

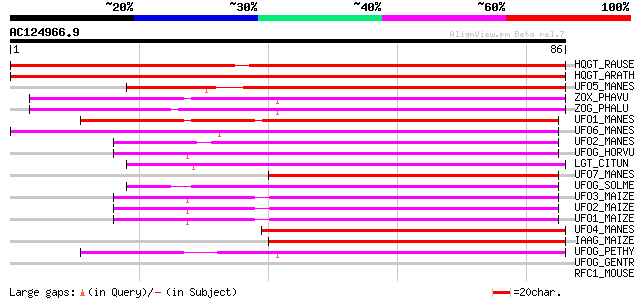
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124966.9 - phase: 0 /pseudo
(86 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.2... 100 9e-22
HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (E... 89 2e-18
UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC 2.4.1... 69 2e-12
ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40) (Ze... 68 4e-12
ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203) (... 68 5e-12
UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC 2.4.1... 64 5e-11
UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC 2.4.1... 57 8e-09
UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC 2.4.1... 57 1e-08
UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 54 7e-08
LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.21... 52 3e-07
UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC 2.4.1... 50 1e-06
UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 50 1e-06
UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 48 5e-06
UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 47 9e-06
UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 47 1e-05
UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC 2.4.1... 46 1e-05
IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (E... 45 4e-05
UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 43 1e-04
UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 40 0.001
RFC1_MOUSE (P35601) Activator 1 140 kDa subunit (Replication fac... 27 7.1
>HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.218)
(Arbutin synthase)
Length = 470
Score = 100 bits (248), Expect = 9e-22
Identities = 48/86 (55%), Positives = 60/86 (68%), Gaps = 2/86 (2%)
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGS 60
+E GP+KA EE G P VYP+GP+I + S D ECL WLD Q SVL++SFGS
Sbjct: 215 LEPGPLKALQEEDQGKPPVYPIGPLIRADSSSKVD--DCECLKWLDDQPRGSVLFISFGS 272
Query: 61 GGTLSHEQIVQLALGLELSNQKFLWV 86
GG +SH Q ++LALGLE+S Q+FLWV
Sbjct: 273 GGAVSHNQFIELALGLEMSEQRFLWV 298
>HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (EC
2.4.1.218) (Arbutin synthase)
Length = 480
Score = 89.0 bits (219), Expect = 2e-18
Identities = 46/86 (53%), Positives = 55/86 (63%)
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGS 60
+E IKA E G P VYPVGP+++ + ECL WLD Q SVLYVSFGS
Sbjct: 218 LEPNAIKALQEPGLDKPPVYPVGPLVNIGKQEAKQTEESECLKWLDNQPLGSVLYVSFGS 277
Query: 61 GGTLSHEQIVQLALGLELSNQKFLWV 86
GGTL+ EQ+ +LALGL S Q+FLWV
Sbjct: 278 GGTLTCEQLNELALGLADSEQRFLWV 303
>UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
5)
Length = 487
Score = 69.3 bits (168), Expect = 2e-12
Identities = 36/69 (52%), Positives = 47/69 (67%), Gaps = 5/69 (7%)
Query: 19 VYPVGPIIDTV-TCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLE 77
V+P+GP+ C + E L WLD+Q SV+YVSFGSGGTLS EQ+++LA GLE
Sbjct: 242 VFPIGPLRRQAGPCG----SNCELLDWLDQQPKESVVYVSFGSGGTLSLEQMIELAWGLE 297
Query: 78 LSNQKFLWV 86
S Q+F+WV
Sbjct: 298 RSQQRFIWV 306
>ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40)
(Zeatin O-beta-D-xylosyltransferase)
Length = 454
Score = 68.2 bits (165), Expect = 4e-12
Identities = 37/85 (43%), Positives = 49/85 (57%), Gaps = 3/85 (3%)
Query: 4 GPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLE--CLSWLDKQQSCSVLYVSFGSG 61
GP E +G V+ +GP + +D+ G C+ WLDKQ+ SV+YVSFG+
Sbjct: 208 GPYVELLERFNGGKEVWALGPFTP-LAVEKKDSIGFSHPCMEWLDKQEPSSVIYVSFGTT 266
Query: 62 GTLSHEQIVQLALGLELSNQKFLWV 86
L EQI +LA GLE S QKF+WV
Sbjct: 267 TALRDEQIQELATGLEQSKQKFIWV 291
>ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203)
(Trans-zeatin O-beta-D-glucosyltransferase)
Length = 459
Score = 67.8 bits (164), Expect = 5e-12
Identities = 36/85 (42%), Positives = 50/85 (58%), Gaps = 3/85 (3%)
Query: 4 GPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLE--CLSWLDKQQSCSVLYVSFGSG 61
GP E +G V+ +GP + + +D+ G C+ WLDKQ+ SV+Y+SFG+
