

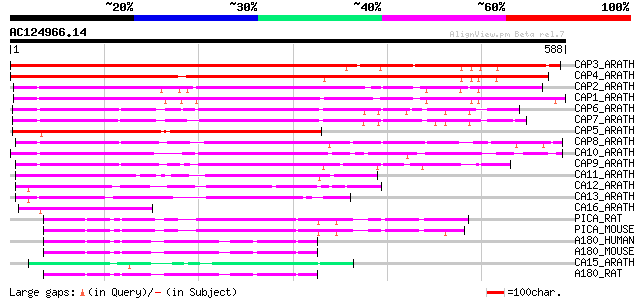
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124966.14 - phase: 0
(588 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CAP3_ARATH (Q8GX47) Putative clathrin assembly protein At4g02650 624 e-178
CAP4_ARATH (Q9SA65) Putative clathrin assembly protein At1g03050 620 e-177
CAP2_ARATH (Q8LF20) Putative clathrin assembly protein At2g25430 462 e-130
CAP1_ARATH (Q8S9J8) Putative clathrin assembly protein At4g32285 457 e-128
CAP6_ARATH (Q8VYT2) Putative clathrin assembly protein At4g25940 255 2e-67
CAP7_ARATH (Q9LVD8) Putative clathrin assembly protein At5g57200 251 4e-66
CAP5_ARATH (Q9ZVN6) Putative clathrin assembly protein At1g05020 249 1e-65
CAP8_ARATH (Q8LBH2) Putative clathrin assembly protein At2g01600 219 1e-56
CA10_ARATH (Q9LHS0) Putative clathrin assembly protein At5g35200 219 2e-56
CAP9_ARATH (P94017) Putative clathrin assembly protein At1g14910 214 5e-55
CA11_ARATH (Q9C502) Putative clathrin assembly protein At1g33340 101 5e-21
CA12_ARATH (Q9C9X5) Putative clathrin assembly protein At1g68110 91 1e-17
CA13_ARATH (Q9FRH3) Putative clathrin assembly protein At1g25240 82 5e-15
CA16_ARATH (Q8L936) Putative clathrin assembly protein At4g40080 79 3e-14
PICA_RAT (O55012) Phosphatidylinositol-binding clathrin assembly... 74 1e-12
PICA_MOUSE (Q7M6Y3) Phosphatidylinositol-binding clathrin assemb... 72 4e-12
A180_HUMAN (O60641) Clathrin coat assembly protein AP180 (Clathr... 72 5e-12
A180_MOUSE (Q61548) Clathrin coat assembly protein AP180 (Clathr... 71 7e-12
CA15_ARATH (Q9LQW4) Putative clathrin assembly protein At1g14686 70 1e-11
A180_RAT (Q05140) Clathrin coat assembly protein AP180 (Clathrin... 70 1e-11
>CAP3_ARATH (Q8GX47) Putative clathrin assembly protein At4g02650
Length = 611
Score = 624 bits (1608), Expect = e-178
Identities = 337/611 (55%), Positives = 435/611 (71%), Gaps = 34/611 (5%)
Query: 1 MSQSTLRRAIGAVKDQTSIGIAKVGS-SASIGDLQVAIVKATKHDENPAEERHIREILSL 59
M S L+RAIGAVKDQTS+G+AKVG S+S+ +L++A+VKAT+HD+ PAE+++IREIL L
Sbjct: 1 MGSSKLKRAIGAVKDQTSVGLAKVGGRSSSLTELEIAVVKATRHDDYPAEDKYIREILCL 60
Query: 60 TCYSRAFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRL 119
T YSR ++S+CV TLS+RL KT +W+VALKTL+LIQRLL DGDRAYEQEIFF+T+RGTRL
Sbjct: 61 TSYSRNYVSACVATLSRRLNKTKNWSVALKTLILIQRLLTDGDRAYEQEIFFATRRGTRL 120
Query: 120 LNMSDFRDKSKSNSWDYSSFVRTYALYLDERLEYRMQYKRGRSGRFAYDEDEEEQSRESK 179
LNMSDFRD S+S+SWDYS+FVRTYALYLDERL+YRMQ +RG+ + + E
Sbjct: 121 LNMSDFRDASQSDSWDYSAFVRTYALYLDERLDYRMQGRRGKKKSGGGGGGDGDSGEEDD 180
Query: 180 RERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVIVA 239
R K IVV+S P+ EMKT+ +F+R+QHLQ LL+RF+ACRPTG AK +R+VIVA
Sbjct: 181 HRGTSNDIRSKAIVVKSKPVAEMKTEKIFNRVQHLQQLLDRFLACRPTGNAKNNRVVIVA 240
Query: 240 LYPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWSKS 299
+YPIVKESFQ Y+++T I+G+ I+RF E+++ + KVY++FCRV KQ+DELD FY W K+
Sbjct: 241 MYPIVKESFQLYYNITEIMGVLIERFMELDIHDSIKVYEIFCRVSKQFDELDPFYGWCKN 300
Query: 300 IGIGRSSEYPEIEKVTTKKLDLMDQFIRDKSLVSQANKLITQEENNEKNEEENEVE---- 355
+ + RSSEYPE+EK+T KKLDLMD+FIRDKS ++ + + +N+ EEE++ E
Sbjct: 301 MAVARSSEYPELEKITQKKLDLMDEFIRDKSALAAQTTKSSSKRSNKSEEEESKTEYIQE 360
Query: 356 --EDMNEIKALPPPEGFNEEQVEEEIKEQDQKEEEKIV---QTEGDLLDLTDSM-TNQDY 409
ED+N IKALP PE +E+ EEE K + +K+ E++V EGDLLDLTD
Sbjct: 361 NQEDLNSIKALPAPE--QKEEEEEEEKMETKKDVEEVVSRQDQEGDLLDLTDEAGVTAGT 418
Query: 410 VGNKLALALFDELPNTTSNTIQALPWHAFDDVS-DWETTLVQSSTNLPNQKPSLGGGFDT 468
VG+ LALALFD + T S W AF+D S DWET LV+S+T L QK LGGGFDT
Sbjct: 419 VGDSLALALFDGVVGTES--ASGPGWEAFNDNSADWETDLVRSATRLSGQKSELGGGFDT 476
Query: 469 LLLDSMYNQ---KPSLQGMNGY---GSASSVAIRS----EATMLALPAPPTSRNGSQ--- 515
LLLD MY +++ Y GSASSVA S A+MLALPAPP + NG++
Sbjct: 477 LLLDGMYQYGAVNAAVKTSTAYGSSGSASSVAFGSAGSPAASMLALPAPPPTANGNRNSP 536
Query: 516 ---DPFAASMLVAPPAYVQMSEMETRQRLLAEEQAIWQQYAKNGMQGQVGFATQQQPNSN 572
DPFAAS+ VAPPAYVQM++ME +QRLL EEQ +W QY ++G QG + F QQ
Sbjct: 537 VMVDPFAASLEVAPPAYVQMNDMEKKQRLLMEEQIMWDQYNRSGRQGHMNFGQNQQ--QQ 594
Query: 573 FYMGGYQQNHY 583
+Y Y Y
Sbjct: 595 YYQLPYSMGPY 605
>CAP4_ARATH (Q9SA65) Putative clathrin assembly protein At1g03050
Length = 599
Score = 620 bits (1600), Expect = e-177
Identities = 340/601 (56%), Positives = 435/601 (71%), Gaps = 37/601 (6%)
Query: 1 MSQSTLRRAIGAVKDQTSIGIAKV-GSSASIGDLQVAIVKATKHDENPAEERHIREILSL 59
M S +RAIGAVKDQTS+G+AKV G SAS+ +L VAIVKAT+H+E PAEE++IREILSL
Sbjct: 1 MGSSKFKRAIGAVKDQTSVGLAKVNGRSASLSELDVAIVKATRHEEFPAEEKYIREILSL 60
Query: 60 TCYSRAFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRL 119
T YSR++I++CV+TLS+RL KT WTVALKTL+LIQRLL +GD+AYEQEIFF+T+RGTRL
Sbjct: 61 TSYSRSYINACVSTLSRRLNKTKCWTVALKTLILIQRLLGEGDQAYEQEIFFATRRGTRL 120
Query: 120 LNMSDFRDKSKSNSWDYSSFVRTYALYLDERLEYRMQYKRGRSGRFAY--DEDEEEQSRE 177
