L ++ E KN
Sbjct: 629 SQAELEASQKEARSLSTELFKLKNAYEETLDHLETLKRENKNLQEEISDLTNQISEGNKN 688
Query: 158 QSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELR---- 213
E++++ K+V E+EK SE +AL + G EE K T +LE+ +L+
Sbjct: 689 LHEIEKVKKQV---EQEK---SEVQLALEEAEGALEHEESK---TLRFQLELSQLKADFE 739
Query: 214 ----RLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASL----LRLTN-- 263
++E+ +RN + S++S L S I K K E L ++L++
Sbjct: 740 RKLAEKDEEMQNIRRNQQRTIDSLQSTLDSEARSRNEAIRLKKKMEGDLNEMEIQLSHAN 799
Query: 264 ---EDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSS 320
+ +K GLQT +L L +N L+ +L SD+ + +++ S
Sbjct: 800 RHAAEATKSARGLQTQIKELQVQLDDLGHLNEDLKEQL------AVSDRRN---NLLQSE 850
Query: 321 GDSISSFSDQCGSANSFNLVKKPKKWPITSSDQLSQVECTNNSIID--KNWIESISEGSN 378
D + + DQ A K + + ++++++ + N S+I+ K IS+ N
Sbjct: 851 LDELRALLDQTERAR-----KLAEHELLEATERVNLLHTQNTSLINQKKKLEGDISQMQN 905
Query: 379 RRRHSISGSNSSEEE 393
SI ++E++
Sbjct: 906 EVEESIQECRNAEQK 920
Score = 32.7 bits (73), Expect = 3.5
Identities = 44/204 (21%), Positives = 84/204 (40%), Gaps = 24/204 (11%)
Query: 135 SLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLE----------EEKSGLSEQLVA 184
SL+QE + +L AE + D L+K LE E++ ++ +L +
Sbjct: 45 SLVQEKNDLLLQLQAEQDTLADAEERCDLLIKSKIQLEAKVKELTERVEDEEEMNSELTS 104
Query: 185 LSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSS 244
R E E KD +LE+ + ++ + + +NLT +++++ +S
Sbjct: 105 KKRKLEDECSELKKD--IDDLEITLAKVEKEKHATENKVKNLTEEMATLDENISKLTKEK 162
Query: 245 ES------DIVAKFKAE---ASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLR 295
+S ++ +AE + L L +QV+ L+ S E + L L
Sbjct: 163 KSLQEAHQQVLDDLQAEEDKVNTLSKAKVKLEQQVDDLEGSLEQEKKVRMDLERAKRKLE 222
Query: 296 TELKNTCST---LDSDKLSSPQSV 316
+LK T + L++DKL + +
Sbjct: 223 GDLKLTQESVMDLENDKLQMEEKL 246
>MYSN_DROME (Q99323) Myosin heavy chain, non-muscle (Zipper protein)
(Myosin II) (Non-muscle MHC)
Length = 2057
Score = 51.6 bits (122), Expect = 7e-06
Identities = 41/157 (26%), Positives = 74/157 (47%), Gaps = 16/157 (10%)
Query: 123 STDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVD----ELVKKVALLEEEKSGL 178
+ DL TEL + S QE+ R + +++ E + +E++ EL +K L++E +
Sbjct: 1317 NADLATELRSVNSSRQENDRRRKQAESQIAELQVKLAEIERARSELQEKCTKLQQEAENI 1376
Query: 179 SEQL--VALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQ 236
+ QL L S ++ S N+E ++ E ++L +E QK L+ +L +ES+
Sbjct: 1377 TNQLEEAELKASAAVK--------SASNMESQLTEAQQLLEEETRQKLGLSSKLRQIESE 1428
Query: 237 LSCSDNSSESDIVAKFKAEASLLRLTN--EDLSKQVE 271
E D AK E L +T +++ K+ E
Sbjct: 1429 KEALQEQLEEDDEAKRNYERKLAEVTTQMQEIKKKAE 1465
Score = 43.9 bits (102), Expect = 0.002
Identities = 55/236 (23%), Positives = 95/236 (39%), Gaps = 40/236 (16%)
Query: 125 DLFTELDHMRSLLQESKEREAKLNAELVECRK-----NQSEVDELVKKVALLEEEKSGLS 179
DL TELD R + + KL +L E N+ + D L L + K L
Sbjct: 1684 DLETELDEERKQRTAAVASKKKLEGDLKEIETTMEMHNKVKEDALKHAKKLQAQVKDALR 1743
Query: 180 EQLVALSRSCGLERQEEDKDGSTQNLELEVVEL--------------------------R 213
+ A + L+ ++ DG + LE EV++L
Sbjct: 1744 DAEEAKAAKEELQALSKEADGKVKALEAEVLQLTEDLASSERARRAAETERDELAEEIAN 1803
Query: 214 RLNKELHM--QKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNE------D 265
NK M +KR L R++++E +L ++SE + KA+ + +LT E +
Sbjct: 1804 NANKGSLMIDEKRRLEARIATLEEELEEEQSNSEVLLDRSRKAQLQIEQLTTELANEKSN 1863
Query: 266 LSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSSG 321
K G E A L + + RT++K T +TL++ K+++ + + + G
Sbjct: 1864 SQKNENGRALLERQNKELKAKLAEIETAQRTKVKATIATLEA-KIANLEEQLENEG 1918
Score = 37.7 bits (86), Expect = 0.11
Identities = 54/298 (18%), Positives = 117/298 (39%), Gaps = 64/298 (21%)
Query: 125 DLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDE----LVKKVALLEEEKSGLSE 180
DL +L+ + Q+ + + +L+A++ + ++ + D+ L+K+ LLEE + LS+
Sbjct: 1038 DLEEQLEEEEAARQKLQLEKVQLDAKIKKYEEDLALTDDQNQKLLKEKKLLEERANDLSQ 1097
Query: 181 QLVALSRSCGL----------------------ERQEEDKDGSTQNLELEVVELRRLNKE 218
L ++Q ++ D S + +E EV +L+ E
Sbjct: 1098 TLAEEEEKAKHLAKLKAKHEATITELEERLHKDQQQRQESDRSKRKIETEVADLKEQLNE 1157
Query: 219 LHMQKRNLTCRLS----------------------------SMESQLSCSDNSSESDIVA 250
+Q + +L+ +ESQL+ E++ A
Sbjct: 1158 RRVQVDEMQAQLAKREEELTQTLLRIDEESATKATAQKAQRELESQLAEIQEDLEAEKAA 1217
Query: 251 KFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKNTCSTLDSDKL 310
+ KAE DLS+++E L+ L+ ++ A + + S EL +L+ + +
Sbjct: 1218 RAKAEK-----VRRDLSEELEALKNELLDSLDTTAAQQELRSKREQELATLKKSLEEETV 1272
Query: 311 SSPQSVVS---SSGDSISSFSDQCGSANSFNLVKKPKKWPI--TSSDQLSQVECTNNS 363
+ + ++S +DQ + V + K + ++D +++ N+S
Sbjct: 1273 NHEGVLADMRHKHSQELNSINDQLENLRKAKTVLEKAKGTLEAENADLATELRSVNSS 1330
Score = 35.8 bits (81), Expect = 0.41
Identities = 56/282 (19%), Positives = 120/282 (41%), Gaps = 36/282 (12%)
Query: 129 ELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRS 188
EL+ + +QE E E A+ + R++ SE E +K LL+ + ++Q + R
Sbjct: 1199 ELESQLAEIQEDLEAEKAARAKAEKVRRDLSEELEALKN-ELLDSLDTTAAQQELRSKRE 1257
Query: 189 CGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDI 248
L ++ + T N E + ++R + + L+S+ QL +N ++
Sbjct: 1258 QELATLKKSLEEETVNHEGVLADMRHKHSQ----------ELNSINDQL---ENLRKAKT 1304
Query: 249 VAKFKAEASLLRLTNEDLSKQVEGLQTSRL----------NEVEELAYLRWVNSCLRTEL 298
V + KA+ + L N DL+ ++ + +SR +++ EL R+EL
Sbjct: 1305 VLE-KAKGT-LEAENADLATELRSVNSSRQENDRRRKQAESQIAELQVKLAEIERARSEL 1362
Query: 299 KNTCSTLD------SDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSD 352
+ C+ L +++L + S++ S S+ Q A L+++ + + S
Sbjct: 1363 QEKCTKLQQEAENITNQLEEAELKASAAVKSASNMESQLTEAQ--QLLEEETRQKLGLSS 1420
Query: 353 QLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSSEEEV 394
+L Q+E ++ ++ +E E ++ + +E+
Sbjct: 1421 KLRQIESEKEALQEQ--LEEDDEAKRNYERKLAEVTTQMQEI 1460
>MYHA_HUMAN (P35580) Myosin heavy chain, nonmuscle type B (Cellular
myosin heavy chain, type B) (Nonmuscle myosin heavy
chain-B) (NMMHC-B)
Length = 1976
Score = 51.6 bits (122), Expect = 7e-06
Identities = 121/596 (20%), Positives = 236/596 (39%), Gaps = 87/596 (14%)
Query: 121 QSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSE 180
Q+ T+LF E + MR+ L K+ ++ +L +S V+E ++ +L+ EK +
Sbjct: 897 QAETELFAEAEEMRARLAAKKQELEEILHDL------ESRVEEEEERNQILQNEKKKMQA 950
Query: 181 QLVALSRSCGLERQEEDKDGSTQNLELEVV----ELRRLNKELHM----------QKRNL 226
+ LE Q ++++G+ Q L+LE V +++++ +E+ + +K+ +
Sbjct: 951 HIQ------DLEEQLDEEEGARQKLQLEKVTAEAKIKKMEEEILLLEDQNSKFIKEKKLM 1004
Query: 227 TCRLSSMESQLSCSDNSSE-------------SDIVAKFKAEASLLRLTNEDLSKQVEGL 273
R++ SQL+ + ++ SD+ + K E R E ++++G
Sbjct: 1005 EDRIAECSSQLAEEEEKAKNLAKIRNKQEVMISDLEERLKKEEK-TRQELEKAKRKLDGE 1063
Query: 274 QTSRLNEVEELA----YLRWVNSCLRTELKNTCSTLDSDKL--SSPQSVVSSSGDSISSF 327
T +++ EL L+ + EL+ + D + L ++ VV I+
Sbjct: 1064 TTDLQDQIAELQAQIDELKLQLAKKEEELQGALARGDDETLHKNNALKVVRELQAQIAEL 1123
Query: 328 SDQCGSAN-SFNLVKKPKKWPITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISG 386
+ S S N +K K+ S++L ++ +D + E +R ++
Sbjct: 1124 QEDFESEKASRNKAEKQKR---DLSEELEALKTELEDTLDTTAAQ--QELRTKREQEVAE 1178
Query: 387 -SNSSEEEVSVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRP 445
+ EEE + Q L+E+ ++ F K+ L N
Sbjct: 1179 LKKALEEETKNHEAQIQDMRQRHATALEELSEQLEQAKRFKANLEKNKQGLETDNKELAC 1238
Query: 446 SCSISSKTKQE-------CSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVELYHSL 498
+ + K E AQVQ + S G+ V+ A + +L + L
Sbjct: 1239 EVKVLQQVKAESEHKRKKLDAQVQ---------ELHAKVSEGDRLRVELAEKASKLQNEL 1289
Query: 499 MKRDS-RRDSSSGGLSDAPDVADVRSSMIGE---IENRSSHLLAIKADIETQGEFVNSL- 553
+ ++ G+ A D A + S + ++ + L + + I E NSL
Sbjct: 1290 DNVSTLLEEAEKKGIKFAKDAASLESQLQDTQELLQEETRQKLNLSSRIRQLEEEKNSLQ 1349
Query: 554 -IREVNDAVYENIDDVVAFV--------KWLDDELGFLVD-ERAVLKHFDWPEKKADTLR 603
+E + +N++ V + K +DD+LG + E A K E + L
Sbjct: 1350 EQQEEEEEARKNLEKQVLALQSQLADTKKKVDDDLGTIESLEEAKKKLLKDAEALSQRLE 1409
Query: 604 EAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALS-EKMERTVYTLLRTRDSL 658
