

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.11 + phase: 0
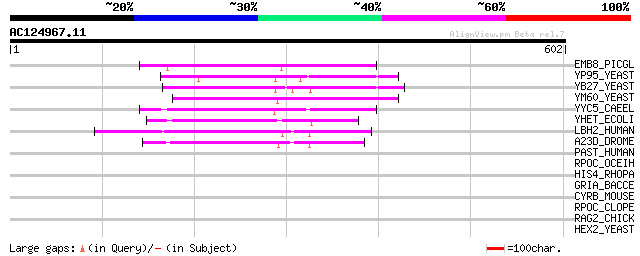
(602 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
EMB8_PICGL (Q40863) Embryogenesis-associated protein EMB8 167 7e-41
YP95_YEAST (Q02891) Hypothetical UPF0017 protein YPL095c 100 1e-20
YB27_YEAST (P38295) Hypothetical UPF0017 protein YBR177c 99 3e-20
YM60_YEAST (Q03649) Hypothetical UPF0017 protein YMR210w 94 8e-19
YYC5_CAEEL (Q18610) Hypothetical UPF0017 protein C44C1.5 in chro... 93 2e-18
YHET_ECOLI (P45524) Hypothetical UPF0017 protein yheT 77 1e-13
LBH2_HUMAN (P08910) Abhydrolase domain containing protein 2 (Pro... 74 1e-12
A23D_DROME (Q24093) UPF0017 protein CG3488 67 1e-10
PAST_HUMAN (Q9Y2W3) Proton-associated sugar transporter A (PAST-... 32 3.7
RPOC_OCEIH (Q8ETY7) DNA-directed RNA polymerase beta' chain (EC ... 32 4.8
HIS4_RHOPA (P60582) 1-(5-phosphoribosyl)-5-[(5-phosphoribosylami... 32 4.8
GRIA_BACCE (O85467) Spore germination protein gerIA 32 4.8
CYRB_MOUSE (P26955) Cytokine receptor common beta chain precurso... 32 6.2
RPOC_CLOPE (Q93R87) DNA-directed RNA polymerase beta' chain (EC ... 31 8.1
RAG2_CHICK (P25022) V(D)J recombination activating protein 2 (RA... 31 8.1
HEX2_YEAST (Q00816) HEX2 protein (SRN1 protein) (REG1 protein) 31 8.1
>EMB8_PICGL (Q40863) Embryogenesis-associated protein EMB8
Length = 457
Score = 167 bits (423), Expect = 7e-41
Identities = 96/266 (36%), Positives = 151/266 (56%), Gaps = 10/266 (3%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELDLEE----ERGLDSTLLIV-PGTPQGSMDDNIRVFV 196
+K +R CL DGG V LDWP+E + E E ++S +LI+ PG GS D ++ +
Sbjct: 113 IKSRRECLRMEDGGTVELDWPLEGEDAELWNGELPVNSPVLILLPGLTGGSDDSYVKHML 172
Query: 197 IDALKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGY 256
+ A K G+ VV N RGCA SP+TTP+ ++A+ + D+C + ++ + + VGW
Sbjct: 173 LRARKHGWHSVVFNSRGCADSPVTTPQFYSASFTKDLCQVVKHVAVRFSESNIYAVGWSL 232
Query: 257 GANMLTKYLAEVGERTPLTAATCIDNPFDLDEATRTF----PYHHVTDQKLTRGLINILQ 312
GAN+L +YL EV PL+ A + NPF+L A F +++V D+ L RGL I
Sbjct: 233 GANILVRYLGEVAGNCPLSGAVSLCNPFNLVIADEDFHKGLGFNNVYDKALARGLRQIFP 292
Query: 313 TNKALFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIP 372
+ LF+G ++ A+SVRDF+ ++ VS+GF + D+Y+ +S+ IK ++
Sbjct: 293 KHTRLFEGIEGEYNIPTVAKARSVRDFDGGLTRVSFGFQSVGDYYSNSSSSLSIKYVQTS 352
Query: 373 VLFIQSDNG-MVPAFSVPRNLIAENP 397
+L IQ+ N + P+ +P I ENP
Sbjct: 353 LLCIQASNDPIAPSRGIPWEDIKENP 378
>YP95_YEAST (Q02891) Hypothetical UPF0017 protein YPL095c
Length = 456
Score = 100 bits (249), Expect = 1e-20