Sbjct: 213 GPYVELLELFNGGKKVWALGPF-NPLAVEKKDSIGFRHPCMEWLDKQEPSSVIYISFGTT 271
Query: 62 GTLSHEQIVQLALGLELSNQKFLWV 86
L EQI Q+A GLE S QKF+WV
Sbjct: 272 TALRDEQIQQIATGLEQSKQKFIWV 296
>UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
1)
Length = 449
Score = 64.3 bits (155), Expect = 5e-11
Identities = 32/74 (43%), Positives = 47/74 (63%), Gaps = 2/74 (2%)
Query: 12 EGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQ 71
E +P +YPVGPI+D V + R+ N E + WLD Q SV+++ FGS G+ S +Q+ +
Sbjct: 212 ESFKDPPIYPVGPILD-VRSNGRNTNQ-EIMQWLDDQPPSSVVFLCFGSNGSFSKDQVKE 269
Query: 72 LALGLELSNQKFLW 85
+A LE S +FLW
Sbjct: 270 IACALEDSGHRFLW 283
>UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
6) (Fragment)
Length = 394
Score = 57.0 bits (136), Expect = 8e-09
Identities = 30/86 (34%), Positives = 46/86 (52%), Gaps = 1/86 (1%)
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTC-SDRDANGLECLSWLDKQQSCSVLYVSFG 59
+E + + ++ S P +YPVGPI+ +D G E + WLD Q SV+++ FG
Sbjct: 140 LESHALNSLKDDQSKIPPIYPVGPILKLSNQENDVGPEGSEIIEWLDDQPPSSVVFLCFG 199
Query: 60 SGGTLSHEQIVQLALGLELSNQKFLW 85
S G +Q ++A LE S +FLW
Sbjct: 200 SMGGFDMDQAKEIACALEQSRHRFLW 225
>UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
2) (Fragment)
Length = 346
Score = 56.6 bits (135), Expect = 1e-08
Identities = 29/69 (42%), Positives = 41/69 (59%), Gaps = 2/69 (2%)
Query: 17 PSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGL 76
P +Y VGPI+D SD E + WLD Q SV+++ FGS G+ S +Q+ ++A L
Sbjct: 113 PPLYHVGPILDVK--SDGRNTHPEIMQWLDDQPEGSVVFLCFGSMGSFSEDQLKEIAYAL 170
Query: 77 ELSNQKFLW 85
E S +FLW
Sbjct: 171 ENSGHRFLW 179
>UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1)
Length = 455
Score = 53.9 bits (128), Expect = 7e-08
Identities = 28/75 (37%), Positives = 42/75 (55%), Gaps = 6/75 (8%)
Query: 17 PSVYPVGPII------DTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIV 70
P+ P+GP T ++ A+ CL+WLD++ + SV YVSFG+ T +++
Sbjct: 237 PNCLPLGPYHLLPGAEPTADTNEAPADPHGCLAWLDRRPARSVAYVSFGTNATARPDELQ 296
Query: 71 QLALGLELSNQKFLW 85
+LA GLE S FLW
Sbjct: 297 ELAAGLEASGAPFLW 311
>LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.210)
(Limonoid glucosyltransferase) (Limonoid GTase) (LGTase)
Length = 511
Score = 52.0 bits (123), Expect = 3e-07
Identities = 28/73 (38%), Positives = 39/73 (53%), Gaps = 5/73 (6%)
Query: 19 VYPVGPIID-----TVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLA 73
+ PVGP+ T+T D EC+ WLDK+ SV+Y+SFG+ L EQ+ ++
Sbjct: 237 IKPVGPLFKNPKAPTLTVRDDCMKPDECIDWLDKKPPSSVVYISFGTVVYLKQEQVEEIG 296
Query: 74 LGLELSNQKFLWV 86
L S FLWV
Sbjct: 297 YALLNSGISFLWV 309
>UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
7) (Fragment)
Length = 287
Score = 50.1 bits (118), Expect = 1e-06
Identities = 23/45 (51%), Positives = 29/45 (64%)
Query: 41 CLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLELSNQKFLW 85
C++WLDKQ+ SV Y+SFGS T ++V LA LE S FLW
Sbjct: 90 CMAWLDKQKPASVAYISFGSVATPPPHELVALAEALEASKVPFLW 134
>UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 433
Score = 49.7 bits (117), Expect = 1e-06
Identities = 26/67 (38%), Positives = 38/67 (55%), Gaps = 3/67 (4%)
Query: 19 VYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLEL 78
V+ +GP+ V S R + C+ WLDKQ+ SV+Y+SFG+ TL +I +A LE
Sbjct: 229 VFNIGPL---VLQSSRKLDESGCIQWLDKQKEKSVVYLSFGTVTTLPPNEIGSIAEALET 285
Query: 79 SNQKFLW 85
F+W
Sbjct: 286 KKTPFIW 292
>UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-W22 allele)
Length = 471
Score = 47.8 bits (112), Expect = 5e-06
Identities = 28/75 (37%), Positives = 39/75 (51%), Gaps = 8/75 (10%)
Query: 17 PSVYPVGPII------DTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIV 70
P+ P GP D T + D +G CL+WL +Q + V YVSFG+ +++
Sbjct: 250 PNCVPFGPYHLLLAEDDADTAAPADPHG--CLAWLGRQPARGVAYVSFGTVACPRPDELR 307
Query: 71 QLALGLELSNQKFLW 85
+LA GLE S FLW
Sbjct: 308 ELAAGLEASGAPFLW 322
>UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-Mc2 allele)