LNMSDFRD S+SNSWDYS+FVRTYALYLDERL++RMQ + G+ G + + DEEEQ +
Sbjct: 121 LNMSDFRDVSRSNSWDYSAFVRTYALYLDERLDFRMQARHGKRGVYCVGGEADEEEQDQA 180
Query: 178 SKRERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVI 237
+ D IVVRS P+ EMKT+ +F R+QHLQ LL+RF+ACRPTG A+ +R+VI
Sbjct: 181 AA-------DLSTAIVVRSQPIAEMKTEQIFIRIQHLQQLLDRFLACRPTGNARNNRVVI 233
Query: 238 VALYPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWS 297
VALYPIVKESFQ Y+D+T I+GI I+RF E+++P+ KVYD+FCRV KQ++ELD FYSW
Sbjct: 234 VALYPIVKESFQIYYDVTEIMGILIERFMELDIPDSIKVYDIFCRVSKQFEELDQFYSWC 293
Query: 298 KSIGIGRSSEYPEIEKVTTKKLDLMDQFIRDKSLV-----SQANKLITQEENNE-KNEEE 351
K++GI RSSEYPEIEK+T KKLDLMD+FIRDKS + S++ K E+++E + EE
Sbjct: 294 KNMGIARSSEYPEIEKITQKKLDLMDEFIRDKSALEHTKQSKSVKSEADEDDDEARTEEV 353
Query: 352 NEVEEDMNEIKALPPPEGFNEEQVEEEIKEQDQKEEEKIVQTEGDLLDLTDSMTNQ-DYV 410
NE +EDMN IKALP P E+ V+ E + +++ EK + GDLLDL ++ +
Sbjct: 354 NEEQEDMNAIKALPEPPPKEEDDVKPEEEAKEEVIIEKKQEEMGDLLDLGNTNGGEAGQA 413
Query: 411 GNKLALALFDELPNTTSNTIQALPWHAF-DDVSDWETTLVQSSTNLPNQKPSLGGGFDTL 469
G+ LALALFD + S + W AF DD +DWET LVQ++TNL QK LGGGFD L
Sbjct: 414 GDSLALALFDGPYASGSGSESGPGWEAFKDDSADWETALVQTATNLSGQKSELGGGFDML 473
Query: 470 LLDSMYNQ---KPSLQGMNGY---GSASSVAI----RSEATMLALPAPPTSRNGS----- 514
LL+ MY +++ Y GSASS+A R ATMLALPAP T+ +
Sbjct: 474 LLNGMYQHGAVNAAVKTSTAYGASGSASSMAFGSAGRPAATMLALPAPSTANGNAGNINS 533
Query: 515 ---QDPFAASMLVAPPAYVQMSEMETRQRLLAEEQAIWQQYAKNGMQGQVGF-ATQQQPN 570
DPFAAS+ VAPPAYVQM++ME +QR+L EEQ +W QY+++G QG + Q QP
Sbjct: 534 PVPMDPFAASLEVAPPAYVQMNDMEKKQRMLMEEQMMWDQYSRDGRQGHMNLRQNQNQPY 593
Query: 571 S 571
S
Sbjct: 594 S 594
>CAP2_ARATH (Q8LF20) Putative clathrin assembly protein At2g25430
Length = 653
Score = 462 bits (1190), Expect = e-130
Identities = 273/636 (42%), Positives = 385/636 (59%), Gaps = 93/636 (14%)
Query: 5 TLRRAIGAVKDQTSIGIAKVGSSASIGDLQVAIVKATKHDENPAEERHIREILSLTCYSR 64
++R+AIGAVKDQTSIGIAKV S+ + DL+VAIVKAT HD++PA E++IREIL+LT SR
Sbjct: 4 SIRKAIGAVKDQTSIGIAKVASNMA-PDLEVAIVKATSHDDDPASEKYIREILNLTSLSR 62
Query: 65 AFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRLLNMSD 124
+I +CV ++S+RL KT W VALK L+L+ RLL +GD +++EI +ST+RGTR+LNMSD
Sbjct: 63 GYILACVTSVSRRLSKTRDWVVALKALMLVHRLLNEGDPIFQEEILYSTRRGTRMLNMSD 122
Query: 125 FRDKSKSNSWDYSSFVRTYALYLDERLEYRMQYKR--------GRSGRFAYDEDEEEQSR 176
FRD++ S+SWD+S+FVRTYA YLD+RLE + ++ G S + ++D + R
Sbjct: 123 FRDEAHSSSWDHSAFVRTYAGYLDQRLELALFERKSGVSVNSGGNSSHHSNNDDRYGRGR 182
Query: 177 ES--------------------------------KRER-YRERD----------RDKEIV 193
+ KR R Y + RD++ V
Sbjct: 183 DDFRSPPPRSYDYENGGGGGSDFRGDNNGYGGVPKRSRSYGDMTEMGGGGGGGGRDEKKV 242
Query: 194 VRSTPLREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVIVALYPIVKESFQTYHD 253
V TPLREM + +F +M HLQ LL+RF++ RPTG AK RM+++ALYP+V+ESF+ Y D
Sbjct: 243 V--TPLREMTPERIFGKMGHLQRLLDRFLSLRPTGLAKNSRMILIALYPVVRESFKLYAD 300
Query: 254 MTSILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWSKSIGIGRSSEYPEIEK 313
+ +L + +D+F +ME + K +D + KQ DEL FY+W K G+ RSSEYPE+++
Sbjct: 301 ICEVLAVLLDKFFDMEYSDCVKAFDAYASAAKQIDELIAFYNWCKETGVARSSEYPEVQR 360
Query: 314 VTTKKLDLMDQFIRDKSLVSQANKLITQEENNEKNEEENEVEEDMNEIKALPPPEGFNEE 373
+T+K L+ +++F+RD++ ++ + + E EE E E DMNEIKALPPPE +
Sbjct: 361 ITSKLLETLEEFVRDRAKRGKSPER-KEIEAPPPVVEEEEPEPDMNEIKALPPPENYTPP 419
Query: 374 QVEEEIKEQDQKEEEKIVQTEGDLLDLTDSMTNQDYVGNKLALALFDELPNTTSNTIQAL 433
E + + EK TE DL++L + D GNK ALALF P
Sbjct: 420 PPPE-----PEPQPEKPQFTE-DLVNLREDEVTADDQGNKFALALFAGPPGNNGK----- 468
Query: 434 PWHAFDD---------------VSDWETTLVQSSTNLPNQKPSLGGGFDTLLLDSMYN-- 476
W AF +DWE LV++++NL Q +LGGGFD LLL+ MY+
Sbjct: 469 -WEAFSSNGVTSAWQNPAAEPGKADWELALVETTSNLEKQTAALGGGFDNLLLNGMYDQG 527
Query: 477 ---QKPSLQGMNGYGSASSVAI----RSEATMLALPAPP-TSRNGSQDPFAASMLVAPPA 528
Q S + G GSASSVA+ ++ +LALPAP T +QDPFAAS+ + PP+
Sbjct: 528 MVRQHVSTSQLTG-GSASSVALPLPGKTNNQVLALPAPDGTVEKVNQDPFAASLTIPPPS 586
Query: 529 YVQMSEMETRQRLLAEEQAIWQQYAKNGMQGQVGFA 564
YVQM+EME +Q LL++EQ +WQQY ++GM+GQ A
Sbjct: 587 YVQMAEMEKKQYLLSQEQQLWQQYQRDGMRGQASLA 622
>CAP1_ARATH (Q8S9J8) Putative clathrin assembly protein At4g32285
Length = 635
Score = 457 bits (1177), Expect = e-128
Identities = 268/645 (41%), Positives = 384/645 (58%), Gaps = 77/645 (11%)
Query: 5 TLRRAIGAVKDQTSIGIAKVGSSASIGDLQVAIVKATKHDENPAEERHIREILSLTCYSR 64
++R+AIG VKDQTSIGIAKV S+ + DL+VAIVKAT HD++ + +++IREILSLT SR
Sbjct: 4 SMRKAIGVVKDQTSIGIAKVASNMA-PDLEVAIVKATSHDDDQSSDKYIREILSLTSLSR 62
Query: 65 AFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRLLNMSD 124
++ +CV ++S+RL KT W VALK L+L+ RLL +GD +++EI ++T+RGTR+LNMSD
Sbjct: 63 GYVHACVTSVSRRLKKTRDWIVALKALMLVHRLLNEGDPLFQEEILYATRRGTRILNMSD 122
Query: 125 FRDKSKSNSWDYSSFVRTYALYLDERLEYRMQYKRGRSG------RFAYDEDEEEQSRES 178
FRD++ S+SWD+S+FVRTYA YLD+RLE + +RGR+G + +D +SR+