E A Y L+K ++ + DD L D+ ++ VA + EK ++ LL S+
Sbjct: 1410 EKALAYDKLEKTKNRLQQELDD--LTVDLDHQRQVASNLEKKQKKFDQLLAEEKSI 1463
Score = 47.0 bits (110), Expect = 2e-04
Identities = 37/153 (24%), Positives = 78/153 (50%), Gaps = 13/153 (8%)
Query: 126 LFTELDHMRSLLQESKEREAKLNAELVECRKNQS----EVDELVKKVALLEEEKSGLSEQ 181
+FT++ + + ++ +E +AK + EL++ ++ Q+ E++E+ +K L EEK+ L+EQ
Sbjct: 837 VFTKVKPLLQVTRQEEELQAK-DEELLKVKEKQTKVEGELEEMERKHQQLLEEKNILAEQ 895
Query: 182 LVALSR--------SCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSM 233
L A + L ++++ + +LE V E N+ L +K+ + + +
Sbjct: 896 LQAETELFAEAEEMRARLAAKKQELEEILHDLESRVEEEEERNQILQNEKKKMQAHIQDL 955
Query: 234 ESQLSCSDNSSESDIVAKFKAEASLLRLTNEDL 266
E QL + + + + K AEA + ++ E L
Sbjct: 956 EEQLDEEEGARQKLQLEKVTAEAKIKKMEEEIL 988
Score = 37.4 bits (85), Expect = 0.14
Identities = 62/282 (21%), Positives = 114/282 (39%), Gaps = 41/282 (14%)
Query: 62 KQLPTKKQQNTTTITTS---DVVTNQKNVNPFVPPHSRVKRSLMGDLSCSQVHPHAFPTH 118
++L TK++Q + + + ++ + H+ L L ++ +
Sbjct: 1166 QELRTKREQEVAELKKALEEETKNHEAQIQDMRQRHATALEELSEQLEQAKRFKANLEKN 1225
Query: 119 RRQSSTD---LFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVA------ 169
++ TD L E+ ++ + ES+ + KL+A++ E SE D L ++A
Sbjct: 1226 KQGLETDNKELACEVKVLQQVKAESEHKRKKLDAQVQELHAKVSEGDRLRVELAEKASKL 1285
Query: 170 ---------LLEE-EKSGLSEQLVALSRSCGLE----------RQEEDKDGSTQNLELEV 209
LLEE EK G+ A S L+ RQ+ + + LE E
Sbjct: 1286 QNELDNVSTLLEEAEKKGIKFAKDAASLESQLQDTQELLQEETRQKLNLSSRIRQLEEEK 1345
Query: 210 VELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQ 269
L+ +E ++NL ++ +++SQL+ + + D+ L + L K
Sbjct: 1346 NSLQEQQEEEEEARKNLEKQVLALQSQLADTKKKVDDDL-----GTIESLEEAKKKLLKD 1400
Query: 270 VEGLQTSRLNEVEELAY--LRWVNSCLRTELKNTCSTLDSDK 309
E L + RL E + LAY L + L+ EL + LD +
Sbjct: 1401 AEAL-SQRLEE-KALAYDKLEKTKNRLQQELDDLTVDLDHQR 1440
Score = 35.4 bits (80), Expect = 0.54
Identities = 44/189 (23%), Positives = 81/189 (42%), Gaps = 20/189 (10%)
Query: 131 DHMRSLLQESKEREAKLNAELVECRKNQSEV----DELVKKVALLEEEKSGLSEQLVALS 186
D + L++ + + EL E R ++ E+ E KK+ LE E L E+L +
Sbjct: 1641 DEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQLQEELASSE 1700
Query: 187 RSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSES 246
R+ QE D+ L E+ L +KR L R++ +E +L + S+
Sbjct: 1701 RARRHAEQERDE------LADEITNSASGKSALLDEKRRLEARIAQLEEELE-EEQSNME 1753
Query: 247 DIVAKFKAEASLLRLTNEDLSKQVEGLQTS--------RLNEVEELAYLRWVNSCLRTEL 298
+ +F+ + N +L+ + Q S R N+ E A L+ + ++++
Sbjct: 1754 LLNDRFRKTTLQVDTLNAELAAERSAAQKSDNARQQLERQNK-ELKAKLQELEGAVKSKF 1812
Query: 299 KNTCSTLDS 307
K T S L++
Sbjct: 1813 KATISALEA 1821
Score = 35.0 bits (79), Expect = 0.71
Identities = 63/320 (19%), Positives = 120/320 (36%), Gaps = 43/320 (13%)
Query: 120 RQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLS 179
+Q DL +LDH R + ++++ K + L E + + E + EK
Sbjct: 1426 QQELDDLTVDLDHQRQVASNLEKKQKKFDQLLAEEKSISARYAEERDRAEAEAREK---- 1481
Query: 180 EQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC 239
+ ALS + LE E K+ E R NK+L + ME +S
Sbjct: 1482 -ETKALSLARALEEALEAKE-----------EFERQNKQLR----------ADMEDLMSS 1519
Query: 240 SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVN-SCLRTEL 298
D+ ++ V + + L E++ Q+E L+ L E+ VN ++ +
Sbjct: 1520 KDDVGKN--VHELEKSKRALEQQVEEMRTQLEELE-DELQATEDAKLRLEVNMQAMKAQF 1576
Query: 299 KNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSDQLSQVE 358
+ T D + ++ + + D+ V KK I D +Q+E
Sbjct: 1577 ERDLQTRDEQNEEKKRLLIKQVRELEAELEDE--RKQRALAVASKKKMEIDLKDLEAQIE 1634
Query: 359 CTNNSIIDK-NWIESISEGSNRRRHSISGSNSSEEEVSVLSKRRQSNCFDSFECLKEIEK 417
N + + + + + + + +S +E+ SK +S + LK +E
Sbjct: 1635 AANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSK-------ESEKKLKSLEA 1687
Query: 418 ESVPMPLFVQQCALEKRALR 437
E + + ++ A +RA R
Sbjct: 1688 EILQLQ---EELASSERARR 1704
Score = 34.3 bits (77), Expect = 1.2
Identities = 41/176 (23%), Positives = 72/176 (40%), Gaps = 29/176 (16%)
Query: 125 DLFTELDHMRS----LLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSE 180
D EL+ R+ + +SKE E KL + E + Q E+ + E+E+ L++
Sbjct: 1656 DYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQLQEELASSERARRHAEQERDELAD 1715
Query: 181 QLV--ALSRSCGLERQ----------EEDKDGSTQNLEL-------EVVELRRLNKELHM 221
++ A +S L+ + EE+ + N+EL +++ LN EL
Sbjct: 1716 EITNSASGKSALLDEKRRLEARIAQLEEELEEEQSNMELLNDRFRKTTLQVDTLNAELAA 1775
Query: 222 QKR------NLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVE 271
++ N +L +L E + +KFKA S L L +Q+E
Sbjct: 1776 ERSAAQKSDNARQQLERQNKELKAKLQELEGAVKSKFKATISALEAKIGQLEEQLE 1831
>HMMR_HUMAN (O75330) Hyaluronan mediated motility receptor
(Intracellular hyaluronic acid binding protein)
(Receptor for hyaluronan-mediated motility) (CD168
antigen)
Length = 724
Score = 51.6 bits (122), Expect = 7e-06
Identities = 37/167 (22%), Positives = 83/167 (49%), Gaps = 4/167 (2%)
Query: 120 RQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLS 179
+++ DL +R LLQE +++++ E K ++ ++ +++ L + L
Sbjct: 78 QKNDKDLKILEKEIRVLLQERGAQDSRIQDLETELEKMEARLNAALREKTSLSANNATLE 137
Query: 180 EQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC 239
+QL+ L+R+ L + + ++G+ +NL + +EL +L + + R + + ME +L
Sbjct: 138 KQLIELTRTNELLKSKFSENGNQKNLRILSLELMKLRNKRETKMRGMMAKQEGMEMKLQV 197
Query: 240 SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNE-VEELA 285
+ S E + E L+ + E + ++ E T +L E +EE++
Sbjct: 198 TQRSLEESQGKIAQLEGKLVSIEKEKIDEKSE---TEKLLEYIEEIS 241
Score = 32.7 bits (73), Expect = 3.5
Identities = 63/279 (22%), Positives = 119/279 (42%), Gaps = 39/279 (13%)
Query: 140 SKEREAKLNAELVECRKNQSEV-DELVKKVALLEEEKSGLSEQLVALSRSCG-LERQEED 197
+ ++ K ++ + +N E DE++ LEE LS+Q+ L+ C LE+++ED
Sbjct: 243 ASDQVEKYKLDIAQLEENLKEKNDEILSLKQSLEENIVILSKQVEDLNVKCQLLEKEKED 302
Query: 198 K-----------DGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDN---- 242
+ QNL+ + + L + +E QK L E +LS S +
Sbjct: 303 HVNRNREHNENLNAEMQNLKQKFI-LEQQEREKLQQKELQIDSLLQQEKELSSSLHQKLC 361
Query: 243 SSESDIVAKFKAEASLLRLTNEDLSK-QVEGLQTSRL-NEVEELAYLRWVN-SCLRTELK 299
S + ++V + L+ T ++L K Q + Q RL ++EE A R L +LK
Sbjct: 362 SFQEEMVKEKNLFEEELKQTLDELDKLQQKEEQAERLVKQLEEEAKSRAEELKLLEEKLK 421
Query: 300 NTCSTLDSDKLSSPQS--VVSSSGDS-ISSFSDQCGSANSFNLVKKPKKWPITSSDQLSQ 356
+ L+ + Q+ ++ DS + S D S+ + ++ ++
Sbjct: 422 GKEAELEKSSAAHTQATLLLQEKYDSMVQSLEDVTAQFESYKAL---------TASEIED 472
Query: 357 VECTNNSIIDKNWIESISEGSNRR--RHSISGSNSSEEE 393
++ N+S+ +K + G N +H I + SS +E
Sbjct: 473 LKLENSSLQEK----AAKAGKNAEDVQHQILATESSNQE 507
>RA50_ARCFU (O29230) DNA double-strand break repair rad50 ATPase
Length = 886
Score = 50.4 bits (119), Expect = 2e-05
Identities = 44/192 (22%), Positives = 90/192 (45%), Gaps = 11/192 (5%)
Query: 98 KRSLMGDLSCSQ-VHPHAFPTHRRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRK 156
K + GD + ++ V H P H + ++ + S++++ + RE ++ ++
Sbjct: 97 KEIVEGDSNITEWVERHLCPAHVFTGA--IYVRQGEIDSIIRDDESRE-RIIRQITRIED 153
Query: 157 NQSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLN 216
++ L + +LE EK L E LS+ ++RQ+E+K + + E+ + L
Sbjct: 154 YENAWKNLGAVIRMLEREKERLKE---FLSQEEQIKRQKEEKKAEIERISEEIKSIESLR 210
Query: 217 KELHMQKRNLTCRLSSMESQLSCSDN--SSESDIVAKFKAEASLLRLTNEDLSKQVEGLQ 274
++L + RNL RL +E S ++ ES ++ + + LR + L + VE ++
Sbjct: 211 EKLSEEVRNLESRLKELEEHKSRLESLRKQESSVLQEVRGLEEKLRELEKQLKEVVERIE 270
Query: 275 --TSRLNEVEEL 284
+ EV+EL
Sbjct: 271 DLEKKAKEVKEL 282
>C190_DROME (Q9VJE5) Restin homolog (Cytoplasmic linker protein 190)
(Microtubule binding protein 190) (d-CLIP-190)
Length = 1690
Score = 49.3 bits (116), Expect = 4e-05
Identities = 119/585 (20%), Positives = 229/585 (38%), Gaps = 59/585 (10%)
Query: 58 DKKTKQLPTKKQQNTTTITTSDVVTNQ-KNVNPFVPPHSRVKRSLMGDLSCSQVHPHAFP 116
+K K+L K+Q T+ + + Q ++ V+ ++ Q+ +
Sbjct: 674 EKTEKELVQSKEQAAKTLNDKEQLEKQISDLKQLAEQEKLVREMTENAINQIQLEKESIE 733
Query: 117 TH---RRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEE 173