Identities = 85/287 (29%), Positives = 129/287 (44%), Gaps = 32/287 (11%)
Query: 164 ELDLEEERGLDSTLLIV-PGTPQGSMDDNIRVFVIDALKRG---FFPVVMNPRGCASSPI 219
EL+ +G L++V G GS + IR D K G F VV+N RGC+ S +
Sbjct: 156 ELEKCHSKGYSYPLVVVLHGLAGGSHEPLIRALSEDLSKVGDGKFQVVVLNARGCSRSKV 215
Query: 220 TTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGANMLTKYLAEVGERTPLTAATC 279
TT R+FTA + D+ + + P + VG +GA MLT YL E G+ PL AA
Sbjct: 216 TTRRIFTALHTGDVREFLNHQKALFPQRKIYAVGTSFGAAMLTNYLGEEGDNCPLNAAVA 275
Query: 280 IDNPFDL----DEATRTFPYHHVTDQKLTRGLINILQTN-------------------KA 316
+ NP+D D+ + +H+ + LT+ L ++ N K
Sbjct: 276 LSNPWDFVHTWDKLAHDWWSNHIFSRTLTQFLTRTVKVNMNELQVPENFEVSHKPTVEKP 335
Query: 317 LFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFI 376
+F + ++ KA + +F+ + S G D +Y KAS+ N + +IKIP L I
Sbjct: 336 VFYTYTR-ENLEKAEKFTDILEFDNLFTAPSMGLPDGLTYYRKASSINRLPNIKIPTLII 394
Query: 377 QSDNGMVPAFSV-PRNLIAENPFTSLLLCSC-LSSSVMDTDTSALSW 421
+ + V +V P ENP +LLC L + D + SW
Sbjct: 395 NATDDPVTGENVIPYKQARENP--CVLLCETDLGGHLAYLDNESNSW 439
>YB27_YEAST (P38295) Hypothetical UPF0017 protein YBR177c
Length = 451
Score = 99.0 bits (245), Expect = 3e-20
Identities = 88/289 (30%), Positives = 134/289 (45%), Gaps = 29/289 (10%)
Query: 166 DLEEERGLDSTLLIV-PGTPQGSMDDNIRVFVIDALKRGFFPVV-MNPRGCASSPITTPR 223
+LEE R +D L+++ G GS + IR + + G F VV +N RGCA S ITT
Sbjct: 156 ELEELREVDLPLVVILHGLAGGSHEPIIRSLAENLSRSGRFQVVVLNTRGCARSKITTRN 215
Query: 224 LFTAADSDDICTAITYINKARPWTTLMGVGWGYGANMLTKYLAEVGERTPLTAATCIDNP 283
LFTA + DI + + P L VG +GA ML YL E G+++PL+AA + NP
Sbjct: 216 LFTAYHTMDIREFLQREKQRHPDRKLYAVGCSFGATMLANYLGEEGDKSPLSAAATLCNP 275
Query: 284 FDL---------DEATRTFPYHHVTDQKLTR------GLINILQTNKALFQGKAKG---- 324
+DL D +RT ++ Q LTR G + + + K
Sbjct: 276 WDLLLSAIRMSQDWWSRTLFSKNIA-QFLTRTVQVNMGELGVPNGSLPDHPPTVKNPSFY 334
Query: 325 ----XDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFIQSDN 380
++ KA KS R+F+E + + GF + ++Y AS+ N + I++P L I S +
Sbjct: 335 MFTPENLIKAKSFKSTREFDEVYTAPALGFPNAMEYYKAASSINRVDTIRVPTLVINSRD 394
Query: 381 GMVPAFSVPRNLIAENPFTSLLLC-SCLSSSVMDTDTSALSWCQLVTVE 428
V P +++ +NP +L C + L + D SW E
Sbjct: 395 DPVVGPDQPYSIVEKNP--RILYCRTDLGGHLAYLDKDNNSWATKAIAE 441
>YM60_YEAST (Q03649) Hypothetical UPF0017 protein YMR210w
Length = 449
Score = 94.4 bits (233), Expect = 8e-19
Identities = 68/252 (26%), Positives = 114/252 (44%), Gaps = 7/252 (2%)
Query: 177 LLIVPGTPQGSMDDNIRVFVID-ALKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICT 235
L+I+ G GS + +R V + K F V N RGC S ITTP L+ ++DI
