

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124963.22 + phase: 0 /pseudo
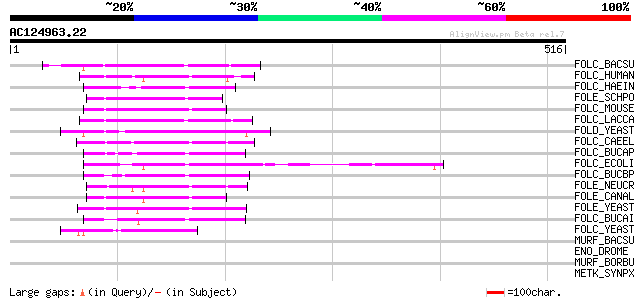
(516 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
FOLC_BACSU (Q05865) Folylpolyglutamate synthase (EC 6.3.2.17) (F... 111 5e-24
FOLC_HUMAN (Q05932) Folylpolyglutamate synthase, mitochondrial p... 100 2e-20
FOLC_HAEIN (P43775) Folylpolyglutamate synthase (EC 6.3.2.17) (F... 89 4e-17
FOLE_SCHPO (O74742) Probable folylpolyglutamate synthase (EC 6.3... 88 6e-17
FOLC_MOUSE (P48760) Folylpolyglutamate synthase, mitochondrial p... 87 1e-16
FOLC_LACCA (P15925) Folylpolyglutamate synthase (EC 6.3.2.17) (F... 86 2e-16
FOLD_YEAST (Q12676) Folylpolyglutamate synthase (EC 6.3.2.17) (F... 82 3e-15
FOLC_CAEEL (Q09509) Putative folylpolyglutamate synthase (EC 6.3... 82 3e-15
FOLC_BUCAP (Q8K9X3) FolC bifunctional protein [Includes: Folylpo... 82 3e-15
FOLC_ECOLI (P08192) FolC bifunctional protein [Includes: Folylpo... 81 7e-15
FOLC_BUCBP (Q89AT2) FolC bifunctional protein [Includes: Folylpo... 79 2e-14
FOLE_NEUCR (O13492) Folylpolyglutamate synthase (EC 6.3.2.17) (F... 76 2e-13
FOLE_CANAL (Q9Y893) Folylpolyglutamate synthase (EC 6.3.2.17) (F... 74 9e-13
FOLE_YEAST (Q08645) Folylpolyglutamate synthase (EC 6.3.2.17) (F... 74 1e-12
FOLC_BUCAI (P57265) FolC bifunctional protein [Includes: Folylpo... 72 3e-12
FOLC_YEAST (P36001) Putative folylpolyglutamate synthase (EC 6.3... 45 6e-04
MURF_BACSU (P96613) UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-... 40 0.015
ENO_DROME (P15007) Enolase (EC 4.2.1.11) (2-phosphoglycerate deh... 33 1.4
MURF_BORBU (Q44777) UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-... 33 1.8
METK_SYNPX (Q7U4S6) S-adenosylmethionine synthetase (EC 2.5.1.6)... 33 2.3
>FOLC_BACSU (Q05865) Folylpolyglutamate synthase (EC 6.3.2.17)
(Folylpoly-gamma-glutamate synthetase) (FPGS)
Length = 430
Score = 111 bits (277), Expect = 5e-24
Identities = 76/226 (33%), Positives = 115/226 (50%), Gaps = 38/226 (16%)
Query: 31 KYGVPTGAGTDSNDGFDLGRMRRLLDRFGNPHSKFKT----------------------E 68
K+GV G LGRM++L+ R G+P K +
Sbjct: 17 KFGVKPG----------LGRMKQLMARLGHPEKKIRAFHVAGTNGKGSTVAFIRSMLQEA 66
Query: 69 GYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCISHFEVF 128
GY+VG ++SP+I T ERI +G P+S + L +++K ++ + E G + FE+
Sbjct: 67 GYTVGTFTSPYIITFNERI-SVNGIPISDEEWTALVNQMKPHVEALDQTEYGQPTEFEIM 125
Query: 129 TAMAFILFAD-EKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLES 187
TA AF+ FA+ KVD + E GLGG D+TN++ VIT++G +H+ LG ++E
Sbjct: 126 TACAFLYFAEFHKVDFVIFETGLGGRFDSTNVVEP---LLTVITSIGHDHMNILGNTIEE 182
Query: 188 IAMAKAGIIKQGCPLVLGGPFLPHIEYIIREKAVNMDSPVVSASDS 233