++ D + LQE K + + + ELVE E +KK+ E
Sbjct: 734 QQLALKQNELEDFQKKQSESEVHLQEIKAQNTQKDFELVES-------GESLKKLQQQLE 786
Query: 174 EKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSM 233
+K+ E+L A LE +++K+ + E E+ +L+ + E + + +L +
Sbjct: 787 QKTLGHEKLQA-----ALEELKKEKETIIKEKEQELQQLQSKSAESESALKVVQVQLEQL 841
Query: 234 ESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSC 293
+ Q + S S VAK E S L+ E+ +++ Q+ N + L N
Sbjct: 842 QQQAAASGEEG-SKTVAKLHDEISQLKSQAEETQSELKSTQS---NLEAKSKQLEAANGS 897
Query: 294 LRTELKNTCSTLD-----SDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPI 348
L E K + L+ ++ Q+ +SS + S + Q +AN+ ++K K
Sbjct: 898 LEEEAKKSGHLLEQITKLKSEVGETQAALSSCHTDVESKTKQLEAANA--ALEKVNKEYA 955
Query: 349 TSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSSEEEVSVLSKRRQSNCFDS 408
S + S ++ I D E +E S+ S S +E++ K S
Sbjct: 956 ESRAEASDLQDKVKEITDTLHAELQAERSSSSALHTKLSKFS-DEIATGHKELTSKADAW 1014
Query: 409 FECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPP 468
+ + + EKE + +Q + L+ S S K QE + +
Sbjct: 1015 SQEMLQKEKELQELRQQLQDSQDSQTKLKAEGERKEKSFEESIKNLQEEVTKAKTENLE- 1073
Query: 469 PPPPPMSFASRGNTAMVKRAPQVVELYHSLMKRDSRRDSSSGGLSDAPDVADVRSSMIGE 528
S G +K + +E+ ++ ++ + S DA +AD++ +++
Sbjct: 1074 --------LSTGTQTTIKDLQERLEITNAELQHKEKMAS-----EDAQKIADLK-TLVEA 1119
Query: 529 IENRSSHLLAIKADIETQGEFVNSLIREVN----------DAVYENIDDVVAFVKWLDDE 578
I+ ++++ A A++ T E + + E N D E + + V +K E
Sbjct: 1120 IQVANANISATNAELSTVLEVLQAEKSETNHIFELFEMEADMNSERLIEKVTGIKEELKE 1179
Query: 579 LGFLVDERAVLKHFDWPEKKADTLREAAFGYQDLKKLESEVSSYK 623
+DER K F+ E+K L++A Q L++ ES+ S K
Sbjct: 1180 THLQLDERQ--KKFEELEEK---LKQAQQSEQKLQQ-ESQTSKEK 1218
Score = 35.4 bits (80), Expect = 0.54
Identities = 58/247 (23%), Positives = 108/247 (43%), Gaps = 26/247 (10%)
Query: 138 QESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSE-QLVALSRSCGLERQEE 196
Q SKE+ ++ L E + + + +ELV+ + E S + E Q L+ S Q E
Sbjct: 1213 QTSKEKLTEIQQSLQELQDSVKQKEELVQNLEEKVRESSSIIEAQNTKLNES---NVQLE 1269
Query: 197 DKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSM-ESQLSCSDNSSESDIVAKF--- 252
+K + + +++E ++ K+L + L+ L + E+ D+ + + + K
Sbjct: 1270 NKTSCLKETQDQLLESQKKEKQLQEEAAKLSGELQQVQEANGDIKDSLVKVEELVKVLEE 1329
Query: 253 KAEASLLRL-----TNEDL------SKQVEG-LQTSRLNEVEELAYLRWVNSCLRTEL-- 298
K +A+ +L TN++L S++ EG LQ L E+L L N L+ L
Sbjct: 1330 KLQAATSQLDAQQATNKELQELLVKSQENEGNLQGESLAVTEKLQQLEQANGELKEALCQ 1389
Query: 299 -KNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSDQLSQV 357
+N L KL +V+ S S + D+ A + + + ++QLSQ+
Sbjct: 1390 KENGLKELQG-KLDESNTVLESQKKSHNEIQDKLEQAQQKERTLQEETSKL--AEQLSQL 1446
Query: 358 ECTNNSI 364
+ N +
Sbjct: 1447 KQANEEL 1453
Score = 35.0 bits (79), Expect = 0.71
Identities = 37/144 (25%), Positives = 65/144 (44%), Gaps = 3/144 (2%)
Query: 139 ESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRS-CGLERQEED 197
+ KE E+++ +L E ++ + V L +++A L+ E E L S S CG+E +
Sbjct: 500 QQKEVESRIAEQLEEEQRLRENVKYLNEQIATLQSELVSKDEALEKFSLSECGIENLRRE 559
Query: 198 KDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEAS 257
+ + E + E + +K RLSS L + +S ES+ V K E
Sbjct: 560 LELLKEENEKQAQEAQAEFTRKLAEKSVEVLRLSSELQNLKATSDSLESERVNK-TDECE 618
Query: 258 LLRLTNEDLSKQVEGLQTSRLNEV 281
+L+ +Q+ L +L+EV
Sbjct: 619 ILQTEVRMRDEQIREL-NQQLDEV 641
Score = 34.3 bits (77), Expect = 1.2
Identities = 36/171 (21%), Positives = 77/171 (44%), Gaps = 17/171 (9%)
Query: 119 RRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGL 178
+++ + + T L+ L +SKE+ AK + + K S++ +L ++ L+ E
Sbjct: 662 QKEGTEEKSTLLEKTEKELVQSKEQAAKTLNDKEQLEKQISDLKQLAEQEKLVREMTENA 721
Query: 179 SEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQK---RNLTCRLSSMES 235
Q+ LE++ ++ + + ELE + ++ E+H+Q+ +N +ES
Sbjct: 722 INQIQ-------LEKESIEQQLALKQNELEDFQKKQSESEVHLQEIKAQNTQKDFELVES 774
Query: 236 --QLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEEL 284
L E + K +A+L E+L K+ E + + E+++L
Sbjct: 775 GESLKKLQQQLEQKTLGHEKLQAAL-----EELKKEKETIIKEKEQELQQL 820
>MYSP_OPIFE (Q9BMQ6) Paramyosin (Fragment)
Length = 638
Score = 48.9 bits (115), Expect = 5e-05
Identities = 49/197 (24%), Positives = 82/197 (40%), Gaps = 6/197 (3%)
Query: 119 RRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGL 178
RR+ T L + +L ++ + E N+ ++E ++D +K E + GL
Sbjct: 90 RRRHQTALNELAAEVENLQKQKGKAEKDKNSLIMEVGNVLGQLDGALKAKQSAESKLEGL 149
Query: 179 SEQLVALSR-SCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQL 237
QL L + L+RQ D + + L E EL N+E Q NL+ SS+ES +
Sbjct: 150 DAQLNRLKGLTDDLQRQLNDLNAAKARLTSENFELLHANQEYEAQVLNLSKSRSSLESAV 209
Query: 238 SCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTE 297
S + + ++F +A L L + LQ E EE + LR S +
Sbjct: 210 DDLKRSLDDEAKSRFNLQAQL-----TSLQMDYDNLQAKYEEESEEASNLRNQVSKFNAD 264
Query: 298 LKNTCSTLDSDKLSSPQ 314
L S + + +S +
Sbjct: 265 LAAMKSKFERELMSKTE 281
>HOK2_HUMAN (Q96ED9) Hook homolog 2 (h-hook2) (hHK2)
Length = 719
Score = 48.5 bits (114), Expect = 6e-05
Identities = 45/156 (28%), Positives = 70/156 (44%), Gaps = 19/156 (12%)
Query: 118 HRRQSSTDLFTELDHMRSLLQE---SKEREAKLNAELVECRKNQSEVDELVKKVALLEEE 174
HR Q+ T L E ++ + E S ER +L A L CR+ E+ EL ++V LEE
Sbjct: 278 HRNQALTSLAQEAQALKDEMDELRQSSERAGQLEATLTSCRRRLGELRELRRQVRQLEER 337
Query: 175 KSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSME 234
+G +E+ RQ ED+ +L ++ RR +EL Q + + + +
Sbjct: 338 NAGHAER----------TRQLEDELRRAGSLRAQLEAQRRQVQELQGQLQEEA--MKAEK 385
Query: 235 SQLSCSDNSSESDIVAKFK----AEASLLRLTNEDL 266
C + + + V K K AE LR NE+L
Sbjct: 386 WLFECRNLEEKYESVTKEKERLLAERDSLREANEEL 421
>SPOF_SCHPO (Q10411) Sporulation-specific protein 15
Length = 1957
Score = 48.1 bits (113), Expect = 8e-05
Identities = 69/323 (21%), Positives = 137/323 (42%), Gaps = 50/323 (15%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECRKNQS-EVDELVKKVALLEEEKSGLSEQLVALS 186
T +D+++ L+ + +E+K++ EL E S ++ L + + + + QL LS
Sbjct: 833 TLIDNVQKLMHKHVNQESKVS-ELKEVNGKLSLDLKNLRSSLNVAISDNDQILTQLAELS 891
Query: 187 RSC-GLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLS------- 238
++ LE++ + ++LE E L N+ELH++ LT +L ES+ S
Sbjct: 892 KNYDSLEQESAQLNSGLKSLEAEKQLLHTENEELHIRLDKLTGKLKIEESKSSDLGKKLT 951
Query: 239 ---------CSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVE-ELAYLR 288
+N S+S + K++ + L +E L+ ++++EVE E L
Sbjct: 952 ARQEEISNLKEENMSQSQAITSVKSKLDETLSKSSKLEADIEHLK-NKVSEVEVERNALL 1010
Query: 289 WVNSCLRTELKNTCSTL------------DSDKLSSPQSVVSSSGDSISSFSDQCGSANS 336
N L +LKN + ++D L S SVVSS +++ S Q +
Sbjct: 1011 ASNERLMDDLKNNGENIASLQTEIEKKRAENDDLQSKLSVVSSEYENLLLISSQTNKSLE 1070
Query: 337 FNLVKKPKKWPITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSSEEEVSV 396
++QL +E ++D+ ++ ++ G +++ + +
Sbjct: 1071 ------------DKTNQLKYIEKNVQKLLDEKDQRNVELEELTSKYGKLGEENAQIKDEL 1118
Query: 397 LSKRRQSN-----CFDSFECLKE 414
L+ R++S C + + LKE
Sbjct: 1119 LALRKKSKKQHDLCANFVDDLKE 1141
Score = 46.2 bits (108), Expect = 3e-04
Identities = 87/449 (19%), Positives = 167/449 (36%), Gaps = 63/449 (14%)
Query: 2 KEEIHNNPSENKTKVSKFSDQNQPPKLQTTKTTNPNNNNHSKPRLWGAHIVKGFS----- 56
K + N + ++K++ +N LQ + + + N S L V F
Sbjct: 132 KSNLLNELKQVRSKLAALEHENGILSLQLSSSNKKDKNTSSVTTLTSEEDVSYFQKKLTN 191
Query: 57 --ADKKTKQLPTKKQQNTTTITTSDVVTNQKNVNPFVPPHSRVKRSLMGDLSCSQ----- 109
++ KQ T + +K+ S +K SL + + ++
Sbjct: 192 MESNFSAKQSEAYDLSRQLLTVTEKLDKKEKDYEKIKEDVSSIKASLAEEQASNKSLRGE 251
Query: 110 --------VHPHAFPTHRRQSSTDLFTELDHMRSLLQES--KEREAKLNAELVECRKNQS 159
V + + RQ+ L E ++ L++ E ++KL EL N S
Sbjct: 252 QERLEKLLVSSNKTVSTLRQTENSLRAECKTLQEKLEKCAINEEDSKLLEELKHNVANYS 311
Query: 160 EVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEED------------KDGSTQN--L 205
D +V K L+E+ + +SE S L + E KD T N L
Sbjct: 312 --DAIVHKDKLIEDLSTRISEFDNLKSERDTLSIKNEKLEKLLRNTIGSLKDSRTSNSQL 369
Query: 206 ELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNED 265