Sbjct: 123 FRDEAHSSSWDHSAFVRTYASYLDQRLELALFERRGRNGGGSSSSHQSNGDDGYNRSRDD 182
Query: 179 KR------------------ERYRERDRDKEIVVRS-----TPLREMKTDDLFSRMQHLQ 215
R +R R EI R TPLREM + +F +M HLQ
Sbjct: 183 FRSPPPRTYDYETGNGFGMPKRSRSFGDVNEIGAREEKKSVTPLREMTPERIFGKMGHLQ 242
Query: 216 LLLERFMACRPTGRAKTHRMVIVALYPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYK 275
LL+RF++CRPTG AK RM+++A+YP+VKESF+ Y D+ +L + +D+F +ME + K
Sbjct: 243 RLLDRFLSCRPTGLAKNSRMILIAMYPVVKESFRLYADICEVLAVLLDKFFDMEYTDCVK 302
Query: 276 VYDVFCRVGKQYDELDLFYSWSKSIGIGRSSEYPEIEKVTTKKLDLMDQFIRDKSLVSQA 335
+D + KQ DEL FY W K G+ RSSEYPE++++T+K L+ +++F+RD+ ++
Sbjct: 303 AFDAYASAAKQIDELIAFYHWCKDTGVARSSEYPEVQRITSKLLETLEEFVRDR---AKR 359
Query: 336 NKLITQEENNEKNEEENEVEE--DMNEIKALPPPEGFNEEQVEEEIKEQDQKEEEKIVQT 393
K ++E VEE DMNEIKALPPPE + Q Q
Sbjct: 360 AKSPERKEIEAPPAPAPPVEEPVDMNEIKALPPPENHTPPPPPAPEPKPQQP------QV 413
Query: 394 EGDLLDLTDSMTNQDYVGNKLALALFDELPNTTSNTIQALPWHAFDD------------- 440
DL++L + + D GNK ALALF P W AF
Sbjct: 414 TDDLVNLREDDVSGDDQGNKFALALFAGPPANNGK------WEAFSSDNNVTSAWQNPAV 467
Query: 441 ---VSDWETTLVQSSTNLPNQKPSLGGGFDTLLLDSMYNQKPSLQGMNGY----GSASSV 493
+DWE LV++++NL +QK ++GGG D LLL+ MY+Q Q ++ GS+SSV
Sbjct: 468 ELGKADWELALVETASNLEHQKAAMGGGLDPLLLNGMYDQGAVRQHVSTSELTGGSSSSV 527
Query: 494 AI----RSEATMLALPAPP-TSRNGSQDPFAASMLVAPPAYVQMSEMETRQRLLAEEQAI 548
A+ + + +LALPAP T + +QDPFAAS+ + PP+YVQM+EM+ +Q LL +EQ +
Sbjct: 528 ALPLPGKVNSHILALPAPDGTVQKVNQDPFAASLTIPPPSYVQMAEMDKKQYLLTQEQQL 587
Query: 549 WQQYAKNGMQGQVGFATQQQPNSNFYMG-----GYQQNHYGNYYH 588
WQQY + GM+GQ A + G G + G YY+
Sbjct: 588 WQQYQQEGMRGQASLAKMNTAQTAMPYGMPPVNGMGPSPMGYYYN 632
>CAP6_ARATH (Q8VYT2) Putative clathrin assembly protein At4g25940
Length = 601
Score = 255 bits (652), Expect = 2e-67
Identities = 185/570 (32%), Positives = 294/570 (51%), Gaps = 69/570 (12%)
Query: 4 STLRRAIGAVKDQTSIGIAKVGSSASIGDLQVAIVKATKHDENPAEERHIREILSLTCY- 62
++ R+A+GA+KD T++ IAKV S DL VAIVKAT H E+ +ERHIR I S T
Sbjct: 5 NSFRKAVGAIKDSTTVSIAKVNSEFK--DLDVAIVKATNHVESAPKERHIRRIFSATSVV 62
Query: 63 -SRAFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRLLN 121
RA ++ C++ L+KRL KT +W VA+K L++I R L +GD + +E+ + RG +L
Sbjct: 63 QPRADVAYCIHALAKRLSKTRNWVVAIKVLIVIHRTLREGDPTFREELLNYSHRG-HILR 121
Query: 122 MSDFRDKSKSNSWDYSSFVRTYALYLDERLE-YRMQYKRGRSGRFAYDEDEEEQSRESKR 180
+S+F+D + +WD S+++RTYAL+L+ERLE YR+ YD + E + S
Sbjct: 122 ISNFKDDTSPLAWDCSAWIRTYALFLEERLECYRV---------LKYDIEAERLPKGSGA 172
Query: 181 ERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVIVAL 240
+ D + R+ R + ++L ++ LQ LL R + C+P G A ++ ++ AL
Sbjct: 173 SS-KNVDFNASQTYRT---RMLSDEELLEQLPALQQLLYRLIGCQPEGSAYSNYLIQYAL 228
Query: 241 YPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWSKSI 300
++KESF+ Y + + +D F EM + K +++ R G+Q + L FY + K +
Sbjct: 229 ALVLKESFKIYCAINDGIINLVDMFFEMSRHDAVKALNIYKRAGQQAENLADFYEYCKGL 288
Query: 301 GIGRSSEYPEIEKVTTKKLDLMDQFIRDKSLVSQANKLITQEENNEKNEEENEVEEDMNE 360
+ R+ ++P + + L M+ +I++ Q+ + + E EK EEE E EE +
Sbjct: 289 ELARNFQFPTLRQPPPSFLATMEDYIKE---APQSGSVQKKLEYQEKEEEEQEEEEAEHS 345
Query: 361 IKALPPPEGFNEEQ--------VEEEIKEQDQKEEEK-----IVQTEGDLLDLTDSMTNQ 407
++ P E N+++ +EEE ++Q++ EEE ++ T+ DLL L +
Sbjct: 346 VQPEEPAEADNQKENSEGDQPLIEEEEEDQEKIEEEDAKPSFLIDTD-DLLGLNEINPKA 404
Query: 408 DYV--GNKLALALF---DELPNTTSNTIQALPWHAFDDVSDWETTLVQSSTNLPNQKP-- 460
+ N LALA++ E P SN + + S WE LV N N P
Sbjct: 405 AEIEDRNALALAIYPPGHEAPG-PSNILSLIETGG----SGWELALVTPQNNNNNNNPRP 459
Query: 461 ----SLGGGFDTLLLDSMYNQKPSLQGMN------GYGSASSVAIRSEATMLALPAPPTS 510
L GGFD LLLDS+Y + + + G+G + A APP
Sbjct: 460 APNTKLAGGFDNLLLDSLYEDDSARRQIQLTNAGYGHGGIDTTA-----------APPNP 508
Query: 511 RNGSQDPFAASMLVAPPAYVQMSEMETRQR 540
QDPFA S +APP VQM+ + +Q+
Sbjct: 509 FQMQQDPFAMSNNIAPPTNVQMAMQQQQQQ 538
>CAP7_ARATH (Q9LVD8) Putative clathrin assembly protein At5g57200
Length = 591
Score = 251 bits (641), Expect = 4e-66
Identities = 186/572 (32%), Positives = 298/572 (51%), Gaps = 66/572 (11%)
Query: 4 STLRRAIGAVKDQTSIGIAKVGSSASIGDLQVAIVKATKHDENPAEERHIREILSLTCY- 62
++ R+A GA+KD T++G+AKV S DL +AIVKAT H E+P +ERH+R+I S T
Sbjct: 5 TSFRKAYGALKDTTTVGLAKVNSEFK--DLDIAIVKATNHVESPPKERHVRKIFSATSVI 62
Query: 63 -SRAFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRLLN 121
RA ++ C++ LSKRL KT +W VA+K L++I R L +GD + +E+ + R +L
Sbjct: 63 QPRADVAYCIHALSKRLSKTRNWVVAMKVLIVIHRTLREGDPTFREELLNYSHR-RHILR 121
Query: 122 MSDFRDKSKSNSWDYSSFVRTYALYLDERLE-YRMQYKRGRSGRFAYDEDEEEQSRESKR 180
+S+F+D + +WD S++VRTYAL+L+ERLE YR+ YD + E + S
Sbjct: 122 ISNFKDDTSPLAWDCSAWVRTYALFLEERLECYRV---------LKYDIEAERLPKASGA 172
Query: 181 ERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVIVAL 240
R R + +DL ++ LQ LL R + C+P G A ++ ++ AL