IA KAGIIK+G P+V P +IR +A +P S D+
Sbjct: 183 IAGEKAGIIKEGIPIVT-AVTQPEALQVIRHEAERHAAPFQSLHDA 227
>FOLC_HUMAN (Q05932) Folylpolyglutamate synthase, mitochondrial
precursor (EC 6.3.2.17) (Folylpoly-gamma-glutamate
synthetase) (FPGS)
Length = 587
Score = 99.8 bits (247), Expect = 2e-20
Identities = 61/169 (36%), Positives = 95/169 (56%), Gaps = 17/169 (10%)
Query: 66 KTEGYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCIS-- 123
++ G G +SSPH+ +RERI +G P+S +L F R+ L++ ++ C+S
Sbjct: 119 RSYGLKTGFFSSPHLVQVRERIR-INGQPISPELFTKYFWRLYHRLEET--KDGSCVSMP 175
Query: 124 -HFEVFTAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALG 182
+F T MAF +F EKVD+AV+E G+GGA D TNII + ++++G +H + LG
Sbjct: 176 PYFRFLTLMAFHVFLQEKVDLAVVEVGIGGAYDCTNIIRKPVVCG--VSSLGIDHTSLLG 233
Query: 183 GSLESIAMAKAGIIKQGCP----LVLGGPFLPHIEYIIREKAVNMDSPV 227
++E IA K GI KQG P L GP ++R++A + P+
Sbjct: 234 DTVEKIAWQKGGIFKQGVPAFTVLQPEGPLA-----VLRDRAQQISCPL 277
>FOLC_HAEIN (P43775) Folylpolyglutamate synthase (EC 6.3.2.17)
(Folylpoly-gamma-glutamate synthetase) (FPGS)
Length = 437
Score = 88.6 bits (218), Expect = 4e-17
Identities = 55/143 (38%), Positives = 80/143 (55%), Gaps = 13/143 (9%)
Query: 69 GYSVGCYSSPHIQTIRERILGRSGD-PVSAKLLNNLFHRIKQDLDQAIKEENGCISHFEV 127
G VG YSSPH+ ER+ ++ D P A + F +D+ E +++FE
Sbjct: 74 GLRVGVYSSPHLLRYNERVRIQNQDLPDEAHTASFAF------IDENKTES---LTYFEF 124
Query: 128 FTAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLES 187
T A LF K+D+ ++E GLGG DATNI+ S AVIT++ +H LG + E+
Sbjct: 125 STLSALHLFKQAKLDVVILEVGLGGRLDATNIVDSH---LAVITSIDIDHTDFLGDTREA 181
Query: 188 IAMAKAGIIKQGCPLVLGGPFLP 210
IA KAGI ++ CP+V+G P +P
Sbjct: 182 IAFEKAGIFRENCPVVIGEPNVP 204
>FOLE_SCHPO (O74742) Probable folylpolyglutamate synthase (EC
6.3.2.17) (Folylpoly-gamma-glutamate synthetase) (FPGS)
Length = 505
Score = 87.8 bits (216), Expect = 6e-17
Identities = 46/127 (36%), Positives = 76/127 (59%), Gaps = 3/127 (2%)
Query: 72 VGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCISHFEVFTAM 131
+G Y+SPH++++ ERI +G P+S +L F + + L+ A+ ++ +F T M
Sbjct: 116 IGMYTSPHLRSVCERIQ-LNGKPISQELFTKYFFDVWERLENAVGSDSEKPMYFRFLTLM 174
Query: 132 AFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLESIAMA 191
A+ +F E VD A+IE G+GG D+TN+I A +T++G +H + LG ++ IA
Sbjct: 175 AWHVFISENVDAAIIEVGIGGEYDSTNLIEKP--YATAVTSLGLDHTSLLGNTIAEIAWQ 232
Query: 192 KAGIIKQ 198
KAGI K+
Sbjct: 233 KAGIYKE 239
>FOLC_MOUSE (P48760) Folylpolyglutamate synthase, mitochondrial
precursor (EC 6.3.2.17) (Folylpoly-gamma-glutamate
synthetase) (FPGS)
Length = 587
Score = 86.7 bits (213), Expect = 1e-16
Identities = 50/134 (37%), Positives = 77/134 (57%), Gaps = 4/134 (2%)