E E+VEL+ N+ +H Q + +LSS E +N S + +++ S +
Sbjct: 370 EEEMVELKESNRTIHSQLTDAESKLSSFE-----QENKSLKGSIDEYQNNLSSKDKMVKQ 424
Query: 266 LSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKN---------TCSTLDSDKLSSPQSV 316
+S Q+E ++S + +LA + ++K+ C S++L ++
Sbjct: 425 VSSQLEEARSSLAHATGKLAEINSERDFQNKKIKDFEKIEQDLRACLNSSSNELKEKSAL 484
Query: 317 VSSSGDSISSFSDQCGSANSFNLVKKPKK--WPITSSDQLSQVECTNNSIIDKNWIESIS 374
+ +++ +Q +K+ KK SS Q Q + N + + ++
Sbjct: 485 IDKKDQELNNLREQ---------IKEQKKVSESTQSSLQSLQRDILNEKKKHEVYESQLN 535
Query: 375 EGSNRRRHSISGSNSSEEEVSVLSKRRQS 403
E + IS S ++S L+ +++
Sbjct: 536 ELKGELQTEISNSEHLSSQLSTLAAEKEA 564
Score = 36.2 bits (82), Expect = 0.32
Identities = 102/544 (18%), Positives = 214/544 (38%), Gaps = 96/544 (17%)
Query: 129 ELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRS 188
EL+ + S + E A++ EL+ RK + +L +EKS EQL
Sbjct: 1096 ELEELTSKYGKLGEENAQIKDELLALRKKSKKQHDLCANFVDDLKEKSDALEQLT----- 1150
Query: 189 CGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDN---SSE 245
L + + + N+ L ++ +L RLS M+ LS SDN
Sbjct: 1151 -----------NEKNELIVSLEQSNSNNEALVEERSDLANRLSDMKKSLSDSDNVISVIR 1199
Query: 246 SDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEEL-----AYLRWVNS----CLRT 296
SD+V + E L+ + LS Q + R + ++ L ++ ++ S C ++
Sbjct: 1200 SDLV-RVNDELDTLKKDKDSLSTQYSEVCQDRDDLLDSLKGCEESFNKYAVSLRELCTKS 1258
Query: 297 ELKNTCS-TLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSD--- 352
E+ S LD + + + + S ++ S + + N N K +T++D
Sbjct: 1259 EIDVPVSEILDDNFVFNAGNFSELSRLTVLSLENYLDAFNQVNFKKMELDNRLTTTDAEF 1318
Query: 353 -----QLSQVECTNNSII-------------DKNWIESISEGSNRRRHSISGSNSSEEEV 394
L +++ ++ + +KN++ +E + G +++E+
Sbjct: 1319 TKVVADLEKLQHEHDDWLIQRGDLEKALKDSEKNFLRKEAEMTENIHSLEEGKEETKKEI 1378
Query: 395 SVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRAL------RIPNPPPRPSCS 448
+ LS R + N + + +++ + + L + EK +L + N + S
Sbjct: 1379 AELSSRLEDNQLATNKLKNQLDHLNQEIRL-KEDVLKEKESLIISLEESLSNQRQKESSL 1437
Query: 449 ISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVELYHSLMKRDSRRDSS 508
+ +K + E + SR N++++++ + +S D
Sbjct: 1438 LDAKNELE---------------HMLDDTSRKNSSLMEKIESI----------NSSLDDK 1472
Query: 509 SGGLSDAPD----VADVRSSMIGEIENRSSHLLAIKADIETQGEFVNSLIREVNDAV--Y 562
S L+ A + + + S + +EN S L K I+ + L E+ + Y
Sbjct: 1473 SFELASAVEKLGALQKLHSESLSLMENIKSQLQEAKEKIQVDESTIQELDHEITASKNNY 1532
Query: 563 E-NIDDVVAFVKWLDDEL----GFLVDERAVLKHFDWPEKKADTLREAAFGYQDLKKLES 617
E ++D + ++ L + + L +E++ +K EK+++ L+ + DL+ +S
Sbjct: 1533 EGKLNDKDSIIRDLSENIEQLNNLLAEEKSAVKRLS-TEKESEILQFNS-RLADLEYHKS 1590
Query: 618 EVSS 621
+V S
Sbjct: 1591 QVES 1594
Score = 35.0 bits (79), Expect = 0.71
Identities = 103/539 (19%), Positives = 214/539 (39%), Gaps = 70/539 (12%)
Query: 56 SADKKTKQLPTKKQQNTTTITTSDVVTNQKN-----VNPFVPPHSRVKRSLMGDLSCSQV 110
+++ + QL T + + T++ ++ KN N F ++ +S+M Q
Sbjct: 547 NSEHLSSQLSTLAAEKEAAVATNNELSESKNSLQTLCNAF---QEKLAKSVMQLKENEQN 603
Query: 111 HPHAFPTHRRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVAL 170
+ ++ + + E +H +++ ++ K+ +KL ++ Q E +K +
Sbjct: 604 FSSLDTSFKKLNESHQELENNH-QTITKQLKDTSSKL-------QQLQLERANFEQKEST 655
Query: 171 LEEEKSGLSEQLVALSRSC-GLERQEEDKDGSTQNLELEVVELRRLNKELHMQK------ 223
L +E + L +L+ L S L +++ED D +N++ +LR+ + L K
Sbjct: 656 LSDENNDLRTKLLKLEESNKSLIKKQEDVDSLEKNIQTLKEDLRKSEEALRFSKLEAKNL 715
Query: 224 RNLTCRLSSMESQLSCSDN---SSESDIVAKFKAEASLLRLTNEDLSKQVEGLQT-SRLN 279
R + L L N SS SD +S L ++ED+ + ++T ++ +
Sbjct: 716 REVIDNLKGKHETLEAQRNDLHSSLSDAKNTNAILSSELTKSSEDVKRLTANVETLTQDS 775
Query: 280 EVEELAYLRWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSFNL 339
+ + ++ VNS + N L D ++ QS ++ +S S C + N+
Sbjct: 776 KAMKQSFTSLVNS--YQSISNLYHELRDDHVNM-QSQNNTLLESESKLKTDCENLTQQNM 832
Query: 340 -----VKKPKKWPITSSDQLSQVECTNN--SIIDKNWIESISEGSNRRRHSISGSNSSEE 392
V+K + ++S+++ N S+ KN S++ +IS ++
Sbjct: 833 TLIDNVQKLMHKHVNQESKVSELKEVNGKLSLDLKNLRSSLNV-------AISDNDQILT 885
Query: 393 EVSVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRPSC-SISS 451
+++ LSK S +E+ES + ++ EK+ L N ++
Sbjct: 886 QLAELSKNYDS-----------LEQESAQLNSGLKSLEAEKQLLHTENEELHIRLDKLTG 934
Query: 452 KTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVELYHSLMKRDSRRDSSSGG 511
K K E S +S N + + V + + S+ ++
Sbjct: 935 KLKIEESKSSDLGKKLTARQEEISNLKEENMSQSQAITSVKSKLDETLSKSSKLEA---- 990
Query: 512 LSDAPDVADVRSSMIGEIENRSSHLLA----IKADIETQGEFVNSLIREVNDAVYENID 566
D+ +++ + E+E + LLA + D++ GE + SL E+ EN D
Sbjct: 991 -----DIEHLKNK-VSEVEVERNALLASNERLMDDLKNNGENIASLQTEIEKKRAENDD 1043
Score = 33.5 bits (75), Expect = 2.1
Identities = 31/153 (20%), Positives = 69/153 (44%), Gaps = 10/153 (6%)
Query: 123 STDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQL 182
+T E++ R + + K+ +L C + S +EL +K AL++++ L+
Sbjct: 439 ATGKLAEINSERDFQNKKIKDFEKIEQDLRACLNSSS--NELKEKSALIDKKDQELNNLR 496
Query: 183 VALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDN 242
+ ++ E S Q+L+ +++ ++ ++ Q L +++++S S++
Sbjct: 497 EQIKEQ---KKVSESTQSSLQSLQRDILNEKKKHEVYESQLNELK---GELQTEISNSEH 550
Query: 243 SSESDIVAKFKAEASLLRLTNEDLSKQVEGLQT 275
S ++ AE TN +LS+ LQT
Sbjct: 551 LSSQ--LSTLAAEKEAAVATNNELSESKNSLQT 581
>MYHB_RABIT (P35748) Myosin heavy chain, smooth muscle isoform (SMMHC)
Length = 1972
Score = 48.1 bits (113), Expect = 8e-05
Identities = 47/185 (25%), Positives = 91/185 (48%), Gaps = 28/185 (15%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECR----KNQSEVDELVKKVALLEEEKSGLSEQLV 183
T+L ++LLQE + E +L AE E R + E++E++ ++ EE+ +QL
Sbjct: 883 TQLSEEKNLLQEQLQAETELYAEAEEMRVRLAAKKQELEEILHEMEARLEEEEDRGQQLQ 942
Query: 184 A-----LSRSCGLERQEEDKDGSTQNLELEVV----ELRRL----------NKELHMQKR 224
A + LE Q E+++ + Q L+LE V ++++L N +L +++
Sbjct: 943 AERKKMAQQMLDLEEQLEEEEAARQKLQLEKVTAEAKIKKLEDDILVMDDQNNKLSKERK 1002
Query: 225 NLTCRLSSMESQLSCSDNSSESDIVAKFKAEASL----LRLTNEDLSKQ-VEGLQTSRLN 279
L R+S + + L+ + +++ K K E+ + +RL E+ S+Q +E L+
Sbjct: 1003 LLEERISDLTTNLAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQELEKLKRKMDG 1062
Query: 280 EVEEL 284
E +L
Sbjct: 1063 EASDL 1067
Score = 43.1 bits (100), Expect = 0.003
Identities = 46/187 (24%), Positives = 88/187 (46%), Gaps = 28/187 (14%)
Query: 126 LFTELDHMRSLLQESKEREAKLNAELVECRKNQ----SEVDELVKKVALLEEEKSGLSEQ 181
LFT++ + + ++ +E +AK + EL + ++ Q SE+ EL +K L EEK+ L EQ
Sbjct: 837 LFTKVKPLLQVTRQEEEMQAKED-ELQKIKERQQKAESELQELQQKHTQLSEEKNLLQEQ 895
Query: 182 LVALSR--------SCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSM 233
L A + L ++++ + +E + E ++L +++ + ++ +
Sbjct: 896 LQAETELYAEAEEMRVRLAAKKQELEEILHEMEARLEEEEDRGQQLQAERKKMAQQMLDL 955
Query: 234 ESQLSCSDNSSESDIVAKFKAEASLLRL---------TNEDLSKQVEGLQ------TSRL 278
E QL + + + + K AEA + +L N LSK+ + L+ T+ L
Sbjct: 956 EEQLEEEEAARQKLQLEKVTAEAKIKKLEDDILVMDDQNNKLSKERKLLEERISDLTTNL 1015
Query: 279 NEVEELA 285
E EE A
Sbjct: 1016 AEEEEKA 1022
Score = 38.9 bits (89), Expect = 0.049
Identities = 46/216 (21%), Positives = 88/216 (40%), Gaps = 19/216 (8%)
Query: 98 KRSLMGDLSCSQVHPHAFPTHRRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKN 157
K+ L GDL ++ + R ++ L M+ +E ++ A + ++N
Sbjct: 1619 KKKLEGDLKDLELQADSAIKGREEAIKQLLKLQAQMKDFQRELEDARASRDEIFATAKEN 1678
Query: 158 QSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNK 217
+ KK LE + L E L A ER + D + L E+
Sbjct: 1679 E-------KKAKSLEADLMQLQEDLAAA------ERARKQADLEKEELAEELASSLSGRN 1725
Query: 218 ELHMQKRNLTCRLSSMESQLSCSDNSSE--SDIVAKFKAEASLLR---LTNEDLSKQVEG 272
L +KR L R++ +E +L + E SD V K +A L T +++ E
Sbjct: 1726 ALQDEKRRLEARIAQLEEELEEEQGNMEAMSDRVRKATQQAEQLSNELATERSTAQKNES 1785
Query: 273 LQTSRLNEVEEL-AYLRWVNSCLRTELKNTCSTLDS 307
+ + +EL + L+ + ++++ K+T + L++
Sbjct: 1786 ARQQLERQNKELKSKLQEMEGAVKSKFKSTIAALEA 1821
Score = 38.1 bits (87), Expect = 0.083
Identities = 46/202 (22%), Positives = 90/202 (43%), Gaps = 20/202 (9%)