Sbjct: 153 LIILHGLTGGSRESYVRAIVHEITTKYDFEACVFNARGCCYSAITTPLLYNGGWTNDIRY 212
Query: 236 AITYINKARPWTTLMGVGWGYGANMLTKYLAEVGERTPLTAATCIDNPFDLDEA----TR 291
+ + K P +G+ GA+++T YL E +RT + A + NPFDL +
Sbjct: 213 CVNDLRKRFPNRKFYMMGFSLGASIMTNYLGEESDRTKIECAISVSNPFDLYNSAYFINS 272
Query: 292 TFPYHHVTDQKLTRGLINILQTNKALFQGKAKGXDVGKALLAK--SVRDFEEAISMVSYG 349
T L L+ +++ + + + DV + L K +VR F+ ++ +G
Sbjct: 273 TPMGSRFYSPALGHNLLRMVRNHLSTLEENPDFKDVIEKHLKKIRTVRQFDNLLTGPMFG 332
Query: 350 FVDIEDFYTKASTRNMIKDIKIPVLFIQSDNGMVPAFSVPRNLIAENPFTSLLLCSCLSS 409
+ + E++Y AS+ I I+ P + + + + + +P + I NP+T LL S
Sbjct: 333 YKNAEEYYKNASSYKRIPGIRTPFIALHAQDDPIVGGDLPIDQIKSNPYTLLLETSTGGH 392
Query: 410 SVMDTDTSALSW 421
D S W
Sbjct: 393 VGWFKDRSGRRW 404
>YYC5_CAEEL (Q18610) Hypothetical UPF0017 protein C44C1.5 in
chromosome X
Length = 375
Score = 93.2 bits (230), Expect = 2e-18
Identities = 70/265 (26%), Positives = 117/265 (43%), Gaps = 17/265 (6%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELDLEEERGLDSTLLIVPGTP--QGSMDDNIRVF--VI 197
L + R + DGG +DW + G D T IV P GS D+ V V
Sbjct: 78 LPFTREIVEFDDGGAAGIDWLIP------EGADDTTPIVVFLPGITGSTHDSSYVLHPVK 131
Query: 198 DALKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYG 257
+A +G+ VV+NPRG + T R + AA D IN+ P +G G+ G
Sbjct: 132 EARDKGWKCVVVNPRGLGGVKLRTTRTYNAATPHDFAFIAKMINERYPDAKKLGCGFSMG 191
Query: 258 ANMLTKYLAEVGERTPLTAATCIDNPFD---LDEATRTFPYHHVTDQKLTRGLINILQTN 314
+L YLA GE L + +P+D ++ F + + + + L+++++
Sbjct: 192 GMILWNYLAMTGENADLDGGMIVSSPWDPLVASDSIECFIPQLIFNSFIAKNLVDMVRPY 251
Query: 315 KALFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVL 374
+ LF+ D + +VR F+ + + YGF +D+Y +A+ + IKIP +
Sbjct: 252 RELFKDMV---DFDEVCRCNTVRGFDRSFVIPMYGFKSCDDYYRQATLATKVDKIKIPCV 308
Query: 375 FIQS-DNGMVPAFSVPRNLIAENPF 398
+ S D+ P +P I E+ +
Sbjct: 309 TLNSVDDYFSPVECIPTLDIMESDY 333
>YHET_ECOLI (P45524) Hypothetical UPF0017 protein yheT
Length = 340
Score = 77.4 bits (189), Expect = 1e-13
Identities = 60/236 (25%), Positives = 107/236 (44%), Gaps = 14/236 (5%)
Query: 149 LSASDGGVVSLDWPVELDLEEERGLDSTLLIVPGTPQGSMDDNIRVFVIDAL-KRGFFPV 207
L DG V L W + + L+V +GS++ +++A KRG+ V
Sbjct: 50 LELPDGDFVDLAWSENPAQAQHK----PRLVVFHGLEGSLNSPYAHGLVEAAQKRGWLGV 105
Query: 208 VMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGANMLTKYLAE 267
VM+ RGC+ P R++ + +++D + ++ + VG+ G NML LA+
Sbjct: 106 VMHFRGCSGEPNRMHRIYHSGETEDASWFLRWLQREFGHAPTAAVGYSLGGNMLACLLAK 165
Query: 268 VGERTPLTAATCIDNPFDLDEATRTFPYHHVT--DQKLTRGLINILQTNKALFQGKAKGX 325
G P+ AA + PF L+ + YH + R L+N+L+ N A G