Query: 69 GYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCI-SHFEV 127
G G + SPH+ +R+RI +G P+S +L F + L++ + + + S+F
Sbjct: 122 GLKTGFFRSPHMVQVRDRIR-INGKPISPELFTKHFWCLYNQLEEFKDDSHVSMPSYFRF 180
Query: 128 FTAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLES 187
T MAF +F EKVD+AV+E G+GGA D TNII + ++++G +H + LG ++E
Sbjct: 181 LTLMAFHVFLQEKVDLAVVEVGIGGAFDCTNIIRKPVVCG--VSSLGIDHTSLLGDTVEK 238
Query: 188 IAMAKAGIIKQGCP 201
IA K GI K G P
Sbjct: 239 IAWQKGGIFKPGVP 252
>FOLC_LACCA (P15925) Folylpolyglutamate synthase (EC 6.3.2.17)
(Folylpoly-gamma-glutamate synthetase) (FPGS)
Length = 428
Score = 85.9 bits (211), Expect = 2e-16
Identities = 53/161 (32%), Positives = 87/161 (53%), Gaps = 6/161 (3%)
Query: 66 KTEGYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGC-ISH 124
+ G +VG Y+SP I ERI+ +P+ L N ++ L++ +++ ++
Sbjct: 62 EASGLTVGLYTSPFIMRFNERIM-IDHEPIPDAALVNAVAFVRAALERLQQQQADFNVTE 120
Query: 125 FEVFTAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGS 184
FE TA+ + F +VD+AVIE G+GG D+TN+I+ +V+T V +H LG +
Sbjct: 121 FEFITALGYWYFRQRQVDVAVIEVGIGGDTDSTNVITP---VVSVLTEVALDHQKLLGHT 177
Query: 185 LESIAMAKAGIIKQGCPLVLGGPFLPHIEYIIREKAVNMDS 225
+ +IA KAGIIK+G P+V G +P ++ K S
Sbjct: 178 ITAIAKHKAGIIKRGIPVVTGN-LVPDAAAVVAAKVATTGS 217
>FOLD_YEAST (Q12676) Folylpolyglutamate synthase (EC 6.3.2.17)
(Folylpoly-gamma-glutamate synthetase) (FPGS)
Length = 427
Score = 82.0 bits (201), Expect = 3e-15
Identities = 60/220 (27%), Positives = 101/220 (45%), Gaps = 30/220 (13%)
Query: 48 LGRMRRLLDRFGNPHSKFKT----------------------EGYSVGCYSSPHIQTIRE 85
L R+ +LL+ GNP + + + Y +G +++PH+ + +
Sbjct: 7 LSRITKLLEHLGNPQNSLRVLHIAGTNGKGSVCTYLSSVLQQKSYQIGKFTTPHLVHVTD 66
Query: 86 RILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCISHFEVFTAMAFILFADEKVDIAV 145
I + P+ + N I+ L+ K + + FE+ T AF F D + V
Sbjct: 67 SIT-INNKPIPLERYQN----IRLQLEALNKSHSLKCTEFELLTCTAFKYFYDVQCQWCV 121
Query: 146 IEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLESIAMAKAGIIKQGCPL-VL 204
IE GLGG DATN+I + A IT + +H + LG +L I+ KAGII +G P V+
Sbjct: 122 IEVGLGGRLDATNVIPGANKACCGITKISLDHESFLGNTLSEISKEKAGIITEGVPFTVI 181
Query: 205 GGPFLPHIEYIIRE--KAVNMDSPVVSASDSGNNLAVKSF 242
G + +++E KA+ + V + +GN + S+
Sbjct: 182 DGTNEASVINVVKERCKALGSELSVTDSQLNGNMIDTNSW 221
>FOLC_CAEEL (Q09509) Putative folylpolyglutamate synthase (EC
6.3.2.17) (Folylpoly-gamma-glutamate synthetase) (FPGS)
Length = 510
Score = 82.0 bits (201), Expect = 3e-15
Identities = 48/165 (29%), Positives = 84/165 (50%), Gaps = 6/165 (3%)
Query: 63 SKFKTEGYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCI 122
S +++G G YSSPH+ +RERI G PVS ++ F + + + + +
Sbjct: 108 SILRSQGLRTGFYSSPHLVHVRERIQV-DGQPVSEQMFAEEFFHVYDIIKR--EHSDNMP 164