Query: 117 THRRQSSTDLFTELDHMRSLLQES--KEREAKLNAEL------VECRKNQSEVDELVKKV 168
T ++ + + +L+ R+ LQE +E EAK N E ++ ++ ++ + V
Sbjct: 1328 TRQKLNVSTKLRQLEDERNSLQEQLDEEMEAKQNLERHISTLNIQLSDSKKKLQDFASTV 1387
Query: 169 ALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTC 228
LEE K +++ +L+ +Q E+K + LE L++ +L + N
Sbjct: 1388 ESLEEGKKRFQKEIESLT------QQYEEKAAAYDKLEKTKNRLQQELDDLVVDLDNQRQ 1441
Query: 229 RLSSMESQLSCSDN--SSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNE--VEEL 284
+S++E + D + E +I +K+ E R E K+ + L +R E +E
Sbjct: 1442 LVSNLEKKQKKFDQLLAEEKNISSKYADERD--RAEAEAREKETKALSLARALEEALEAK 1499
Query: 285 AYLRWVNSCLRTELKNTCSTLD 306
L N L+ E+++ S+ D
Sbjct: 1500 EELERTNKMLKAEMEDLVSSKD 1521
Score = 35.8 bits (81), Expect = 0.41
Identities = 45/203 (22%), Positives = 82/203 (40%), Gaps = 32/203 (15%)
Query: 140 SKEREAKLNAELVECRKNQSEVDELVKKVALLEEE-KSGLSEQLVALSRS----CGLERQ 194
SKER+ L + + N +E +E K + L+ + +S +SE V L + LE+
Sbjct: 998 SKERKL-LEERISDLTTNLAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQELEKL 1056
Query: 195 EEDKDGSTQNLELEVVELRRLNKELHM---------------------QKRNLTCRLSSM 233
+ DG +L ++ +L+ EL M QK N ++ +
Sbjct: 1057 KRKMDGEASDLHEQIADLQAQIAELKMQLAKKEEELQAALARLEDETSQKNNALKKIREL 1116
Query: 234 ESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSC 293
E +S +S+ A+ KAE DL +++E L+T + ++ A + + +
Sbjct: 1117 EGHISDLQEDLDSERAARNKAEKQ-----KRDLGEELEALKTELEDTLDTTATQQELRAK 1171
Query: 294 LRTELKNTCSTLDSDKLSSPQSV 316
E+ LD + S V
Sbjct: 1172 REQEVTVLKKALDEETRSHEAQV 1194
>MYHB_CHICK (P10587) Myosin heavy chain, gizzard smooth muscle
Length = 1978
Score = 47.4 bits (111), Expect = 1e-04
Identities = 42/158 (26%), Positives = 69/158 (43%), Gaps = 21/158 (13%)
Query: 123 STDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQL 182
+T+L E + ++L + + E+ ++ V +K + EL K LE E S L EQ+
Sbjct: 1017 TTNLAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQELEKIKRKLEGESSDLHEQI 1076
Query: 183 VALSRSCG-----LERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQL 237
L L ++EE+ + LE E QK N ++ +ES +
Sbjct: 1077 AELQAQIAELKAQLAKKEEELQAALARLEDET-----------SQKNNALKKIRELESHI 1125
Query: 238 SCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQT 275
S ES+ A+ KAE DLS+++E L+T
Sbjct: 1126 SDLQEDLESEKAARNKAEKQ-----KRDLSEELEALKT 1158
Score = 45.8 bits (107), Expect = 4e-04
Identities = 43/169 (25%), Positives = 85/169 (49%), Gaps = 27/169 (15%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECR----KNQSEVDELVKKVALLEEEKSGLSEQLV 183
T+L ++LLQE + E +L AE E R + E++E++ ++ EE+ S+QL
Sbjct: 888 TQLCEEKNLLQEKLQAETELYAEAEEMRVRLAAKKQELEEILHEMEARIEEEEERSQQLQ 947
Query: 184 ALSRS-----CGLERQEEDKDGSTQNLELEVV----ELRRL----------NKELHMQKR 224
A + LE Q E+++ + Q L+LE V +++++ N +L +++
Sbjct: 948 AEKKKMQQQMLDLEEQLEEEEAARQKLQLEKVTADGKIKKMEDDILIMEDQNNKLTKERK 1007
Query: 225 NLTCRLSSMESQLSCSDNSSESDIVAKFKAEASL----LRLTNEDLSKQ 269
L R+S + + L+ + +++ K K E+ + +RL E+ S+Q
Sbjct: 1008 LLEERVSDLTTNLAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQ 1056
Score = 40.0 bits (92), Expect = 0.022
Identities = 48/202 (23%), Positives = 90/202 (43%), Gaps = 20/202 (9%)
Query: 117 THRRQSSTDLFTELDHMRSLLQES--KEREAKLNAE------LVECRKNQSEVDELVKKV 168
T ++ + T +L+ ++ LQE +E EAK N E ++ ++ ++ E V
Sbjct: 1333 TRQKLNVTTKLRQLEDDKNSLQEQLDEEVEAKQNLERHISTLTIQLSDSKKKLQEFTATV 1392
Query: 169 ALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTC 228
+EE K L ++ +L+ +Q E+K S LE L++ +L + N
Sbjct: 1393 ETMEEGKKKLQREIESLT------QQFEEKAASYDKLEKTKNRLQQELDDLVVDLDNQRQ 1446
Query: 229 RLSSMESQLSCSDN--SSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNE--VEEL 284
+S++E + D + E +I +K+ E R E K+ + L +R E +E
Sbjct: 1447 LVSNLEKKQKKFDQMLAEEKNISSKYADERD--RAEAEAREKETKALSLARALEEALEAK 1504
Query: 285 AYLRWVNSCLRTELKNTCSTLD 306
L N L+ E+++ S+ D
Sbjct: 1505 EELERTNKMLKAEMEDLVSSKD 1526
Score = 37.0 bits (84), Expect = 0.19
Identities = 60/294 (20%), Positives = 115/294 (38%), Gaps = 29/294 (9%)
Query: 4 EIHNNPSENKTKVSKFSDQNQPPKLQTTKTTNPNNNNHSKPRLWGAHIV---KGFSADKK 60
E+ +E K +++K ++ Q + T+ NN K R +HI + ++K
Sbjct: 1078 ELQAQIAELKAQLAKKEEELQAALARLEDETSQKNNALKKIRELESHISDLQEDLESEKA 1137
Query: 61 TKQLPTKKQQN---------TTTITTSDVVTNQKNVNPFVPPHSRV-KRSLMGDLSCSQV 110
+ K++++ T T D Q+ + V KR+L + +
Sbjct: 1138 ARNKAEKQKRDLSEELEALKTELEDTLDTTATQQELRAKREQEVTVLKRALEEETRTHEA 1197
Query: 111 HPHAFPTHRRQSSTDLFTELDHMRSL---LQESKEREAKLNAELVECRKNQSEVDELVKK 167
Q+ +L +L+ + L ++K+ K NA+L +E+ L +
Sbjct: 1198 QVQEMRQKHTQAVEELTEQLEQFKRAKANLDKTKQTLEKDNADLA------NEIRSLSQA 1251
Query: 168 VALLEEEKSGLSEQLVAL-SRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNL 226
+E +K L QL L S+ ER + + L++EV + L E + L
Sbjct: 1252 KQDVEHKKKKLEVQLQDLQSKYSDGERVRTELNEKVHKLQIEVENVTSLLNEAESKNIKL 1311
Query: 227 TCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTN------EDLSKQVEGLQ 274
T ++++ SQL + + + K L +L + E L ++VE Q
Sbjct: 1312 TKDVATLGSQLQDTQELLQEETRQKLNVTTKLRQLEDDKNSLQEQLDEEVEAKQ 1365
Score = 36.2 bits (82), Expect = 0.32
Identities = 26/130 (20%), Positives = 62/130 (47%), Gaps = 2/130 (1%)
Query: 137 LQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEE 196
LQ +KER+ K AEL E + +++ E +K L E+ ++ A L +++
Sbjct: 866 LQRTKERQQKAEAELKELEQKHTQLCE--EKNLLQEKLQAETELYAEAEEMRVRLAAKKQ 923
Query: 197 DKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEA 256
+ + +E + E +++L +K+ + ++ +E QL + + + + K A+
Sbjct: 924 ELEEILHEMEARIEEEEERSQQLQAEKKKMQQQMLDLEEQLEEEEAARQKLQLEKVTADG 983
Query: 257 SLLRLTNEDL 266
+ ++ ++ L
Sbjct: 984 KIKKMEDDIL 993
Score = 33.5 bits (75), Expect = 2.1
Identities = 36/180 (20%), Positives = 76/180 (42%), Gaps = 25/180 (13%)
Query: 121 QSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKK------------- 167
+ +++ T D MR +Q++++ +L E +KN++ +L ++
Sbjct: 1752 EEHSNIETMSDRMRKAVQQAEQLNNELATERATAQKNENARQQLERQNKELRSKLQEMEG 1811
Query: 168 ---------VALLEEEKSGLSEQLVALSR-SCGLERQEEDKDGSTQNLELEVVELRRLNK 217
+A LE + + L EQL +R + KD ++ L+V + R+ +
Sbjct: 1812 AVKSKFKSTIAALEAKIASLEEQLEQEAREKQAAAKTLRQKDKKLKDALLQVEDERKQAE 1871
Query: 218 ELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLT--NEDLSKQVEGLQT 275
+ Q RL ++ QL ++ S+ + K + L T N+ L ++V L++
Sbjct: 1872 QYKDQAEKGNLRLKQLKRQLEEAEEESQRINANRRKLQRELDEATESNDALGREVAALKS 1931
Score = 32.3 bits (72), Expect = 4.6
Identities = 59/305 (19%), Positives = 126/305 (40%), Gaps = 46/305 (15%)
Query: 138 QESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEED 197
+E++++ L AEL++ +++ + + K+ L EK ++E+L S + G +++
Sbjct: 1681 RENEKKAKNLEAELIQLQEDLAAAERARKQADL---EKEEMAEELA--SANSGRTSLQDE 1735
Query: 198 KDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEAS 257
K + LE + +L E H ++ R+ Q +N +A +A A
Sbjct: 1736 K----RRLEARIAQLEEELDEEHSNIETMSDRMRKAVQQAEQLNNE-----LATERATAQ 1786
Query: 258 LLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKNTCSTLDSDKLSSPQSVV 317
+ L +Q + L+ S+L E+E ++++ K+T + L++ S + +
Sbjct: 1787 KNENARQQLERQNKELR-SKLQEME---------GAVKSKFKSTIAALEAKIASLEEQLE 1836
Query: 318 SSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSDQLSQVECTNNSIIDKNWIESISEGS 377
+ + A + L +K KK D L QVE + + + +G+
Sbjct: 1837 QEAREK---------QAAAKTLRQKDKK----LKDALLQVEDERKQA--EQYKDQAEKGN 1881
Query: 378 NRRRHSISGSNSSEEEVSVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALR 437
R + +EEE ++ R+ + +E+++ + ++ A K LR
Sbjct: 1882 LRLKQLKRQLEEAEEESQRINANRR-------KLQRELDEATESNDALGREVAALKSKLR 1934
Query: 438 IPNPP 442
N P
Sbjct: 1935 RGNEP 1939
>MYHA_BOVIN (Q27991) Myosin heavy chain, nonmuscle type B (Cellular
myosin heavy chain, type B) (Nonmuscle myosin heavy
chain-B) (NMMHC-B)
Length = 1976
Score = 47.0 bits (110), Expect = 2e-04
Identities = 37/153 (24%), Positives = 78/153 (50%), Gaps = 13/153 (8%)
Query: 126 LFTELDHMRSLLQESKEREAKLNAELVECRKNQS----EVDELVKKVALLEEEKSGLSEQ 181
+FT++ + + ++ +E +AK + EL++ ++ Q+ E++E+ +K L EEK+ L+EQ