Sbjct: 166 EGNDLPVDAAVIVSAPFMLEACS----YHMEKGFSRVYQRYLLNLLKANAARKLAAYPGT 221
Query: 326 ---DVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFIQS 378
++ + + +R+F++ I+ +G+ D D+Y + S M+ I P L I +
Sbjct: 222 LPINLAQLKSVRRIREFDDLITARIHGYADAIDYYRQCSAMPMLNRIAKPTLIIHA 277
>LBH2_HUMAN (P08910) Abhydrolase domain containing protein 2
(Protein PHPS1-2)
Length = 425
Score = 73.6 bits (179), Expect = 1e-12
Identities = 74/320 (23%), Positives = 139/320 (43%), Gaps = 24/320 (7%)
Query: 93 LFASPTPFNRFVLLRCPSISFKDNERLIKDEKHYGRIRVNKREKDLEEELKY-QRVCLSA 151
L+ + +RF+L CP ++ + LI + + + + + + Y R ++
Sbjct: 44 LYFQDSGLSRFLLKSCPLLTKEYIPPLIWGKSGHIQTALYGKMGRVRSPHPYGHRKFITM 103
Query: 152 SDGGVVSLDWPVELDLEEERGLDSTLLIVPGTPQGSMDDNIRVFVIDALKRGFFPVVMNP 211
SDG + D L E G D T++I PG S IR FV A K G+ V+N
Sbjct: 104 SDGATSTFDLFEPL-AEHCVGDDITMVICPGIANHSEKQYIRTFVDYAQKNGYRCAVLNH 162
Query: 212 RGCASS-PITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGANMLTKYLAEV-G 269
G + +T+PR+FT + + + YI K P T L+ VG+ G N++ KYL E
Sbjct: 163 LGALPNIELTSPRMFTYGCTWEFGAMVNYIKKTYPLTQLVVVGFSLGGNIVCKYLGETQA 222
Query: 270 ERTPLTAATCIDNPFDLDEATRTFP--------YHHVTDQKLTRGLINILQTNKALFQGK 321
+ + + + A TF Y+ + + + IL +ALF
Sbjct: 223 NQEKVLCCVSVCQGYSALRAQETFMQWDQCRRFYNFLMADNMKK---IILSHRQALFGDH 279
Query: 322 AK------GXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLF 375
K D+ + A S+ ++ + +G+ ++++Y + S + I +P++
Sbjct: 280 VKKPQSLEDTDLSRLYTATSLMQIDDNVMRKFHGYNSLKEYYEEESCMRYLHRIYVPLML 339
Query: 376 IQ-SDNGMV--PAFSVPRNL 392
+ +D+ +V ++P++L
Sbjct: 340 VNAADDPLVHESLLTIPKSL 359
>A23D_DROME (Q24093) UPF0017 protein CG3488
Length = 398
Score = 67.4 bits (163), Expect = 1e-10
Identities = 62/256 (24%), Positives = 117/256 (45%), Gaps = 23/256 (8%)
Query: 145 QRVCLSASDGGVVSLDWPVELDLEEERGLDSTLLIVPGTPQGSMDDNIRVFVIDALKRGF 204
+RV +S DG ++ D L+ +E+ D T+ I PG S IR FV A G+
Sbjct: 84 ERVYMSLKDGSTLTYDLYQPLNEQED---DITVAICPGIANSSESVYIRTFVHLAQCNGY 140
Query: 205 FPVVMNPRGCASS-PITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGANMLTK 263
V+N G S +T+ R+FT ++D + ++++ + ++ VG+ G N++TK
Sbjct: 141 RCAVLNHIGALRSVQVTSTRIFTYGHTEDFAAMVEHLHQKYRQSRIVAVGFSLGGNLVTK 200
Query: 264 YLAEVGERTP--LTAATCIDNPFDLDEAT---------RTFPYHHVTDQKLTRGLINILQ 312
Y+ E + P + I ++ E T R F + +T+ + IL+
Sbjct: 201 YMGEDQKTKPDKVIGGISICQGYNAVEGTKWLLNWQNFRRFYLYIMTENVKS----IILR 256
Query: 313 TNKALFQGKAK---GXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDI 369
L + K + + + A ++ + +EA + Y F ++ Y +S+ I
Sbjct: 257 HRHILLSDEVKARHNLNEREIIAAATLPELDEAYTRRVYNFPSTQELYKWSSSLFYFDTI 316