Query: 123 SHFEVFTAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALG 182
++F+ T +AF +F V + ++E G+GG D TN++ + +TT+ +H++ LG
Sbjct: 165 AYFKFLTLLAFRIFVKLNVQVMILEVGIGGEYDCTNVVEKPKVCG--VTTLDYDHMSILG 222
Query: 183 GSLESIAMAKAGIIKQGCPLVLGGPFLPHIEYIIREKAVNMDSPV 227
L IA KAGI K+ P P E ++ +A++ P+
Sbjct: 223 NKLSEIAWHKAGIFKESVP-AFYSPTTTEAEEVLIARAISKHVPL 266
>FOLC_BUCAP (Q8K9X3) FolC bifunctional protein [Includes:
Folylpolyglutamate synthase (EC 6.3.2.17)
(Folylpoly-gamma-glutamate synthetase) (FPGS);
Dihydrofolate synthase (EC 6.3.2.12)]
Length = 418
Score = 82.0 bits (201), Expect = 3e-15
Identities = 56/152 (36%), Positives = 85/152 (55%), Gaps = 11/152 (7%)
Query: 69 GYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCISHFEVF 128
GY VG Y+SPH+ ERI +G +S K +++F + D ++ N +++FE
Sbjct: 69 GYQVGLYTSPHLINYSERIKV-NGLYLSEK--DHIFSFLIIDAEKG----NVSLTYFEFI 121
Query: 129 TAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLESI 188
T A LF+ +DI ++E GLGG DATNII S +VIT +G +H + LG SI
Sbjct: 122 TLSALFLFSQYSLDIIILEVGLGGRLDATNIIDSD---LSVITNIGIDHTSCLGTDRISI 178
Query: 189 AMAKAGIIKQGCPLVLGGPFLP-HIEYIIREK 219
K+GI ++G V+G +P ++ I +EK
Sbjct: 179 GREKSGIFRKGKIAVIGEKNIPISVDEIAKEK 210
>FOLC_ECOLI (P08192) FolC bifunctional protein [Includes:
Folylpolyglutamate synthase (EC 6.3.2.17)
(Folylpoly-gamma-glutamate synthetase) (FPGS);
Dihydrofolate synthase (EC 6.3.2.12)]
Length = 422
Score = 80.9 bits (198), Expect = 7e-15
Identities = 89/342 (26%), Positives = 142/342 (41%), Gaps = 70/342 (20%)
Query: 69 GYSVGCYSSPHIQTIRERILGRSGD-PVSAKLLNNLFHRIKQDLDQAIKEENGCIS--HF 125
GY VG YSSPH+ ER+ + + P SA + I+ G IS +F
Sbjct: 75 GYKVGVYSSPHLVRYTERVRVQGQELPESAHTASFA----------EIESARGDISLTYF 124
Query: 126 EVFTAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSL 185
E T A LF ++D+ ++E GLGG DATNI+ + AV+T++ +H LG
Sbjct: 125 EYGTLSALWLFKQAQLDVVILEVGLGGRLDATNIVDAD---VAVVTSIALDHTDWLGPDR 181
Query: 186 ESIAMAKAGIIKQGCPLVLGGPFLPH-IEYIIREKAVNMDSPVVSASDSGNNLAVKSFSI 244
ESI KAGI + P ++G P +P I + +EK + V + S + +FS
Sbjct: 182 ESIGREKAGIFRSEKPAIVGEPEMPSTIADVAQEKGALLQRRGVEWNYSVTD-HDWAFSD 240
Query: 245 LNGKPCQICDIEIQTVKDLKLAMDPDPRCACGPVSCMI*SCKCLVLTNFKMQQLLLV*HS 304
+G T+++L L + P P A +
Sbjct: 241 AHG-----------TLENLPLPLVPQPNAATALAA------------------------- 264
Query: 305 VFVI*VDGEFLMNL*GVG*SVHTSLEGVSS*NLKKPRPWDFLEQQYCLI*SVAHTKESAK 364
+ G+ S + +G++S L P + + + +I VAH +A+
Sbjct: 265 -----------LRASGLEVSENAIRDGIASAIL--PGRFQIVSESPRVIFDVAHNPHAAE 311
Query: 365 ALMNTIKMASPKARLAFVVAMASDKDHAG---FAREILSDAY 403
L +K R+ V+ M DKD AG + + ++ D Y
Sbjct: 312 YLTGRMKALPKNGRVLAVIGMLHDKDIAGTLAWLKSVVDDWY 353