Sbjct: 837 VFTKVKPLLQVTRQEEELQAK-DEELLKVKEKQTKVEGELEEMERKHQQLLEEKNILAEQ 895
Query: 182 LVALSR--------SCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSM 233
L A + L ++++ + +LE V E N+ L +K+ + + +
Sbjct: 896 LQAETELFAEAEEMRARLAAKKQELEEILHDLESRVEEEEERNQILQNEKKKMQAHIQDL 955
Query: 234 ESQLSCSDNSSESDIVAKFKAEASLLRLTNEDL 266
E QL + + + + K AEA + ++ E L
Sbjct: 956 EEQLDEEEGARQKLQLEKVTAEAKIKKMEEEIL 988
Score = 46.2 bits (108), Expect = 3e-04
Identities = 116/601 (19%), Positives = 232/601 (38%), Gaps = 84/601 (13%)
Query: 121 QSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSE 180
Q+ T+LF E + MR+ L K+ ++ +L +S V+E ++ +L+ EK +
Sbjct: 897 QAETELFAEAEEMRARLAAKKQELEEILHDL------ESRVEEEEERNQILQNEKKKMQA 950
Query: 181 QLVALSRSCGLERQEEDKDGSTQNLELEVV----ELRRLNKELHM----------QKRNL 226
+ LE Q ++++G+ Q L+LE V +++++ +E+ + +K+ +
Sbjct: 951 HIQ------DLEEQLDEEEGARQKLQLEKVTAEAKIKKMEEEILLLEDQNSKFIKEKKLM 1004
Query: 227 TCRLSSMESQLSCSDNSSE-------------SDIVAKFKAEASLLRLTNEDLSKQVEGL 273
R++ SQL+ + ++ SD+ + K E R E ++++G
Sbjct: 1005 EDRIAECSSQLAEEEEKAKNLAKIRNKQEVMISDLEERLKKEEK-TRQELEKAKRKLDGE 1063
Query: 274 QTSRLNEVEELA----YLRWVNSCLRTELKNTCSTLDSDKL--SSPQSVVSSSGDSISSF 327
T +++ EL L+ + EL+ + D + L ++ VV I+
Sbjct: 1064 TTDLQDQIAELQAQIDELKIQVAKKEEELQGALARGDDETLHKNNALKVVRELQAQIAEL 1123
Query: 328 SDQCGSAN-SFNLVKKPKKWPITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISG 386
+ S S N +K K+ S++L ++ +D + E +R ++
Sbjct: 1124 QEDFESEKASRNKAEKQKR---DLSEELEALKTELEDTLDTTAAQ--QELRTKREQEVAE 1178
Query: 387 -SNSSEEEVSVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRP 445
+ EEE + Q L+E+ ++ F K+ L N
Sbjct: 1179 LKKALEEETKSHEAQIQDMRQRHATALEELSEQLEQAKRFKANLEKNKQGLETDNKELAC 1238
Query: 446 SCSISSKTKQE-------CSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVELYHSL 498
+ + K E AQVQ + S G+ V+ A + +L + L
Sbjct: 1239 EVKVLQQVKAESEHKRKKLDAQVQ---------ELHAKVSEGDRLRVELAEKANKLQNEL 1289
Query: 499 MKRDS-RRDSSSGGLSDAPDVADVRSSMIGEIE----------NRSSHLLAI---KADIE 544
+ ++ G+ A D A + S + E N SS + + ++ ++
Sbjct: 1290 DNVSTLLEEAEKKGIKFAKDAAGLESQLQDTQELLQEETRQKLNLSSRIRQLEEERSSLQ 1349
Query: 545 TQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVD-ERAVLKHFDWPEKKADTLR 603
Q E R + + + K +DD+LG + + E A K E + L
Sbjct: 1350 EQQEEEEEARRSLEKQLQALQAQLTDTKKKVDDDLGTIENLEEAKKKLLKDVEVLSQRLE 1409
Query: 604 EAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNCK 663
E A Y L+K ++ + DD + D + + L +K ++ L ++ R +
Sbjct: 1410 EKALAYDKLEKTKTRLQQELDDLLVDLDHQRQIVSNLEKKQKKFDQLLAEEKNISARYAE 1469
Query: 664 E 664
E
Sbjct: 1470 E 1470
Score = 37.7 bits (86), Expect = 0.11
Identities = 39/191 (20%), Positives = 83/191 (43%), Gaps = 22/191 (11%)
Query: 117 THRRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECR---------KNQSEV---DEL 164
T +Q DL +LDH R ++ ++++ K + L E + ++++E ++
Sbjct: 1423 TRLQQELDDLLVDLDHQRQIVSNLEKKQKKFDQLLAEEKNISARYAEERDRAEAEAREKE 1482
Query: 165 VKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKR 224
K ++L + L + A ++ L ED S ++ V EL + + L Q
Sbjct: 1483 TKALSLARALEEALEAREEAERQNKQLRADMEDLMSSKDDVGKNVHELEKSKRALEQQVE 1542
Query: 225 NLTCRLSSMESQLSCSDNS------SESDIVAKFKAEASLLRLTNED----LSKQVEGLQ 274
+ +L +E +L ++++ + + A+F+ + NE+ L KQV L+
Sbjct: 1543 EMRTQLEELEDELQATEDAKLRLEVNMQAMKAQFERDLQTRDEQNEEKKRLLIKQVRELE 1602
Query: 275 TSRLNEVEELA 285
+E ++ A
Sbjct: 1603 AELEDERKQRA 1613
Score = 35.4 bits (80), Expect = 0.54
Identities = 44/189 (23%), Positives = 81/189 (42%), Gaps = 20/189 (10%)
Query: 131 DHMRSLLQESKEREAKLNAELVECRKNQSEV----DELVKKVALLEEEKSGLSEQLVALS 186
D + L++ + + EL E R ++ E+ E KK+ LE E L E+L +
Sbjct: 1641 DEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQLQEELASSE 1700
Query: 187 RSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSES 246
R+ QE D+ L E+ L +KR L R++ +E +L + S+
Sbjct: 1701 RARRHAEQERDE------LADEIANSASGKSALLDEKRRLEARIAQLEEELE-EEQSNME 1753
Query: 247 DIVAKFKAEASLLRLTNEDLSKQVEGLQTS--------RLNEVEELAYLRWVNSCLRTEL 298
+ +F+ + N +L+ + Q S R N+ E A L+ + ++++
Sbjct: 1754 LLNDRFRKTTLQVDTLNTELAAERSAAQKSDNARQQLERQNK-ELKAKLQELEGAVKSKF 1812
Query: 299 KNTCSTLDS 307
K T S L++
Sbjct: 1813 KATISALEA 1821
Score = 35.0 bits (79), Expect = 0.71
Identities = 43/178 (24%), Positives = 73/178 (40%), Gaps = 33/178 (18%)
Query: 125 DLFTELDHMRS----LLQESKEREAKLN---AELVECRKN-----------QSEVDELVK 166
D EL+ R+ + +SKE E KL AE+++ ++ + E DEL
Sbjct: 1656 DYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQLQEELASSERARRHAEQERDELAD 1715
Query: 167 KVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLEL-------EVVELRRLNKEL 219
++A KS L ++ L + + EE+ + N+EL +++ LN EL
Sbjct: 1716 EIANSASGKSALLDEKRRLEAR--IAQLEEELEEEQSNMELLNDRFRKTTLQVDTLNTEL 1773
Query: 220 HMQKR------NLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVE 271
++ N +L +L E + +KFKA S L L +Q+E
Sbjct: 1774 AAERSAAQKSDNARQQLERQNKELKAKLQELEGAVKSKFKATISALEAKIGQLEEQLE 1831
>MLP2_YEAST (P40457) MLP2 protein (Myosin-like protein 2)
Length = 1679
Score = 46.6 bits (109), Expect = 2e-04
Identities = 89/424 (20%), Positives = 164/424 (37%), Gaps = 43/424 (10%)
Query: 53 KGFSADKKTKQLPTKKQQNTTTITTSDV---VTNQKNVNPFVPPHSRVKRSLMGDLSCSQ 109
K S + K L K ++T ++ ++ +T QKN+N + SL D S
Sbjct: 242 KQLSQSVEEKVLEMKNLKDTASVEKAEFSKEMTLQKNMNDLLRSQLT---SLEKDCSLRA 298
Query: 110 VHPHAFPTHRRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVA 169
+ + + R TD+ EL + L++SK +L +++C K +E +
Sbjct: 299 IEKNDDNSCRNPEHTDVIDELIDTKLRLEKSKNECQRLQNIVMDCTKE----EEATMTTS 354
Query: 170 LLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCR 229
+ L + L R L ++ K LE ++EL EL K R
Sbjct: 355 AVSPTVGKLFSDIKVLKRQ--LIKERNQKFQLQNQLEDFILELEHKTPELISFKE----R 408
Query: 230 LSSMESQLSCSDNSSESDIVAKFKAEASL--LRLTNEDLSKQVEGLQTSRLNEVEELAYL 287
S+E +L S E+ + K K E + LR + L RL+ ++ L
Sbjct: 409 TKSLEHELKRSTELLETVSLTKRKQEREITSLRQKINGCEANIHSLVKQRLDLARQVKLL 468
Query: 288 RWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSS-----GDSISSFSDQCGSANSFNLVKK 342
S ++ T S L D+L S + ++ SS DS + +++ ++ N +++
Sbjct: 469 LLNTSA----IQETASPLSQDELISLRKILESSNIVNENDSQAIITERLVEFSNVNELQE 524
Query: 343 PKKWPITS----SDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSSEEEVSVLS 398
+ +D+L E + + K ++I E + + + E +++L
Sbjct: 525 KNVELLNCIRILADKLENYEGKQDKTLQKVENQTIKEAKDAIIELENINAKMETRINILL 584
Query: 399 KRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECS 458
+ R DS++ L E+ A ++ +R +SS TK E S
Sbjct: 585 RER-----DSYKLLASTEENKANTNSVTSMEAAREKKIR------ELEAELSS-TKVENS 632
Query: 459 AQVQ 462
A +Q
Sbjct: 633 AIIQ 636
>PUMA_PARUN (O61308) 227 kDa spindle- and centromere-associated
protein (PUMA1)
Length = 1955
Score = 46.2 bits (108), Expect = 3e-04
Identities = 73/322 (22%), Positives = 140/322 (42%), Gaps = 38/322 (11%)
Query: 99 RSLMGDLSCSQVHPHAFPTHRRQSSTDLFTELDHMRSLLQE-SKEREAKLNAELVECRKN 157
R L +L + A T S + TE+ + +L + KE+++ ++++ +K
Sbjct: 682 RRLNDELDTNVKTGQAKVTSLENSIISVQTEVTKLTTLNDKLQKEKQSIMSSK----QKA 737
Query: 158 QSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGS-----TQNLELEVVEL 212
++VD L +K+ LE+E L E+ AL + RQ ++ S ++L+ + E+
Sbjct: 738 DTDVDLLKEKLRKLEQECDKLKEENKALHEDEQIARQMCKEEASRIHLLERDLKDAMTEV 797
Query: 213 RRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIV-----------AKFKAEASLLRL 261
L K+L + RL S+ SD S+I K+KA+ L+
Sbjct: 798 EELKKQLQKMDEENSERLESVLRTKISSDTVDTSEIAEYTEVKVKELREKYKADLERLQS 857
Query: 262 TNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKNTC--STLDSDKLSSPQSVVSS 319
+DL ++V+ L+ +ELA + + RTE+ + L+SD+L + + V
Sbjct: 858 NKDDLERRVQILE-------DELAERQRIVERQRTEMNDLKLEYQLESDRLRAEMATVEL 910
Query: 320 SGDS-ISSFSDQ-CGSANSFNLVKKPKKWPITSSDQLSQVECTNNSIIDK---NWIES-- 372
S + DQ A+S+ + + + I+ +++ + +++ + W E
Sbjct: 911 KYQSEVEDERDQRSRDADSWKVTSEELRSKISFMEKMLEEAKHRETVLREEATEWEEKHD 970
Query: 373 -ISEGSNRRRHSISGSNSSEEE 393
IS S + R+ I S EE