Sbjct: 173 ASKTHRTR------------MLSGEDLLEQLPALQQLLYRLIGCQPEGAAYSNYLIQYAL 220
Query: 241 YPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWSKSI 300
++KESF+ Y + + +D F EM + K +++ R G+Q + L FY + K +
Sbjct: 221 ALVLKESFKIYCAINDGIINLVDMFFEMSRHDAVKALNIYKRAGQQAENLAEFYDYCKGL 280
Query: 301 GIGRSSEYPEIEKVTTKKLDLMDQFIRDKSLVSQANKLITQEENNEKNEEENEVEEDMNE 360
+ R+ ++P + + L M+++I++ Q+ + + E EK EEE E EE+ E
Sbjct: 281 ELARNFQFPTLRQPPPSFLATMEEYIKE---APQSGSVQKKLEYQEKEEEEQEQEEEQPE 337
Query: 361 IKALPPPEGFNEE-------QVEEEIKEQDQKEEEK---IVQTEGDLLDLTDSMTNQDYV 410
A + N E + EEE KE+ + EE K ++ T+ DLL L + +
Sbjct: 338 EPAEEENQNENTENDQPLIEEEEEEPKEEIEVEEAKPSPLIDTD-DLLGLHEINPKAAEI 396
Query: 411 --GNKLALALFDELPNTT--SNTIQALPWHAFDDVSDWETTLV---QSSTNLPNQKP--- 460
N +LA++ T+ SN++ + S WE LV ++ N N +P
Sbjct: 397 EQNNAFSLAIYPPGHETSAPSNSLSLIEAGG----SGWELALVTPQNNNNNNNNPRPVIA 452
Query: 461 -SLGGGFDTLLLDSMYNQKPSLQGMN----GYGSASSVAIRSEATMLALPAPPTSRNGSQ 515
LGGGFD LLLDS+Y + + + GYG ++ + A+ + P Q
Sbjct: 453 TKLGGGFDNLLLDSLYEDDTARRQIQLTNAGYGFGATAIPGALAS-----SNPNPFGVQQ 507
Query: 516 DPFAASMLVAPPAYVQMSEMETRQRLLAEEQA 547
DPFA S +APP VQM+ M+ +Q ++ Q+
Sbjct: 508 DPFAMSNNMAPPTNVQMA-MQQQQMMMMNNQS 538
>CAP5_ARATH (Q9ZVN6) Putative clathrin assembly protein At1g05020
Length = 653
Score = 249 bits (637), Expect = 1e-65
Identities = 136/333 (40%), Positives = 211/333 (62%), Gaps = 11/333 (3%)
Query: 4 STLRRAIGAVKDQTSIGIAKVGSSASIGD----LQVAIVKATKHDEN-PAEERHIREILS 58
S L++AIGAVKDQTSI +AKV + A+ G L+VAI+KAT HDE P ++R + EIL
Sbjct: 3 SKLKKAIGAVKDQTSISLAKVANGATGGGDLTTLEVAILKATSHDEEVPIDDRLVTEILG 62
Query: 59 LTCYSRAFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTR 118
+ ++ +SC + +R+ +T +W VALK+LVL+ R+ DGD + +E+ + +RG +
Sbjct: 63 IISSKKSHAASCAAAIGRRIGRTRNWIVALKSLVLVLRIFQDGDPYFPREVLHAMKRGAK 122
Query: 119 LLNMSDFRDKSKSNSWDYSSFVRTYALYLDERLEYRMQYKRGRSGRFAYDEDEEEQSRES 178
+LN+S FRD S S WD+++FVRT+ALYLDERL+ + K R R+ + E+ R S
Sbjct: 123 ILNLSSFRDDSNSCPWDFTAFVRTFALYLDERLDCFLTGKLQR--RYT---NREQTGRIS 177
Query: 179 KRERYRERDRDKEIVVRSTP-LREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVI 237
R R K + P +R+MK L ++ + Q LL+R +A RPTG AK +R+V
Sbjct: 178 TNSTTRSRFNPKAGIKSHEPAVRDMKPVMLLDKITYWQKLLDRAIATRPTGDAKANRLVK 237
Query: 238 VALYPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWS 297
++LY +++ESF Y D++ L + +D F ++ + R KQ++EL+ FY S
Sbjct: 238 MSLYAVMQESFDLYRDISDGLALLLDSFFHLQYQSCINAFQACVRASKQFEELNAFYDLS 297
Query: 298 KSIGIGRSSEYPEIEKVTTKKLDLMDQFIRDKS 330
KSIGIGR+SEYP I+K++ + L+ + +F++D+S
Sbjct: 298 KSIGIGRTSEYPSIQKISLELLETLQEFLKDQS 330
>CAP8_ARATH (Q8LBH2) Putative clathrin assembly protein At2g01600
Length = 571
Score = 219 bits (559), Expect = 1e-56
Identities = 183/602 (30%), Positives = 285/602 (46%), Gaps = 69/602 (11%)
Query: 7 RRAIGAVKDQTSIGIAKVGSSASIGDLQVAIVKATKHDENPAEERHIREILSLTCYSRAF 66
R+A GA+KD T +G+ +V S + DL VAIVKAT H E P ++RH+R+I + T +RA
Sbjct: 8 RKAYGALKDSTKVGLVRVNSEYA--DLDVAIVKATNHVECPPKDRHLRKIFAATSVTRAR 65
Query: 67 --ISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRLLNMSD 124
++ C++ LS+RL KT +WTVALKTL++I RLL +GD + +E+ +QRG R+L +S+
Sbjct: 66 ADVAYCIHALSRRLHKTRNWTVALKTLIVIHRLLREGDPTFREELLNFSQRG-RILQLSN 124
Query: 125 FRDKSKSNSWDYSSFVRTYALYLDERLEYRMQYKRGRSGRFAYDEDEEE--QSRESKRER 182
F+D S +WD S++VRTYAL+L+ERLE K YD + E +S + +
Sbjct: 125 FKDDSSPIAWDCSAWVRTYALFLEERLECFRVLK--------YDTEAERLPKSNPGQDKG 176
Query: 183 Y-RERDRDKEIVVRSTPLREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVIVALY 241
Y R RD D E +L ++ LQ LL R + CRP G A + ++ AL
Sbjct: 177 YSRTRDLDGE--------------ELLEQLPALQQLLYRLIGCRPEGAANHNHVIQYALA 222
Query: 242 PIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWSKSIG 301
++KESF+ Y + + ID+F EM E +++ R G+Q L FY K +
Sbjct: 223 LVLKESFKVYCAINDGIINLIDKFFEMAKHEAITSLEIYKRAGQQARSLSDFYEACKGLE 282
Query: 302 IGRSSEYPEIEKVTTKKLDLMDQFIRDKSLVSQANK---LITQEENNEKNEEENEVEEDM 358
+ R+ ++P + + L M+++I++ V L+T ++ E+ E +
Sbjct: 283 LARNFQFPVLREPPQSFLTTMEEYIKEAPRVVDVPAEPLLLTYRPDDGLTTEDTEPSHEE 342
Query: 359 NEIKALPPPE-GFNEEQVEEEIKEQDQKEEEKIVQTEGDLLDLTDSMTNQDYVGNKLALA 417
E+ LP + E+ E + + T+ DL L + + ++ ALA
Sbjct: 343 REM--LPSDDVVVVSEETEPSPPPPPSANAQNFIDTD-DLWGLNTGAPDTSVIEDQNALA 399
Query: 418 LFDELPNTTSNTIQALPWHAFDDVSDWETTLV--QSSTNLPNQKPSLGGGFDTLLLDSMY 475
L + T + +D + WE LV SS + + L GG DTL L S+Y
Sbjct: 400 LAIVSTDADPPTPHFGQPNNYDP-TGWELALVTAPSSDISASTERKLAGGLDTLTLSSLY 458
Query: 476 NQKPSLQGMNGYGSASSVAIRSEATMLALPAPPTSRNGSQDPFAASMLVAPPAYVQMSEM 535
+ I S+ + PAP + S DPFA+S APP Q
Sbjct: 459 D--------------DGAYIASQRPVYGAPAP--NPFASHDPFASSNGTAPPPQQQAVNN 502
Query: 536 ---ETRQRLLAEEQAIWQQYAKNGMQGQVGFA---------TQQQPNSNFYMGGYQQNHY 583
+Q + Q +Q + F QQPN++ Y G + N +
Sbjct: 503 PFGAYQQTYQHQPQPTYQHQSNPPTNNSNPFGDFGEFPVNPVSQQPNTSGY-GDFSVNQH 561
Query: 584 GN 585
N