>FOLC_BUCBP (Q89AT2) FolC bifunctional protein [Includes:
Folylpolyglutamate synthase (EC 6.3.2.17)
(Folylpoly-gamma-glutamate synthetase) (FPGS);
Dihydrofolate synthase (EC 6.3.2.12)]
Length = 426
Score = 79.3 bits (194), Expect = 2e-14
Identities = 50/156 (32%), Positives = 83/156 (53%), Gaps = 11/156 (7%)
Query: 69 GYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCISHFEVF 128
GY VG Y+SPH+ ER+ ++ L +L+H I D + + ++ FE
Sbjct: 74 GYRVGLYTSPHLMRYTERVR------INGFELEHLYH-ISAFNDVKYFQNDVLLTRFEFI 126
Query: 129 TAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLESI 188
T A ILF +DI ++E GLGG DATNI+S+ +VIT + +H LG + SI
Sbjct: 127 TLSALILFKSYNLDIIILEVGLGGRLDATNILSAD---VSVITNIDIDHSKILGVNRSSI 183
Query: 189 AMAKAGIIKQGCPLVLGGPFLPHI-EYIIREKAVNM 223
++ K+GI ++ ++ P + +Y+ ++K V +
Sbjct: 184 SVEKSGIFRKNKIAIVADNNFPKVAQYLAKKKKVRL 219
>FOLE_NEUCR (O13492) Folylpolyglutamate synthase (EC 6.3.2.17)
(Folylpoly-gamma-glutamate synthetase) (FPGS)
Length = 528
Score = 76.3 bits (186), Expect = 2e-13
Identities = 50/155 (32%), Positives = 86/155 (55%), Gaps = 9/155 (5%)
Query: 72 VGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLD--QAIKEENGCIS---HFE 126
+G ++SPH+ +RERI S P+S +L F + L+ Q K+E S +
Sbjct: 129 IGLFTSPHLIAVRERIRIDS-KPISEELFARYFFEVWDRLETSQLAKDEVELGSKPIYAR 187
Query: 127 VFTAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLE 186
T M++ ++ E VD+A+ E G+GG DATN++ ++ I+T+G +H+ LG +++
Sbjct: 188 YLTLMSYHVYLSEGVDVAIYETGIGGEYDATNVVDRPVVSG--ISTLGIDHVFVLGDTVD 245
Query: 187 SIAMAKAGIIKQGCPLVLGGPFLPHIEYIIREKAV 221
IA KAGI+K G P +P +++++AV
Sbjct: 246 KIAWHKAGIMKTGSP-AFTIEQVPSATQVLKDRAV 279
>FOLE_CANAL (Q9Y893) Folylpolyglutamate synthase (EC 6.3.2.17)
(Folylpoly-gamma-glutamate synthetase) (FPGS)
Length = 514
Score = 73.9 bits (180), Expect = 9e-13
Identities = 43/140 (30%), Positives = 71/140 (50%), Gaps = 13/140 (9%)
Query: 72 VGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCIS-------- 123
+G Y+SPH++++RERI +G P++ + F + + C +
Sbjct: 102 IGLYTSPHLKSVRERIR-INGQPINQEKFAKYFFEVWDKFTTTKSDPQECPTLQPCDQVK 160
Query: 124 --HFEVFTAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAAL 181
+F+ T ++F +F E VD A+ E G+GG D+TNII + I+ +G +H L
Sbjct: 161 PMYFKYLTILSFHVFLQEGVDTAIYEVGVGGTYDSTNIIDKPTVTG--ISALGIDHTFML 218
Query: 182 GGSLESIAMAKAGIIKQGCP 201
G ++ SI K GI K+G P
Sbjct: 219 GNNIASITENKTGIFKKGVP 238
>FOLE_YEAST (Q08645) Folylpolyglutamate synthase (EC 6.3.2.17)
(Folylpoly-gamma-glutamate synthetase) (FPGS)
Length = 548
Score = 73.6 bits (179), Expect = 1e-12
Identities = 49/165 (29%), Positives = 78/165 (46%), Gaps = 11/165 (6%)
Query: 64 KFKTEGYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKE------ 117