Sbjct: 971 IISNESLKLRNEIERIRSDAEE 992
Score = 41.6 bits (96), Expect = 0.008
Identities = 37/179 (20%), Positives = 78/179 (42%), Gaps = 31/179 (17%)
Query: 118 HRRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEE--- 174
H + DL +L++ R+ L KN ++++ +K++ L+ +E
Sbjct: 568 HLEDTKLDLLKDLENQRTRYDAVTNELDTLQTTFETSTKNIAQLEANIKEINLMRDEISK 627
Query: 175 -KSGLSEQLVALSRSCGLE--RQEE--------DKDGSTQNLEL-----EVVELRRLNKE 218
K L+++L ++ +E R+E+ +D Q L++ EV+ LRRLN E
Sbjct: 628 EKDSLAQKLADVTHKLEIETVRREDIQRSCVGHSEDVEKQRLQIIEYEREVMALRRLNDE 687
Query: 219 LHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSR 277
L + +++S+E+ + + E + L N+ L K+ + + +S+
Sbjct: 688 LDTNVKTGQAKVTSLENS------------IISVQTEVTKLTTLNDKLQKEKQSIMSSK 734
Score = 32.3 bits (72), Expect = 4.6
Identities = 45/203 (22%), Positives = 86/203 (42%), Gaps = 28/203 (13%)
Query: 129 ELDHM-RSLLQESKE----REAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLV 183
E DH+ R +++KE +E + EL R Q E+DE KK+ + E E++ +S +
Sbjct: 1147 EHDHLERDYREKTKEVDRLKEVEKTFELKVNRARQ-ELDEFSKKLIVTETERNAISGEAQ 1205
Query: 184 ALSRSCGLERQE-EDKDGSTQNLELEVVELRRLNKE-----LHMQKR------NLTCRLS 231
L + L +++ + K E+ R+++E +H + +L RL
Sbjct: 1206 KLDKEVQLVKEQLQYKSDEFHKALDELANAHRISEEGRVNAIHQLEARRFEIDDLKSRLE 1265
Query: 232 SMESQLSC---------SDNSSESDIVAKFKAEASLLRLTNEDLS-KQVEGLQTSRLNEV 281
+ E +L+ + + +D + +F+A S + E + +E ++ +
Sbjct: 1266 NSEQRLATLQQEYVNADKERGALNDAMRRFQATISRSVVAEEHVDVSTIETQMQKLMSRI 1325
Query: 282 EELAYLRWVNSCLRTELKNTCST 304
E + R LKN CST
Sbjct: 1326 EAIERERNEYRDSLNRLKNRCST 1348
>MYSP_ECHGR (P35417) Paramyosin
Length = 863
Score = 46.2 bits (108), Expect = 3e-04
Identities = 47/174 (27%), Positives = 73/174 (41%), Gaps = 8/174 (4%)
Query: 117 THRRQSSTDLFTELDHMRSLLQESKEREAKLNAELV-ECRKNQSEVDELVKKVALLEEEK 175
T RR+ +T + +E+ LQ+ K R K ++L+ E ++D +K A E +
Sbjct: 103 TLRRKHNT-MISEISSEVENLQKQKGRAEKDKSQLMLEIDNVLGQLDGALKAKASAESKL 161
Query: 176 SGLSEQLVALSR-SCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSME 234
GL QL L + L+RQ D + + L E EL R+N+E Q N + S +E
Sbjct: 162 EGLDSQLTRLKALTDDLQRQMADVNSAKSRLAAENFELVRVNQEYEAQVVNFSKTKSVLE 221
Query: 235 SQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLR 288
QL + + D + + L L + LQ E E LR
Sbjct: 222 GQLDDLKRAMDEDARNRLNLQTQL-----SSLQMDYDNLQARYEEEAEAAGNLR 270
>MYSP_TAESO (P35418) Paramyosin (Antigen B) (AgB)
Length = 863
Score = 45.8 bits (107), Expect = 4e-04
Identities = 46/174 (26%), Positives = 74/174 (42%), Gaps = 8/174 (4%)
Query: 117 THRRQSSTDLFTELDHMRSLLQESKEREAKLNAELV-ECRKNQSEVDELVKKVALLEEEK 175
T RR+ +T + +E+ LQ+ K R K ++L+ E ++D +K A E +
Sbjct: 103 TLRRKHNT-MISEISSEVENLQKQKGRAEKDKSQLMLEIDNVLGQLDGALKAKASAESKL 161
Query: 176 SGLSEQLVALSR-SCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSME 234
GL QL L + L+RQ D + + L E EL R+N+E Q + +++E
Sbjct: 162 EGLDSQLTRLKALTDDLQRQMADANSAKSRLAAENFELVRVNQEYEAQVVTFSKTKAALE 221
Query: 235 SQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLR 288
SQL + + D + + L L + LQ E E LR
Sbjct: 222 SQLDDLKRAMDEDARNRLSLQTQL-----SSLQMDYDNLQARYEEEAEAAGNLR 270
>MYHB_MOUSE (O08638) Myosin heavy chain, smooth muscle isoform (SMMHC)
Length = 1972
Score = 45.8 bits (107), Expect = 4e-04
Identities = 43/169 (25%), Positives = 84/169 (49%), Gaps = 27/169 (15%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECR----KNQSEVDELVKKVALLEEEKSGLSEQLV 183
T+L ++LLQE + E +L AE E R + E++E++ ++ EE+ +QL
Sbjct: 883 TQLAEEKTLLQEQLQAETELYAESEEMRVRLAAKKQELEEILHEMEARLEEEEDRRQQLQ 942
Query: 184 A-----LSRSCGLERQEEDKDGSTQNLELEVV----ELRRL----------NKELHMQKR 224
A + LE Q E+++ + Q L+LE V ++++L N +L +++
Sbjct: 943 AERKKMAQQMLDLEEQLEEEEAARQKLQLEKVTAEAKIKKLEDDILVMDDQNSKLSKERK 1002
Query: 225 NLTCRLSSMESQLSCSDNSSESDIVAKFKAEASL----LRLTNEDLSKQ 269
L R+S + + L+ + +++ K K E+ + +RL E+ S+Q
Sbjct: 1003 LLEERVSDLTTNLAEEEEKAKNLTKLKSKHESMISELEVRLKKEEKSRQ 1051
Score = 43.1 bits (100), Expect = 0.003
Identities = 45/195 (23%), Positives = 83/195 (42%), Gaps = 13/195 (6%)
Query: 123 STDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQL 182
+T+L E + ++L + + E+ ++ V +K + EL K LE + S EQ+
Sbjct: 1012 TTNLAEEEEKAKNLTKLKSKHESMISELEVRLKKEEKSRQELEKLKRKLEGDASDFHEQI 1071
Query: 183 VALSRSCG-LERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSD 241
L L+ Q K+ Q L RL++E+ QK N ++ +E +S
Sbjct: 1072 ADLQAQIAELKMQLAKKEEELQ------AALARLDEEI-AQKNNALKKIRELEGHISDLQ 1124
Query: 242 NSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKNT 301
+S+ A+ KAE DL +++E L+T + ++ A + + + E+
Sbjct: 1125 EDLDSERAARNKAEKQ-----KRDLGEELEALKTELEDTLDSTATQQELRAKREQEVTVL 1179
Query: 302 CSTLDSDKLSSPQSV 316
LD + S V
Sbjct: 1180 KKALDEETRSHEAQV 1194
Score = 40.8 bits (94), Expect = 0.013
Identities = 38/170 (22%), Positives = 82/170 (47%), Gaps = 14/170 (8%)
Query: 126 LFTELDHMRSLLQESKEREAK---LNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQL 182
LFT++ + + ++ +E +AK + +K ++E+ EL +K L EEK+ L EQL
Sbjct: 837 LFTKVKPLLQVTRQEEEMQAKEEEMQKITERQQKAETELKELEQKHTQLAEEKTLLQEQL 896
Query: 183 VALSR--------SCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSME 234
A + L ++++ + +E + E ++L +++ + ++ +E
Sbjct: 897 QAETELYAESEEMRVRLAAKKQELEEILHEMEARLEEEEDRRQQLQAERKKMAQQMLDLE 956
Query: 235 SQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEEL 284
QL + + + + K AEA + +L ++ L V Q S+L++ +L
Sbjct: 957 EQLEEEEAARQKLQLEKVTAEAKIKKLEDDIL---VMDDQNSKLSKERKL 1003
Score = 40.0 bits (92), Expect = 0.022
Identities = 45/202 (22%), Positives = 91/202 (44%), Gaps = 20/202 (9%)
Query: 117 THRRQSSTDLFTELDHMRSLLQES--KEREAKLNAEL------VECRKNQSEVDELVKKV 168
T ++ + + +L+ R+ LQ+ +E EAK N E ++ ++ ++ + +
Sbjct: 1328 TRQKLNVSTKLRQLEDERNSLQDQLDEEMEAKQNLERHVSTLNIQLSDSKKKLQDFASTI 1387
Query: 169 ALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTC 228
++EE K L +++ GL +Q E+K + LE L++ +L + N
Sbjct: 1388 EVMEEGKKRLQKEME------GLSQQYEEKAAAYDKLEKTKNRLQQELDDLVVDLDNQRQ 1441
Query: 229 RLSSMESQLSCSDN--SSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNE--VEEL 284
+S++E + D + E +I +K+ E R E K+ + L +R E +E
Sbjct: 1442 LVSNLEKKQKKFDQLLAEEKNISSKYADERD--RAEAEAREKETKALSLARALEEALEAK 1499
Query: 285 AYLRWVNSCLRTELKNTCSTLD 306
L N L+ E+++ S+ D
Sbjct: 1500 EELERTNKMLKAEMEDLVSSKD 1521
Score = 39.3 bits (90), Expect = 0.037
Identities = 64/311 (20%), Positives = 122/311 (38%), Gaps = 34/311 (10%)
Query: 98 KRSLMGDLSCSQVHPHAFPTHRRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKN 157
K+ L GDL ++ + R ++ L M+ +E + A + ++N
Sbjct: 1619 KKKLEGDLKDLELQADSAIKGREEAIKQLRKLQAQMKDFQRELDDARASRDEIFATSKEN 1678
Query: 158 QSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNK 217
+ KK LE + L E L A ER + D + L E+
Sbjct: 1679 E-------KKAKSLEADLMQLQEDLAAA------ERARKQADLEKEELAEELASSLSGRN 1725
Query: 218 ELHMQKRNLTCRLSSMESQLSCSDNSSE--SDIVAKFKAEASLLR---LTNEDLSKQVEG 272
L +KR L R++ +E +L + E SD V K +A L T +++ E
Sbjct: 1726 TLQDEKRRLEARIAQLEEELEEEQGNMEAMSDRVRKATLQAEQLSNELATERSTAQKNES 1785
Query: 273 LQTSRLNEVEEL-AYLRWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQC 331
+ + +EL + L+ V ++ +LK+T + L++ + V + +
Sbjct: 1786 ARQQLERQNKELRSKLQEVEGAVKAKLKSTVAALEAKIAQLEEQVEQEAREK-------- 1837
Query: 332 GSANSFNLVKKPKKWPITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSSE 391
A + +L +K KK + L QVE + + + + E +G+ + + +E
Sbjct: 1838 -QAATKSLKQKDKK----LKEVLLQVE--DERKMAEQYKEQAEKGNTKVKQLKRQLEEAE 1890
Query: 392 EEVSVLSKRRQ 402
EE ++ R+
Sbjct: 1891 EESQCINANRR 1901
>MYSS_RABIT (P02562) Myosin heavy chain, skeletal muscle (Fragments)
Length = 1084
Score = 45.4 bits (106), Expect = 5e-04
Identities = 106/514 (20%), Positives = 189/514 (36%), Gaps = 78/514 (15%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVAL-LEEEKSGLSEQLVALS 186
T+L+ L+ S E+E K+ +L ++ +L ++ ++ +E +K L E+L L
Sbjct: 183 TKLEQQVDDLEGSLEQEKKIRMDLERAKRKLEGDLKLAQETSMDIENDKQQLDEKLKKLE 242