Sbjct: 562 NN 563
>CA10_ARATH (Q9LHS0) Putative clathrin assembly protein At5g35200
Length = 544
Score = 219 bits (557), Expect = 2e-56
Identities = 180/596 (30%), Positives = 280/596 (46%), Gaps = 91/596 (15%)
Query: 2 SQSTLRRAIGAVKDQTSIGIAKVGSSASIGDLQVAIVKATKHDENPAEERHIREILSLTC 61
SQS+LRR +GA+KD T++ +AKV S +L +AIVKAT H E P++ER+IR I
Sbjct: 7 SQSSLRRYLGAIKDTTTVSLAKVNSDYK--ELDIAIVKATNHVERPSKERYIRAIFMAIS 64
Query: 62 YSR--AFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRL 119
+R A ++ C++ L++RL +T +W VALKTL++I R L + D+ + +E+ ++ + +
Sbjct: 65 ATRPRADVAYCIHALARRLSRTHNWAVALKTLIVIHRALREVDQTFHEEVINYSRSRSHM 124
Query: 120 LNMSDFRDKSKSNSWDYSSFVRTYALYLDERLEYRMQYKRGRSGRFAYDEDEEEQSRESK 179
LNMS F+D S N+W YS++VR YAL+L+ERLE
Sbjct: 125 LNMSHFKDDSGPNAWAYSAWVRFYALFLEERLEC-------------------------- 158
Query: 180 RERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVIVA 239
+R D E+ T +++ T DL ++ LQ LL R + C+P G A + ++ +A
Sbjct: 159 ---FRVLKYDVEVDPPRT--KDLDTPDLLEQLPALQELLFRVLDCQPEGAAVQNHIIQLA 213
Query: 240 LYPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWSKS 299
L ++ ES + Y +T + +D+F +M+ + K D++ R KQ L F+ KS
Sbjct: 214 LSMVISESTKIYQALTDGIDNLVDKFFDMQRNDAVKALDMYRRAVKQAGRLSEFFEVCKS 273
Query: 300 IGIGRSSEYPEIEKVTTKKLDLMDQFIRDKSLVSQANKLITQEENNEKNEEENEVEEDMN 359
+ +GR + +IE+ T L M++++++ L + K E+ EK E+
Sbjct: 274 VNVGRGERFIKIEQPPTSFLQAMEEYVKEAPLAAGVKK----EQVVEKLTAPKEILAIEY 329
Query: 360 EIKALPPPEGFNEEQVEEEIKEQDQKEEEKIVQTEGDLLDLTD--SMTNQDYVGNKLALA 417
EI PP+ E+ E + E EK V+ + DLL + D M ++ N LALA
Sbjct: 330 EI----PPKVVEEKPAS---PEPVKAEAEKPVEKQPDLLSMDDPAPMVSELEEKNALALA 382
Query: 418 LFD---ELPNTTSNTIQALPWHAFDDVSDWETTLV--QSSTNLPNQKPSLGGGFDTLLLD 472
+ E P++T++ + + WE LV SS L GG D L LD
Sbjct: 383 IVPVSVEQPHSTTDFTNG-------NSTGWELALVTAPSSNEGAAADSKLAGGLDKLTLD 435
Query: 473 SMYNQ--KPSLQGMNGYGSASSVAIRSEATMLALPAPPTSRNGSQDPFAASMLVAPPAYV 530
S+Y + S Q Y + + M PF AS VA P
Sbjct: 436 SLYEDAIRVSQQQNRSYNPWEQNPVHNGHMM-------------HQPFYASNGVAAPQPF 482
Query: 531 QMSEMETRQRLLAEEQAIWQQYAKNG-MQGQVGFATQQQPNSNFYMGGYQQNHYGN 585
QM+ Q Q+ G M G V QQQ + N +GN
Sbjct: 483 QMANQ--------NHQTFGYQHQNAGMMMGPVQQPYQQQQQN-------MNNPFGN 523
>CAP9_ARATH (P94017) Putative clathrin assembly protein At1g14910
Length = 692
Score = 214 bits (545), Expect = 5e-55
Identities = 171/538 (31%), Positives = 269/538 (49%), Gaps = 60/538 (11%)
Query: 7 RRAIGAVKDQTSIGIAKVGSSASIGDLQVAIVKATKHDENPAEERHIREILSLTCY--SR 64
RRA GA+KD T +G+ +V S + +L VAIVKAT H E P ++RH+R+I T R
Sbjct: 8 RRAYGALKDTTKVGLVRVNSDYA--ELDVAIVKATNHVECPPKDRHLRKIFLATSAIRPR 65
Query: 65 AFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRLLNMSD 124
A ++ C++ LS+RL KT +WTVALK L++I RLL DGD + +E+ +Q+G R++ +S+
Sbjct: 66 ADVAYCIHALSRRLHKTRNWTVALKALLVIHRLLRDGDPTFREELLNFSQKG-RIMQISN 124
Query: 125 FRDKSKSNSWDYSSFVRTYALYLDERLEYRMQYKRGRSGRFAYDEDEEEQSRESKRERYR 184
F+D S +WD S +VRTYAL+L+ERLE K YD + E + S +
Sbjct: 125 FKDDSSPVAWDCSGWVRTYALFLEERLECFRVLK--------YDIEAERLPKVSPGQ--- 173
Query: 185 ERDRDKEIVVRSTPLREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVIVALYPIV 244
E+ K R++ + L ++ LQ LL R + C+P G AK + ++ AL ++
Sbjct: 174 EKGYSKT--------RDLDGEKLLEQLPALQQLLHRLIGCKPEGAAKHNHIIQYALSLVL 225
Query: 245 KESFQTYHDMTSILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWSKSIGIGR 304
KESF+ Y + + +++F EM E K +++ R G Q L FY K + + R
Sbjct: 226 KESFKVYCAINEGIINLVEKFFEMPRHEAIKALEIYKRAGLQAGNLSAFYEVCKGLELAR 285
Query: 305 SSEYPEIEKVTTKKLDLMDQFIRDKSL---VSQANKLITQEENNEKNEEENEVEEDMNEI 361
+ ++P + + L M++++RD V+ L+T ++ E +V E
Sbjct: 286 NFQFPVLREPPQSFLTTMEEYMRDAPQMVDVTSGPLLLTYTPDDGLTSE--DVGPSHEEH 343
Query: 362 KALPPPEGFNEEQVEEEIKEQDQKEEE---KIVQTEGDLLDLTDSMTNQDYVGNKLALAL 418
+ P + E ++ Q E + T+ DLL L D + + ++ ALAL
Sbjct: 344 ETSSPSDSAVVPSEETQLSSQSPPSVETPQNFIDTD-DLLGLHDDTPDPLAILDQNALAL 402
Query: 419 FDELPNTTSNTIQALPW---HAFD-DVSDWETTLVQSSTN--LPNQKPSLGGGFDTLLLD 472
SN + + P+ A D D S WE LV + +N + L GG DTL L+
Sbjct: 403 -----ALVSNDVDSSPFSFGQARDLDPSGWELALVTTPSNDISAATERQLAGGLDTLTLN 457
Query: 473 SMYNQKPSLQGMNGYGSASSVAIRSEATMLALPAPPTSRNGSQDPFAASMLVAPPAYV 530
S+Y+ A+R+ A A P ++ QD FA S V+PP+ V
Sbjct: 458 SLYDDG---------------ALRA-AQQPAYGVPASNPFEVQDLFAFSDSVSPPSAV 499
>CA11_ARATH (Q9C502) Putative clathrin assembly protein At1g33340
Length = 374
Score = 101 bits (252), Expect = 5e-21
Identities = 95/392 (24%), Positives = 166/392 (42%), Gaps = 40/392 (10%)
Query: 7 RRAIGAVKDQTSIGIAKVGS--SASIGDLQVAIVKATKHDENPAEERHIREILSLTCYSR 64
R+ +G KD SIG A V + + D++VA+V+AT HD+ P +++ + EIL L +
Sbjct: 10 RQVLGLAKDHASIGRAIVQNYNEKAFFDIEVAVVRATSHDDCPVDDKTMHEILFLVSNTP 69
Query: 65 AFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRLLNMSD 124
I +S+RL KT VA KTL+L RLL R+ EQ++ + G + S