++K + +G Y+SPH++++RERI +G+P+S + F + LD
Sbjct: 144 QYKEQLPRIGLYTSPHLKSVRERIR-INGEPISEEKFAKYFFEVWDRLDSTTSSLDKFPH 202
Query: 118 --ENGCISHFEVFTAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGE 175
+F+ T ++F F E V E G+GG D+TNII + +T +G
Sbjct: 203 MIPGSKPGYFKFLTLLSFHTFIQEDCKSCVYEVGVGGELDSTNIIEKPIVCG--VTLLGI 260
Query: 176 EHLAALGGSLESIAMAKAGIIKQGCPLVLGGPFLPHIEYIIREKA 220
+H LG ++E IA K GI K G P P I++E+A
Sbjct: 261 DHTFMLGDTIEEIAWNKGGIFKSGAPAFTVEKQPPQGLTILKERA 305
Score = 30.8 bits (68), Expect = 8.8
Identities = 17/58 (29%), Positives = 31/58 (53%), Gaps = 4/58 (6%)
Query: 455 KEQPVSSESNLTDGKTILATESSLKDCLRMANEILNRRRDEKGVIVITGSLHIVSSVL 512
+E V + + + D + +S+++ ANE++ DE I +TGSLH+V +L
Sbjct: 486 QESLVKNWNKIDDNRAKTHVTASIEE----ANELIETLYDEPADIFVTGSLHLVGGLL 539
>FOLC_BUCAI (P57265) FolC bifunctional protein [Includes:
Folylpolyglutamate synthase (EC 6.3.2.17)
(Folylpoly-gamma-glutamate synthetase) (FPGS);
Dihydrofolate synthase (EC 6.3.2.12)]
Length = 411
Score = 72.0 bits (175), Expect = 3e-12
Identities = 52/156 (33%), Positives = 77/156 (49%), Gaps = 19/156 (12%)
Query: 69 GYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEE---NGCI-SH 124
GY VG Y+SPH+ ER+ +N ++ +D E NG + ++
Sbjct: 69 GYQVGLYTSPHLINFVERVR-----------INGFVLHEEEHIDSFQNVELVRNGVLLTY 117
Query: 125 FEVFTAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGS 184
FE T A ILF +D +++ GLGG DATNII S ++IT +G +H + LG
Sbjct: 118 FEFITLAALILFKRYSLDCIILKVGLGGRLDATNIIDSD---ISIITNIGIDHTSILGRD 174
Query: 185 LESIAMAKAGIIKQGCPLVLGGPFLPHIEY-IIREK 219
SIA K G+ ++ V+G +P Y I +EK
Sbjct: 175 RISIAREKCGVFRKNKISVIGETDIPCSMYQIAKEK 210
>FOLC_YEAST (P36001) Putative folylpolyglutamate synthase (EC
6.3.2.17) (Folylpoly-gamma-glutamate synthetase) (FPGS)
Length = 430
Score = 44.7 bits (104), Expect = 6e-04
Identities = 38/154 (24%), Positives = 67/154 (42%), Gaps = 31/154 (20%)
Query: 48 LGRMRRLLDRFGNPH-----------------SKFKT----------EGYSVGCYSSPHI 80
L R+ RLL+ GNP SKF T + +G Y++ +
Sbjct: 10 LPRINRLLEHVGNPQDSLSILHIAGTNGKETVSKFLTSILQHPGQQRQRVLIGRYTTSSL 69
Query: 81 QTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCISHFEVFTAMAFILFADEK 140
+E + + + +S L+ + RI+++L +A ++ E+ T++A + FA +
Sbjct: 70 LNAKEEDISINNEAIS--LIE--YSRIEKELIEADSSLKLQCNNLELLTSVALVYFAKKN 125
Query: 141 VDIAVIEAGLGGARDATNIISSSGLAAAVITTVG 174
+IE GL G +D +II+ IT VG
Sbjct: 126 CQWCIIETGLAGKQDPGSIIAGQSRVCCAITNVG 159
>MURF_BACSU (P96613)
UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine
ligase (EC 6.3.2.10) (UDP-MurNAc-pentapeptide
synthetase) (D-alanyl-D-alanine-adding enzyme)
Length = 457
Score = 40.0 bits (92), Expect = 0.015
Identities = 35/117 (29%), Positives = 52/117 (43%), Gaps = 11/117 (9%)