Query: 370 KIPVLFIQS-DNGMVP 384
K P++FI + D+ ++P
Sbjct: 317 KKPMIFINAKDDPLIP 332
>PAST_HUMAN (Q9Y2W3) Proton-associated sugar transporter A (PAST-A)
(Deleted in neuroblastoma 5 protein) (DNb-5) (Fragment)
Length = 509
Score = 32.3 bits (72), Expect = 3.7
Identities = 17/53 (32%), Positives = 26/53 (48%)
Query: 34 SFPVPPPSPFENLFNTLISQCSSVNSIDFIAPSLGFASGSALFFSRFKSSQNS 86
S P+ PPSP + + IS+ SS+ I A S G A+ ++ F +S
Sbjct: 96 SSPISPPSPLTPKYGSFISRDSSLTGISEFASSFGTANIDSVLIDCFTGGHDS 148
>RPOC_OCEIH (Q8ETY7) DNA-directed RNA polymerase beta' chain (EC
2.7.7.6) (RNAP beta' subunit) (Transcriptase beta'
chain) (RNA polymerase beta' subunit)
Length = 1203
Score = 32.0 bits (71), Expect = 4.8
Identities = 29/90 (32%), Positives = 42/90 (46%), Gaps = 3/90 (3%)
Query: 256 YGANMLTKYLAEVGERTPLTAATCIDNPFDLDEATRTFPYHHVTDQKLTRGLINILQTNK 315
YG N+ T EVGE + AA I P RTF V +T+GL I + +
Sbjct: 903 YGRNLATGSDVEVGEAVGIIAAQSIGEP-GTQLTMRTFHTGGVAGDDITQGLPRIQELFE 961
Query: 316 ALF-QGKAKGXDV-GKALLAKSVRDFEEAI 343
A +G+A ++ G L K V+D +E +
Sbjct: 962 ARNPKGQAVITEIDGTVLEFKEVKDKQEIV 991
>HIS4_RHOPA (P60582)
1-(5-phosphoribosyl)-5-[(5-
phosphoribosylamino)methylideneamino]
imidazole-4-carboxamide isomerase (EC 5.3.1.16)
(Phosphoribosylformimino-5-aminoimidazole carboxamide
ribotide isomerase)
Length = 245
Score = 32.0 bits (71), Expect = 4.8
Identities = 23/65 (35%), Positives = 31/65 (47%), Gaps = 5/65 (7%)
Query: 454 GLLKGRH-----PLLTDIDLTIIPSKGLTLVEDARTDKNPKVGKLLELARSDAYNGYSID 508
GLLKG + L I + +I S GL +ED RT +P+ KL A +D
Sbjct: 172 GLLKGINWDATIALAEAISIPVIASGGLASIEDVRTLLSPRAKKLEGAIAGRALYDGRLD 231
Query: 509 PSEDL 513
P+E L
Sbjct: 232 PTEAL 236
>GRIA_BACCE (O85467) Spore germination protein gerIA
Length = 663
Score = 32.0 bits (71), Expect = 4.8
Identities = 26/90 (28%), Positives = 40/90 (43%), Gaps = 10/90 (11%)
Query: 480 EDARTDK--NPKVGKLLELARSDAYNGYSIDPSEDLLEGSKNDAGLHFGPQQDVQQNFEQ 537
ED+ DK NPK G + ++ + DPS+D + K + QD QQ+ +Q
Sbjct: 78 EDSSQDKQQNPKQG---DSSQDKQQSAKQKDPSQDKQQNPKQEDS-----SQDKQQSAKQ 129
Query: 538 GDMSLQVKDGPLQQTSSSGRALVGEEDAAS 567
GD S + Q SS + ++D S
Sbjct: 130 GDSSQDKQQSAKQGDSSQDKQQNAKQDEPS 159
Score = 31.6 bits (70), Expect = 6.2
Identities = 22/92 (23%), Positives = 41/92 (43%), Gaps = 3/92 (3%)
Query: 483 RTDKNPKVGKLLELARSDAYNGYSIDPSEDLLEGSKNDAGLHFGPQ---QDVQQNFEQGD 539
R K KL E + ++ D +++ K++ G + + QD QQ+ +QGD
Sbjct: 7 RKKKKSNTSKLNETDNQEQHSNNQEDDNKEQTRSMKHNKGKNNEQKDSSQDKQQSAKQGD 66
Query: 540 MSLQVKDGPLQQTSSSGRALVGEEDAASVDSE 571
S + P Q+ SS + ++ +S D +
Sbjct: 67 SSQDKQQNPKQEDSSQDKQQNPKQGDSSQDKQ 98