Sbjct: 70 GSIPFLAEQISRRLAKTRDCLVAGKTLLLFHRLLRGSSRSIEQQLHIAHTSGHLQIGCSW 129
Query: 125 FRDKSKSNSWDYSSFVRTYALYLDERLEYRMQYKRGRSGRFAYDEDEEEQSRESKRERYR 184
F S S+ F++ Y YL ER+ + + ++G+ E S +K RY+
Sbjct: 130 FMMSLDSRSF---VFLQNYVAYLQERVGWII----NQAGKL-----EPVMSGGTKFSRYK 177
Query: 185 ERDRDKEIVVRSTPLREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVIVALYPIV 244
E+ D +F + Q + + + C P +V A I+
Sbjct: 178 EKSMDL----------------VFHILPKCQEFIAQVLKCSPVDAWPIDNLVQAATGNIL 221
Query: 245 KESFQTYHDMTSILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWSKSIGIGR 304
KESFQ Y + + + ++ P + + +Q +L + Y + +
Sbjct: 222 KESFQVYMTYSDGMTALVSMLFDLSRPARDLACGMLRKASQQIQDLRILYDKCRGFAGMK 281
Query: 305 SSEYPEIEKVTTKKLDLMDQFI-----RDKSLVSQANKLITQEENNEKNEEENEVEEDMN 359
S +YP ++ ++ + +++ R SL + IT NE +E++
Sbjct: 282 SLDYPSVQAISMDHIVALEECSSYGGKRGFSLSTNLRDAIT---CNELKQEQHSASFSST 338
Query: 360 EIKALPPPEGFNEEQV--EEEIKEQDQKEEEK 389
+LP + V + E E+ K+ EK
Sbjct: 339 SPFSLPVETKISMVWVVFDNEDGEESDKQTEK 370
>CA12_ARATH (Q9C9X5) Putative clathrin assembly protein At1g68110
Length = 379
Score = 90.5 bits (223), Expect = 1e-17
Identities = 89/394 (22%), Positives = 172/394 (43%), Gaps = 52/394 (13%)
Query: 7 RRAIGAVKDQTS---IGIAKVGSSASIGDLQVAIVKATKHDENPAEERHIREILSLTCYS 63
+RA A+KD+ S +G ++ SS DL+ AI+KAT HD++ + + + S
Sbjct: 5 KRAAAAIKDRKSLLAVGFSRRNSSYRNADLEAAIIKATSHDDSSVDYSNAHRVYKWIRSS 64
Query: 64 RAFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRL-LNM 122
+ + V +S R+ T SW VALK+L+L+ +L + E RL ++
Sbjct: 65 PLNLKTLVYAISSRVNHTRSWIVALKSLMLLHGVLCCKVPSVVGEF-------RRLPFDL 117
Query: 123 SDFRD--KSKSNSWDYSSFVRTYALYLDERLEYRMQYKRGRSGRFAYDEDEEEQSRESKR 180
SDF D S +W ++ FVRTY +L Y +Q +
Sbjct: 118 SDFSDGHSCLSKTWGFNVFVRTYFAFLHH-----------------YSSFLSDQIHRLRG 160
Query: 181 ERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVIVAL 240
R ++ + V++ R+Q LQ LL+ + RP +++ A+
Sbjct: 161 NNRRSLEKTSDSVIQE-----------LERIQKLQSLLDMILQIRPVADNMKKTLILEAM 209
Query: 241 YPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWSKSI 300
+V ES Y + + + + E K+ + + Q ++L +++ + K
Sbjct: 210 DCLVIESINIYGRICGAVMKVLPLAGKSEAATVLKIVN---KTTSQGEDLIVYFEFCKGF 266
Query: 301 GIGRSSEYPEIEKVTTKKLDLMDQFIRDKSLVSQANKLITQEENNEKNEEENEVEEDMNE 360
G+ + E P+ ++ ++++ +++ I V + KL E++ EK +E+ V + +
Sbjct: 267 GVSNAREIPQFVRIPEEEVEAIEKMI---DTVQEKPKL---EKDEEKEDEKAMVV--LEQ 318
Query: 361 IKALPPPEGFNEEQVEEEIKEQDQKEEEKIVQTE 394
K L E E++ + D+K++ +I + E
Sbjct: 319 PKKLQTIITDKWEIFEDDYRCFDRKDKWEIFEDE 352
>CA13_ARATH (Q9FRH3) Putative clathrin assembly protein At1g25240
Length = 376
Score = 81.6 bits (200), Expect = 5e-15
Identities = 87/362 (24%), Positives = 148/362 (40%), Gaps = 63/362 (17%)
Query: 7 RRAIGAVKDQTS---IGIAKVGSSASIGDLQVAIVKATKHDENPAEERHIREILSLTCYS 63
+RA GA+KD+ + IG ++ S + DL AI+ AT HD++ + + + S
Sbjct: 5 KRASGALKDRKTLFTIGFSRKTSFRN-PDLDSAIIHATSHDDSSVDYHNAHRVYKWIRSS 63
Query: 64 RAFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRL-LNM 122
A + V+ LS R+ +T SW VALK L+L+ +L S Q RL ++
Sbjct: 64 PANLKPLVHALSSRVNRTRSWIVALKALMLVHGVLCCK--------VTSLQEIRRLPFDL 115
Query: 123 SDFRD--KSKSNSWDYSSFVRTYALYLDERLEYRMQYKRGRSGRFAYDEDEEEQSRESKR 180
SDF D S +W +++F+R Y +LD+ + R R + D +E
Sbjct: 116 SDFSDGHSRPSKTWGFNAFIRAYFSFLDQYSFFLSDQIRRRHKKPQLDSVNQE------- 168
Query: 181 ERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVIVAL 240
R++ LQ LL + RP +++ A+
Sbjct: 169 ---------------------------LERIEKLQSLLHMLLQIRPMADNMKKTLILEAM 201
Query: 241 YPIVKESFQTYHDMTS-ILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWSKS 299
+V E F Y + S I + I E + + Q ++L L++ + K
Sbjct: 202 DCVVIEIFDIYGRICSAIAKLLIKIHPAAGKAEAVIALKIVKKATSQGEDLALYFEFCKE 261
Query: 300 IGIGRSSEYPEIEKVTTKKLDLMDQFIRDKSLVSQANKLITQEENNEKNEEENEVEEDMN 359
G+ + + P+ VT + D + K+I E E ++E+EVEE+ +
Sbjct: 262 FGVSNAHDIPKF--VTIPEED-----------IKAIEKVINGVEEEEVKKKEDEVEEEKS 308
Query: 360 EI 361
I
Sbjct: 309 II 310
>CA16_ARATH (Q8L936) Putative clathrin assembly protein At4g40080
Length = 365
Score = 79.0 bits (193), Expect = 3e-14
Identities = 52/147 (35%), Positives = 79/147 (53%), Gaps = 5/147 (3%)
Query: 10 IGAVKDQTSIGIAKVGSSASIG---DLQVAIVKATKHDEN-PAEERHIREILSLTCYSRA 65
IG +KD+ S A + SS + +++++AT HD + P RH+ ILS SRA
Sbjct: 11 IGRIKDKASQSKAALVSSNTKSKTLSFHLSVLRATTHDPSTPPGNRHLAVILSAGTGSRA 70
Query: 66 FISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEI-FFSTQRGTRLLNMSD 124
SS V ++ +RL T VALK+L++I ++ G + ++ F G L +S
Sbjct: 71 TASSAVESIMERLHTTGDACVALKSLIIIHHIVKHGRFILQDQLSVFPASGGRNYLKLSA 130
Query: 125 FRDKSKSNSWDYSSFVRTYALYLDERL 151
FRD+ W+ SS+VR YALYL+ L
Sbjct: 131 FRDEKSPLMWELSSWVRWYALYLEHLL 157
>PICA_RAT (O55012) Phosphatidylinositol-binding clathrin assembly
protein (Clathrin assembly lymphoid myeloid leukemia
protein) (rCALM)
Length = 640