Query: 134 ILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLESIAMAKA 193
+L E +IAV+E G+ + + + AAVIT +GE H+ L GS E IA AK
Sbjct: 151 VLAMPENTEIAVLEMGMSAKGEIDLLSRLANPDAAVITNIGESHMQDL-GSREGIAEAKL 209
Query: 194 GII----KQGCPLVLGGPFLPHIEYIIREKAVNMDS------PVVSASDSGNNLAVK 240
II + G + +G L Y + K + VS S+ G + +K
Sbjct: 210 EIINGLKEDGVLIYIGDEPLLQNAYSCQTKTYGTGTHNDYQLQDVSQSEEGTHFTIK 266
>ENO_DROME (P15007) Enolase (EC 4.2.1.11) (2-phosphoglycerate
dehydratase) (2-phospho-D-glycerate hydro-lyase)
Length = 433
Score = 33.5 bits (75), Expect = 1.4
Identities = 26/95 (27%), Positives = 45/95 (47%), Gaps = 13/95 (13%)
Query: 158 NIISSSGLAAAVITTVGEEHLAALGGSL---ESIAMAKAGIIKQGCPLVLGGPFLPHIEY 214
+++ + + +I G E+ + G + S+A+AKAG K+G PL HI
Sbjct: 83 DVVDQASIDNFMIKLDGTENKSKFGANAILGVSLAVAKAGAAKKGVPL------YKHIAD 136
Query: 215 IIREKAVNMDSP----VVSASDSGNNLAVKSFSIL 245
+ K + + P + S +GN LA++ F IL
Sbjct: 137 LAGNKEIILPVPAFNVINGGSHAGNKLAMQEFMIL 171
>MURF_BORBU (Q44777)
UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine
ligase (EC 6.3.2.10) (UDP-MurNAc-pentapeptide
synthetase) (D-alanyl-D-alanine-adding enzyme)
Length = 464
Score = 33.1 bits (74), Expect = 1.8
Identities = 30/116 (25%), Positives = 51/116 (43%), Gaps = 4/116 (3%)
Query: 134 ILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLESIAMAKA 193
IL + + AV E G+ + + +IT + H+ A L++IA K+
Sbjct: 163 ILRVEGNEEYAVFEVGVSYVGEMDLLSQILKPEIVIITNISYAHMQAFK-ELQAIAFEKS 221
Query: 194 GIIKQGCPLVLGGPFLPHIEYIIREKAVNMDSPVVS-ASDSGNNLAVKSFSILNGK 248
II + + + + Y+ EK + +P V NL++KSFS L+GK
Sbjct: 222 KIIGKNIEIFVVNEMNDYCVYL--EKRAKIANPNVKIVYFDFENLSIKSFSFLDGK 275
>METK_SYNPX (Q7U4S6) S-adenosylmethionine synthetase (EC 2.5.1.6)
(Methionine adenosyltransferase) (AdoMet synthetase)
(MAT)
Length = 419
Score = 32.7 bits (73), Expect = 2.3
Identities = 20/46 (43%), Positives = 22/46 (47%), Gaps = 4/46 (8%)
Query: 241 SFSILNGKPCQICDIEIQTVKDLKLAMDPDPRCACGPV----SCMI 282
S S+ G P +ICD V D LA DP R AC V CMI
Sbjct: 8 SESVTEGHPDKICDQVSDAVLDALLAQDPSSRVACETVVNTGLCMI 53
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.327 0.140 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,530,131
Number of Sequences: 164201
Number of extensions: 2273713
Number of successful extensions: 7683
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 7614
Number of HSP's gapped (non-prelim): 54
length of query: 516
length of database: 59,974,054
effective HSP length: 115
effective length of query: 401
effective length of database: 41,090,939
effective search space: 16477466539
effective search space used: 16477466539
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC124963.22