Query: 187 RSCGLERQEEDKDGSTQNLEL-----EVVELRRLNK-ELHMQKRNLTCRLSSMESQLSCS 240
L+ + ED+ NL+ E +E R ++ + Q+ +L+ L + +L +
Sbjct: 243 FMTNLQSKIEDEQALMTNLQRIEELEEEIEAERASRAKAEKQRSDLSRELEEISERLEEA 302
Query: 241 DNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKN 300
++ + I K EA + K L+ + L A LR ++ EL
Sbjct: 303 GGATSAQIEMNKKREA--------EFEKMRRDLEEATLQHEATAAALRKKHADSVAELGE 354
Query: 301 TCSTLD--SDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSDQLSQVE 358
L KL +S + D ++ + A NL K + T DQLS+V+
Sbjct: 355 QIDNLQRVKQKLEKEKSELKMEIDDLAGNMETVSKAKG-NLEKMCR----TLEDQLSEVK 409
Query: 359 CTNNSIIDKNWIESISEGSNRRRHSISGSNS---SEEEVSVLSKRRQSNCF-DSFECLK- 413
+ I +S R H+ SG S E++ V R F E LK
Sbjct: 410 TKEEE--HQRLINELS-AQKARLHTESGEFSRQLDEKDAMVSQLSRGGQAFTQQIEGLKR 466
Query: 414 EIEKESVPMPLFVQQCALEKRALRIPNPPPRPSCSI---SSKTKQECSAQVQPPPPPPPP 470
++E+E+ K AL R C + + +QE A++Q
Sbjct: 467 QLEEET-----------KAKSALAHALQSSRRDCDLLREQYEEEQEAKAELQ-RAMSKAN 514
Query: 471 PPPMSFASRGNTAMVKRAPQVVELYHSLMKRDSRRDSSSGGLSDAPDVADVRSSMIGEIE 530
+ ++ T ++R ++ E L +R L DA + + +S +E
Sbjct: 515 SEVSQWRTKCETDAIQRTEELEEAKKKLAQR----------LQDAEEHVEAVNSKCASLE 564
Query: 531 NRSSHLLAIKADIETQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLK 590
L D+ E N+ + D N D V+A
Sbjct: 565 KTKQRLQNEAEDLMIDVERSNATCARM-DKKQRNFDKVLA-------------------- 603
Query: 591 HFDWPEKKADTLREAAFGYQDLKKLESEVSSYKD 624
+W K +T E ++ + L +EV K+
Sbjct: 604 --EWKHKYEETQAELEASQKESRSLSTEVFKVKN 635
Score = 42.0 bits (97), Expect = 0.006
Identities = 46/206 (22%), Positives = 83/206 (39%), Gaps = 16/206 (7%)
Query: 84 QKNVNPFVPPHSRVKRSLMGDLSCSQVHPHAFPTHRRQSSTDLFTELDHMRSLLQESKER 143
Q+N + + +L SQ + T + LDH+ +L +E+K
Sbjct: 595 QRNFDKVLAEWKHKYEETQAELEASQKESRSLSTEVFKVKNAYEESLDHLETLKRENKNL 654
Query: 144 EAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQ 203
+ +++ + ++ + EL K +++EKS L AL + G EE K Q
Sbjct: 655 QQEISDLTEQIAESAKHIHELEKVKKQIDQEKSELQ---AALEEAEGSLEHEEGKILRIQ 711
Query: 204 NLELEVV------ELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEAS 257
LEL V ++ ++E+ KRN + SM+S L S + K K E
Sbjct: 712 -LELNQVKSEIDRKIAEKDEEIDQLKRNHLRVVESMQSTLDAEIRSRNDALRIKKKMEGD 770
Query: 258 L------LRLTNEDLSKQVEGLQTSR 277
L L N ++ ++ L+ ++
Sbjct: 771 LNEMEIQLNHANRQAAEAIKNLRNTQ 796
Score = 35.8 bits (81), Expect = 0.41
Identities = 44/194 (22%), Positives = 84/194 (42%), Gaps = 18/194 (9%)
Query: 121 QSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVD-ELVKKVALLEEEKSGLS 179
Q+ D + + + L++ + EAK+ E+ E +++ E++ EL K LE+E S L
Sbjct: 51 QAEADSLADAEERQDLIKTKIQLEAKIK-EVTERAEDEEEINAELTAKKRKLEDECSELK 109
Query: 180 EQLVALSRSCG-LERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLS 238
+ + L + +E+++ + +NL E+ L +L +K+ L Q +
Sbjct: 110 KDIDDLELTLAKVEKEKHATENKVKNLTEEMAGLDETIAKLTKEKKAL-----QEAHQQT 164
Query: 239 CSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTEL 298
D +E D + + L L +QV+ L+ S E + L L +L
Sbjct: 165 LDDLQAEED-------KVNTLTKAKTKLEQQVDDLEGSLEQEKKIRMDLERAKRKLEGDL 217
Query: 299 K---NTCSTLDSDK 309
K T +++DK
Sbjct: 218 KLAQETSMDIENDK 231
>MYHB_HUMAN (P35749) Myosin heavy chain, smooth muscle isoform (SMMHC)
Length = 1972
Score = 45.4 bits (106), Expect = 5e-04
Identities = 42/169 (24%), Positives = 84/169 (48%), Gaps = 27/169 (15%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECR----KNQSEVDELVKKVALLEEEKSGLSEQLV 183
++L ++LLQE + E +L AE E R + E++E++ ++ EE+ +QL
Sbjct: 883 SQLTEEKNLLQEQLQAETELYAEAEEMRVRLAAKKQELEEILHEMEARLEEEEDRGQQLQ 942
Query: 184 A-----LSRSCGLERQEEDKDGSTQNLELEVV----ELRRL----------NKELHMQKR 224
A + LE Q E+++ + Q L+LE V ++++L N +L +++
Sbjct: 943 AERKKMAQQMLDLEEQLEEEEAARQKLQLEKVTAEAKIKKLEDEILVMDDQNNKLSKERK 1002
Query: 225 NLTCRLSSMESQLSCSDNSSESDIVAKFKAEASL----LRLTNEDLSKQ 269
L R+S + + L+ + +++ K K E+ + +RL E+ S+Q
Sbjct: 1003 LLEERISDLTTNLAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQ 1051
Score = 42.4 bits (98), Expect = 0.004
Identities = 45/195 (23%), Positives = 82/195 (41%), Gaps = 13/195 (6%)
Query: 123 STDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQL 182
+T+L E + ++L + + E+ ++ V +K + EL K LE + S EQ+
Sbjct: 1012 TTNLAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQELEKLKRKLEGDASDFHEQI 1071
Query: 183 VALSRSCG-LERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSD 241
L L+ Q K+ Q L RL+ E+ QK N ++ +E +S
Sbjct: 1072 ADLQAQIAELKMQLAKKEEELQ------AALARLDDEI-AQKNNALKKIRELEGHISDLQ 1124
Query: 242 NSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKNT 301
+S+ A+ KAE DL +++E L+T + ++ A + + + E+
Sbjct: 1125 EDLDSERAARNKAEKQ-----KRDLGEELEALKTELEDTLDSTATQQELRAKREQEVTVL 1179
Query: 302 CSTLDSDKLSSPQSV 316
LD + S V
Sbjct: 1180 KKALDEETRSHEAQV 1194
Score = 42.4 bits (98), Expect = 0.004
Identities = 35/152 (23%), Positives = 74/152 (48%), Gaps = 11/152 (7%)
Query: 126 LFTELDHMRSLLQESKEREAK---LNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQL 182
LFT++ + + ++ +E +AK L +K ++E+ EL +K + L EEK+ L EQL
Sbjct: 837 LFTKVKPLLQVTRQEEEMQAKEDELQKTKERQQKAENELKELEQKHSQLTEEKNLLQEQL 896
Query: 183 VALSR--------SCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSME 234
A + L ++++ + +E + E ++L +++ + ++ +E
Sbjct: 897 QAETELYAEAEEMRVRLAAKKQELEEILHEMEARLEEEEDRGQQLQAERKKMAQQMLDLE 956
Query: 235 SQLSCSDNSSESDIVAKFKAEASLLRLTNEDL 266
QL + + + + K AEA + +L +E L
Sbjct: 957 EQLEEEEAARQKLQLEKVTAEAKIKKLEDEIL 988
Score = 37.4 bits (85), Expect = 0.14
Identities = 46/216 (21%), Positives = 87/216 (39%), Gaps = 19/216 (8%)
Query: 98 KRSLMGDLSCSQVHPHAFPTHRRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKN 157
K+ L GDL ++ + R ++ L M+ +E ++ A + ++N
Sbjct: 1619 KKKLEGDLKDLELQADSAIKGREEAIKQLRKLQAQMKDFQRELEDARASRDEIFATAKEN 1678
Query: 158 QSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNK 217
+ KK LE + L E L A ER + D + L E+
Sbjct: 1679 E-------KKAKSLEADLMQLQEDLAAA------ERARKQADLEKEELAEELASSLSGRN 1725
Query: 218 ELHMQKRNLTCRLSSMESQLSCSDNSSE--SDIVAKFKAEASLLR---LTNEDLSKQVEG 272
L +KR L R++ +E +L + E SD V K +A L T +++ E
Sbjct: 1726 ALQDEKRRLEARIAQLEEELEEEQGNMEAMSDRVRKATQQAEQLSNELATERSTAQKNES 1785
Query: 273 LQTSRLNEVEEL-AYLRWVNSCLRTELKNTCSTLDS 307
+ + +EL + L + ++++ K+T + L++
Sbjct: 1786 ARQQLERQNKELRSKLHEMEGAVKSKFKSTIAALEA 1821
Score = 36.2 bits (82), Expect = 0.32
Identities = 45/202 (22%), Positives = 88/202 (43%), Gaps = 20/202 (9%)
Query: 117 THRRQSSTDLFTELDHMRSLLQES--KEREAKLNAEL------VECRKNQSEVDELVKKV 168
T ++ + + +L+ R+ LQ+ +E EAK N E ++ ++ ++ + V
Sbjct: 1328 TRQKLNVSTKLRQLEEERNSLQDQLDEEMEAKQNLERHISTLNIQLSDSKKKLQDFASTV 1387
Query: 169 ALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTC 228
LEE K +++ L +Q E+K + LE L++ +L + N
Sbjct: 1388 EALEEGKKRFQKEIE------NLTQQYEEKAAAYDKLEKTKNRLQQELDDLVVDLDNQRQ 1441
Query: 229 RLSSMESQLSCSDN--SSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNE--VEEL 284
+S++E + D + E +I +K+ E R E K+ + L +R E +E
Sbjct: 1442 LVSNLEKKQRKFDQLLAEEKNISSKYADERD--RAEAEAREKETKALSLARALEEALEAK 1499
Query: 285 AYLRWVNSCLRTELKNTCSTLD 306
L N L+ E+++ S+ D
Sbjct: 1500 EELERTNKMLKAEMEDLVSSKD 1521
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.128 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 83,515,878
Number of Sequences: 164201
Number of extensions: 3660341
Number of successful extensions: 32511
Number of sequences better than 10.0: 754
Number of HSP's better than 10.0 without gapping: 257
Number of HSP's successfully gapped in prelim test: 520
Number of HSP's that attempted gapping in prelim test: 24779
Number of HSP's gapped (non-prelim): 3556
length of query: 732
length of database: 59,974,054
effective HSP length: 118
effective length of query: 614
effective length of database: 40,598,336
effective search space: 24927378304
effective search space used: 24927378304
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC124968.5