Score = 73.6 bits (179), Expect = 1e-12
Identities = 92/469 (19%), Positives = 191/469 (40%), Gaps = 75/469 (15%)
Query: 37 IVKATKHDENPAEERHIREILSLTCYSRAFISSCVNTLSKRLIKTSSWTVALKTLVLIQR 96
+ KAT H+ +++H+ ++ T I ++L +R SSW V K+L+
Sbjct: 26 VCKATTHEIMGPKKKHLDYLIQCTNEMNVNIPQLADSLFERTTN-SSWVVVFKSLITTHH 84
Query: 97 LLADGDRAYEQEIFFSTQRGTRLLNMSDFRDKSKSNSWDYSSFVRTYALYLDER-LEYRM 155
L+ G+ + Q + R T L N+S+F DKS +D S+F+R Y+ YL+E+ + YR
Sbjct: 85 LMVYGNERFIQ---YLASRNT-LFNLSNFLDKSGLQGYDMSTFIRRYSRYLNEKAVSYR- 139
Query: 156 QYKRGRSGRFAYDEDEEEQSRESKRERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQ 215
+ A+D + ++ + +R M T+ L + +Q
Sbjct: 140 --------QVAFDFTKVKRGADG-------------------VMRTMNTEKLLKTVPIIQ 172
Query: 216 LLLERFMACRPTGRAKTHRMVIVALYPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYK 275
++ + T+ ++ A + K++ + + + ++++ +M+ + +
Sbjct: 173 NQMDALLDFNVNSNELTNGVINAAFMLLFKDAIRLFAAYNEGIINLLEKYFDMKKNQCKE 232
Query: 276 VYDVFCRVGKQYDELDLFYSWSKSIGIGRSSEYPEIEKVTTKKLDLMDQF--------IR 327
D++ + + + F ++ +GI R + P++ + + LD ++Q I+
Sbjct: 233 GLDIYKKFLTRMTRISEFLKVAEQVGIDR-GDIPDLSQAPSSLLDALEQHLASLEGKKIK 291
Query: 328 DKSLVSQANKLITQEEN----------NEKNEEENEVEEDMNEIKALPPPEGFNEEQVEE 377
D + S+A L + ++ E++ +EE+ +KAL
Sbjct: 292 DSTAASRATTLSNAVSSLASTGLSLTKVDEREKQAALEEEQARLKAL------------- 338
Query: 378 EIKEQDQKEEEKIVQTEGDLLDLTDSMTNQDYVGNKLALALFDELPNTTSNTIQALPWHA 437
KEQ KE K T L S + G A A+ ++SN+ LP
Sbjct: 339 --KEQRLKELAKKPHTS---LTTAASPVSTSAGGIMTAPAIDIFSTPSSSNSTSKLP--- 390
Query: 438 FDDVSDWETTLVQSSTNLPNQKPSLGGGFDTLLLDSMYNQKPSLQGMNG 486
+D+ D + S + + P + + + D++ + P L +G
Sbjct: 391 -NDLLDLQQPTFHPSVHAMSAAPQVASTWGDAVDDAIPSLNPFLTKSSG 438
>PICA_MOUSE (Q7M6Y3) Phosphatidylinositol-binding clathrin assembly
protein (Clathrin assembly lymphoid myeloid leukemia)
(CALM)
Length = 660
Score = 72.0 bits (175), Expect = 4e-12
Identities = 95/467 (20%), Positives = 191/467 (40%), Gaps = 78/467 (16%)
Query: 37 IVKATKHDENPAEERHIREILSLTCYSRAFISSCVNTLSKRLIKTSSWTVALKTLVLIQR 96
+ KAT H+ +++H+ ++ T I ++L +R SSW V K+L+
Sbjct: 26 VCKATTHEIMGPKKKHLDYLIQCTNEMNVNIPQLADSLFERTTN-SSWVVVFKSLITTHH 84
Query: 97 LLADGDRAYEQEIFFSTQRGTRLLNMSDFRDKSKSNSWDYSSFVRTYALYLDER-LEYRM 155
L+ G+ + Q + R T L N+S+F DKS +D S+F+R Y+ YL+E+ + YR
Sbjct: 85 LMVYGNERFIQ---YLASRNT-LFNLSNFLDKSGLQGYDMSTFIRRYSRYLNEKAVSYR- 139
Query: 156 QYKRGRSGRFAYDEDEEEQSRESKRERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQ 215
+ A+D + ++ + +R M T+ L + +Q
Sbjct: 140 --------QVAFDFTKVKRGADG-------------------VMRTMNTEKLLKTVPIIQ 172
Query: 216 LLLERFMACRPTGRAKTHRMVIVALYPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYK 275
++ + T+ ++ A + K++ + + + ++++ +M+ + +
Sbjct: 173 NQMDALLDFNVNSNELTNGVINAAFMLLFKDAIRLFAAYNEGIINLLEKYFDMKKNQCKE 232
Query: 276 VYDVFCRVGKQYDELDLFYSWSKSIGIGRSSEYPEIEKVTTKKLDLMDQF--------IR 327
D++ + + + F ++ +GI R + P++ + + LD ++Q I+
Sbjct: 233 GLDIYKKFLTRMTRISEFLKVAEQVGIDR-GDIPDLSQAPSSLLDALEQHLASLEGKKIK 291
Query: 328 DKSLVSQANKLITQEEN----------NEKNEEENEVEEDMNEIKALPPPEGFNEEQVEE 377
D + S+A L + ++ E++ +EE+ +KAL
Sbjct: 292 DSTAASRATTLSNAVSSLASTGLSLTKVDEREKQAALEEEQARLKAL------------- 338
Query: 378 EIKEQDQKEEEKIVQTEGDLLDLTDSMTNQDYVGNKLALALFDELPNTTSNTIQALPWHA 437
KEQ KE K T L S + G A A+ ++SN+ LP
Sbjct: 339 --KEQRLKELAKKPHTS---LTTAASPVSTSAGGIMTAPAIDIFSTPSSSNSTSKLP--- 390
Query: 438 FDDVSDWETTLVQSSTNLPNQKP---SLGGGFDTLLLDSMYNQKPSL 481
+D+ D + S + + P S G + LD++ + PSL
Sbjct: 391 -NDLLDLQQPTFHPSVHAMSAAPQGASTWGDPFSATLDAVEDAIPSL 436
>A180_HUMAN (O60641) Clathrin coat assembly protein AP180 (Clathrin
coat associated protein AP180) (91 kDa
synaptosomal-associated protein)
Length = 907
Score = 71.6 bits (174), Expect = 5e-12
Identities = 65/291 (22%), Positives = 129/291 (43%), Gaps = 33/291 (11%)
Query: 36 AIVKATKHDENPAEERHIREILSLTCYSRAFISSCVNTLSKRLIKTSSWTVALKTLVLIQ 95
A+ KAT H+ +++H+ ++ T + I +TL +R SSW V K LV
Sbjct: 25 AVCKATTHEVMGPKKKHLDYLIQATNETNVNIPQMADTLFERATN-SSWVVVFKALVTTH 83
Query: 96 RLLADGDRAYEQEIFFSTQRGTRLLNMSDFRDKSKSNSWDYSSFVRTYALYLDERLEYRM 155
L+ G+ + Q + R T L N+S+F DKS S+ +D S+F+R Y+ YL+E+
Sbjct: 84 HLMVHGNERFIQ---YLASRNT-LFNLSNFLDKSGSHGYDMSTFIRRYSRYLNEK----- 134
Query: 156 QYKRGRSGRFAYDEDEEEQSRESKRERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQ 215
F+Y + + +R K R E +++S P+ + + D L H
Sbjct: 135 --------AFSYRQMAFDFARVKKGADGVMRTMAPEKLLKSMPILQGQIDALLEFDVHPN 186
Query: 216 LLLERFMACRPTGRAKTHRMVIVALYPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYK 275
L + + L+ + + F Y+D L +++F EM+ +
Sbjct: 187 ELTNGVI-----------NAAFMLLFKDLIKLFACYNDGVINL---LEKFFEMKKGQCKD 232
Query: 276 VYDVFCRVGKQYDELDLFYSWSKSIGIGRSSEYPEIEKVTTKKLDLMDQFI 326
+++ R + + F ++ +GI + + P++ + + ++ ++Q +
Sbjct: 233 ALEIYKRFLTRMTRVSEFLKVAEQVGIDK-GDIPDLTQAPSSLMETLEQHL 282