Length = 471
Score = 47.0 bits (110), Expect = 9e-06
Identities = 28/75 (37%), Positives = 39/75 (51%), Gaps = 8/75 (10%)
Query: 17 PSVYPVGPII------DTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIV 70
P+ P GP D T + D +G CL+WL +Q + V YVSFG+ +++
Sbjct: 250 PNCVPFGPYHLLLAEDDADTAAPADPHG--CLAWLGRQPARGVAYVSFGTVACPRPDELR 307
Query: 71 QLALGLELSNQKFLW 85
+LA GLE S FLW
Sbjct: 308 ELAAGLEASAAPFLW 322
>UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-McC allele)
Length = 471
Score = 46.6 bits (109), Expect = 1e-05
Identities = 28/75 (37%), Positives = 39/75 (51%), Gaps = 8/75 (10%)
Query: 17 PSVYPVGPII------DTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIV 70
P+ P GP D T + D +G CL+WL +Q + V YVSFG+ +++
Sbjct: 250 PNCVPFGPYHLLLAEDDADTAAPADPHG--CLAWLGRQPARGVAYVSFGTVACPRPDELR 307
Query: 71 QLALGLELSNQKFLW 85
+LA GLE S FLW
Sbjct: 308 ELAAGLEDSGAPFLW 322
>UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC
2.4.1.91) (UDP-glucose flavonoid
3-O-glucosyltransferase 4) (Fragment)
Length = 241
Score = 46.2 bits (108), Expect = 1e-05
Identities = 22/47 (46%), Positives = 29/47 (60%)
Query: 40 ECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLELSNQKFLWV 86
E L WLD + SV+Y GS L+ Q+ +L LGLE +NQ F+WV
Sbjct: 21 ELLKWLDLWEPGSVIYACLGSISGLTSWQLAELGLGLESTNQPFIWV 67
>IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (EC
2.4.1.121) (IAA-Glu synthetase) ((Uridine
5'-diphosphate-glucose:indol-3-ylacetyl)-beta-D-glucosyl
transferase)
Length = 471
Score = 44.7 bits (104), Expect = 4e-05
Identities = 22/46 (47%), Positives = 28/46 (60%)
Query: 41 CLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLELSNQKFLWV 86
C WLD + SV YVSFGS +L + Q +LA GL + + FLWV
Sbjct: 268 CTKWLDTKPDRSVAYVSFGSLASLGNAQKEELARGLLAAGKPFLWV 313
>UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Anthocyanin rhamnosyl transferase)
Length = 473
Score = 43.1 bits (100), Expect = 1e-04
Identities = 27/77 (35%), Positives = 42/77 (54%), Gaps = 7/77 (9%)
Query: 12 EGSGNPSVYPVGPIIDTVTCSDRDANGLE--CLSWLDKQQSCSVLYVSFGSGGTLSHEQI 69
E N V+ +GP++ D + LE +WL+K + +V+Y SFGS L+ +Q+
Sbjct: 237 EAQFNKPVFLIGPVVP-----DPPSGKLEEKWATWLNKFEGGTVIYCSFGSETFLTDDQV 291
Query: 70 VQLALGLELSNQKFLWV 86
+LALGLE + F V
Sbjct: 292 KELALGLEQTGLPFFLV 308
>UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 453
Score = 39.7 bits (91), Expect = 0.001
Identities = 25/84 (29%), Positives = 39/84 (45%), Gaps = 2/84 (2%)
Query: 2 EMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSG 61
E+ PI + ++ +GP+ + + N ECL WL Q+ SV+Y+SFG+
Sbjct: 222 EIDPIITNHLRSTNQLNILNIGPLQTLSSSIPPEDN--ECLKWLQTQKESSVVYLSFGTV 279
Query: 62 GTLSHEQIVQLALGLELSNQKFLW 85
++ LA LE FLW
Sbjct: 280 INPPPNEMAALASTLESRKIPFLW 303
>RFC1_MOUSE (P35601) Activator 1 140 kDa subunit (Replication factor
C large subunit) (A1 140 kDa subunit) (RF-C 140 kDa
subunit) (Activator 1 large subunit) (A1-P145)
(Differentiation specific element binding protein)
(ISRE-binding protein)
Length = 1131
Score = 27.3 bits (59), Expect = 7.1
Identities = 17/53 (32%), Positives = 26/53 (48%), Gaps = 5/53 (9%)
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSV 53
++ P+KA +E S P V T +R +N ECL W+DK + S+
Sbjct: 532 LKNSPMKAVKKEASTCPRGLDVKE-----THGNRSSNKEECLLWVDKYKPASL 579
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.135 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,356,663
Number of Sequences: 164201
Number of extensions: 416685
Number of successful extensions: 613
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 591
Number of HSP's gapped (non-prelim): 20
length of query: 86
length of database: 59,974,054
effective HSP length: 62
effective length of query: 24
effective length of database: 49,793,592
effective search space: 1195046208
effective search space used: 1195046208
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC124966.9