>CYRB_MOUSE (P26955) Cytokine receptor common beta chain precursor
(CDw131 antigen) (GM-CSF/IL-3/IL-5 receptor common
beta-chain)
Length = 896
Score = 31.6 bits (70), Expect = 6.2
Identities = 26/86 (30%), Positives = 38/86 (43%), Gaps = 9/86 (10%)
Query: 162 PVELDLEEERGLDS--TLLIVPGTPQGSMDDNIRVFVIDALKRGFFPVVMNPRGCASSPI 219
PVEL +EE+ D+ TL I G P+GSM + V D PV+ P G S+ +
Sbjct: 677 PVELSMEEQEARDNPVTLPISSGGPEGSMMASDYVTPGD-------PVLTLPTGPLSTSL 729
Query: 220 TTPRLFTAADSDDICTAITYINKARP 245
+A S +C + + P
Sbjct: 730 GPSLGLPSAQSPSLCLKLPRVPSGSP 755
>RPOC_CLOPE (Q93R87) DNA-directed RNA polymerase beta' chain (EC
2.7.7.6) (RNAP beta' subunit) (Transcriptase beta'
chain) (RNA polymerase beta' subunit)
Length = 1178
Score = 31.2 bits (69), Expect = 8.1
Identities = 19/52 (36%), Positives = 23/52 (43%), Gaps = 1/52 (1%)
Query: 256 YGANMLTKYLAEVGERTPLTAATCIDNPFDLDEATRTFPYHHVTDQKLTRGL 307
YG NM T Y +GE + AA I P RTF V +T+GL
Sbjct: 880 YGMNMATSYKINIGEAVGIVAAQSIGEP-GTQLTMRTFHTGGVAGADITQGL 930
>RAG2_CHICK (P25022) V(D)J recombination activating protein 2
(RAG-2)
Length = 528
Score = 31.2 bits (69), Expect = 8.1
Identities = 17/37 (45%), Positives = 23/37 (61%), Gaps = 1/37 (2%)
Query: 116 NERLIKDEKHYGRIRVNKREKDLEEELKYQRVCLSAS 152
N++LI D ++ +R NK E+D EEEL Q C AS
Sbjct: 335 NKQLISDANYFYILRCNKAEEDEEEELTAQ-TCSQAS 370
>HEX2_YEAST (Q00816) HEX2 protein (SRN1 protein) (REG1 protein)
Length = 1014
Score = 31.2 bits (69), Expect = 8.1
Identities = 35/131 (26%), Positives = 55/131 (41%), Gaps = 15/131 (11%)
Query: 480 EDARTDKNPKVGKLLELARSDAYNGYSIDPSEDLLEGSKNDAGLHFGPQQDVQQNFEQG- 538
+D D N G L S +G S + DL + S + A PQQ NF G
Sbjct: 750 DDQEVDDNHDEGLSLRRTLSLGKSG-STNSLYDLAQPSLSSAT----PQQKNPTNFTGGK 804
Query: 539 -----DMSLQVKDGPLQQTSSSGRALVG--EEDAASVDSEHGHVMQTAQVVTN--MLDVT 589
D L V+ PL++ SSSG + E+ +S + E + +Q+V +
Sbjct: 805 TDVDKDAQLAVRPYPLKRNSSSGNFIFNSDSEEESSSEEEQRPLPANSQLVNRSVLKGSV 864
Query: 590 MPGTLTEEQKK 600
P ++ ++KK
Sbjct: 865 TPANISSQKKK 875
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,432,190
Number of Sequences: 164201
Number of extensions: 3088652
Number of successful extensions: 6953
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 6921
Number of HSP's gapped (non-prelim): 26
length of query: 602
length of database: 59,974,054
effective HSP length: 116
effective length of query: 486
effective length of database: 40,926,738
effective search space: 19890394668
effective search space used: 19890394668
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC124967.11