>A180_MOUSE (Q61548) Clathrin coat assembly protein AP180 (Clathrin
coat associated protein AP180) (91 kDa
synaptosomal-associated protein) (Phosphoprotein F1-20)
Length = 901
Score = 71.2 bits (173), Expect = 7e-12
Identities = 65/291 (22%), Positives = 129/291 (43%), Gaps = 33/291 (11%)
Query: 36 AIVKATKHDENPAEERHIREILSLTCYSRAFISSCVNTLSKRLIKTSSWTVALKTLVLIQ 95
A+ KAT H+ +++H+ ++ T + I +TL +R SSW V K LV
Sbjct: 25 AVCKATTHEVMGPKKKHLDYLIQATNETNVNIPQMADTLFERATN-SSWVVVFKALVTTH 83
Query: 96 RLLADGDRAYEQEIFFSTQRGTRLLNMSDFRDKSKSNSWDYSSFVRTYALYLDERLEYRM 155
L+ G+ + Q + R T L N+S+F DKS S+ +D S+F+R Y+ YL+E+
Sbjct: 84 HLMVHGNERFIQ---YLASRNT-LFNLSNFLDKSGSHGYDMSTFIRRYSRYLNEK----- 134
Query: 156 QYKRGRSGRFAYDEDEEEQSRESKRERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQ 215
F+Y + + +R K R E +++S P+ + + D L H
Sbjct: 135 --------AFSYRQMAFDFARVKKGADGVMRTMVPEKLLKSMPILQGQIDALLEFDVHPN 186
Query: 216 LLLERFMACRPTGRAKTHRMVIVALYPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYK 275
L + + L+ + + F Y+D L +++F EM+ +
Sbjct: 187 ELTNGVI-----------NAAFMLLFKDLIKLFACYNDGVINL---LEKFFEMKKGQCKD 232
Query: 276 VYDVFCRVGKQYDELDLFYSWSKSIGIGRSSEYPEIEKVTTKKLDLMDQFI 326
+++ R + + F ++ +GI + + P++ + + ++ ++Q +
Sbjct: 233 ALEIYKRFLTRMTRVSEFLKVAEQVGIDK-GDIPDLTQAPSSLMETLEQHL 282
>CA15_ARATH (Q9LQW4) Putative clathrin assembly protein At1g14686
Length = 339
Score = 70.5 bits (171), Expect = 1e-11
Identities = 75/350 (21%), Positives = 139/350 (39%), Gaps = 50/350 (14%)
Query: 21 IAKVGSSASIGD--LQVAIVKATKHDENPAEERHIREILSLTCYSRAFISSCVNTLSKRL 78
+ K G S D L A+VKAT HDE + + I S + + V+ +S R+
Sbjct: 10 VLKDGPSLIAADDILTAAVVKATSHDELSIDTESAQFIYRHVLSSPSSLKPLVSLISSRV 69
Query: 79 IKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRLLNMSDF---RDKSKSNSWD 135
+T SW VALK L+L+ E G ++S F + S S
Sbjct: 70 KRTRSWAVALKGLMLMHGFFLCKSTVAE-------SIGRLPFDLSSFGEGNSRIMSKSGG 122
Query: 136 YSSFVRTYALYLDERLEYRMQYKRGRSGRFAYDEDEEEQSRESKRERYRERDRDKEIVVR 195
++ FVR Y +LD R + R RY E + +++R
Sbjct: 123 FNLFVRAYFAFLDRR---------------------SILFHDGNRHRYNE---ESSVLIR 158
Query: 196 STPLREMKTDDLFSRMQHLQLLLERFMACRPTGRAKTHRMVIVALYPIVKESFQTYHDM- 254
+R+M Q++++ + +P G ++ A+ +V E + Y +
Sbjct: 159 LVIIRKM------------QIIVDSLIRIKPIGENMMIPVINEAMENVVSEIMEIYGWIC 206
Query: 255 TSILGIFIDRFTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWSKSIGIGRSSEYPEIEKV 314
I + + +++ E + + KQ EL ++ + K +G+ + E P ++
Sbjct: 207 RRIAEVLPNVHSKIGKTEADLALKIVAKSMKQGGELKKYFEFCKDLGVSNAQEIPNFVRI 266
Query: 315 TTKKLDLMDQFIRDKSLVSQANKLITQEENNEKNEEENEVEEDMNEIKAL 364
+ +D+ +R ++ S E E+ EEE E+E ++++ L
Sbjct: 267 PEADVIHLDELVR-TAMESSEESAERTEIAEEEEEEEEEIETKLSDLITL 315
>A180_RAT (Q05140) Clathrin coat assembly protein AP180 (Clathrin
coat associated protein AP180) (91 kDa
synaptosomal-associated protein)
Length = 915
Score = 70.5 bits (171), Expect = 1e-11
Identities = 65/291 (22%), Positives = 128/291 (43%), Gaps = 33/291 (11%)
Query: 36 AIVKATKHDENPAEERHIREILSLTCYSRAFISSCVNTLSKRLIKTSSWTVALKTLVLIQ 95
A+ KAT H+ +++H+ ++ T + I +TL +R SSW V K LV
Sbjct: 25 AVCKATTHEVMGPKKKHLDYLIQATNETNVNIPQMADTLFERATN-SSWVVVFKALVTTH 83
Query: 96 RLLADGDRAYEQEIFFSTQRGTRLLNMSDFRDKSKSNSWDYSSFVRTYALYLDERLEYRM 155
L+ G+ + Q + R T L N+S+F DKS S+ +D S+F+R Y+ YL+E+
Sbjct: 84 HLMVHGNERFIQ---YLASRNT-LFNLSNFLDKSGSHGYDMSTFIRRYSRYLNEK----- 134
Query: 156 QYKRGRSGRFAYDEDEEEQSRESKRERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQ 215
F+Y + + +R K R E +++S P+ + + D L H
Sbjct: 135 --------AFSYRQMAFDFARVKKGADGVMRTMVPEKLLKSMPILQGQIDALLEFDVHPN 186
Query: 216 LLLERFMACRPTGRAKTHRMVIVALYPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYK 275
L + + L+ + + F Y+D L +++F EM+ +
Sbjct: 187 ELTNGVI-----------NAAFMLLFKDLIKLFACYNDGVINL---LEKFFEMKKGQCKD 232
Query: 276 VYDVFCRVGKQYDELDLFYSWSKSIGIGRSSEYPEIEKVTTKKLDLMDQFI 326
+++ R + + F + +GI + + P++ + + ++ ++Q +
Sbjct: 233 ALEIYKRFLTRMTRVSEFLKVADEVGIDK-GDIPDLTQAPSSLMETLEQHL 282
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.131 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 67,614,589
Number of Sequences: 164201
Number of extensions: 2930782
Number of successful extensions: 20104
Number of sequences better than 10.0: 422
Number of HSP's better than 10.0 without gapping: 142
Number of HSP's successfully gapped in prelim test: 288
Number of HSP's that attempted gapping in prelim test: 14965
Number of HSP's gapped (non-prelim): 2610
length of query: 588
length of database: 59,974,054
effective HSP length: 116
effective length of query: 472
effective length of database: 40,926,738
effective search space: 19317420336
effective search space used: 19317420336
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 69 (31.2 bits)
Medicago: description of AC124966.14


