

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124963.21 + phase: 0 /pseudo
(739 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
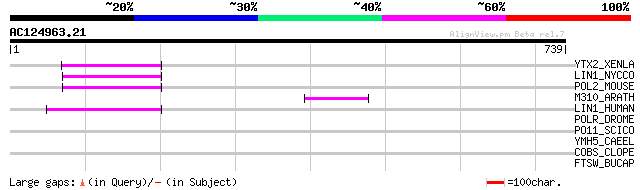
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 58 1e-07
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 55 7e-07
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 52 6e-06
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 52 6e-06
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 49 6e-05
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 36 0.42
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 34 1.2
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 34 1.6
COBS_CLOPE (Q8XLK5) Cobalamin synthase (EC 2.-.-.-) 32 6.0
FTSW_BUCAP (Q8K9T3) Cell division protein ftsW 32 7.9
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 57.8 bits (138), Expect = 1e-07
Identities = 39/133 (29%), Positives = 64/133 (47%)
Query: 70 RRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVVD 129
R + ++ +SL+ + YKI+AK ++ RL+ V+ VI QS V GR I D + + +++
Sbjct: 520 RLIKNWRPVSLLSTDYKIVAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLH 579
Query: 130 DAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVNG 189
A R L +D E+A+D VD YL + F +K +A + +N
Sbjct: 580 FARRTGLSLAFLSLDQEKAFDRVDHQYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINW 639
Query: 190 SPTDEFPLKRGCK 202
S T RG +
Sbjct: 640 SLTAPLAFGRGVR 652
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 55.1 bits (131), Expect = 7e-07
Identities = 36/133 (27%), Positives = 66/133 (49%), Gaps = 1/133 (0%)
Query: 71 RLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVVDD 130
R ++ ISL+ KIL K+L NR++ I +I Q F+ G Q I + V+
Sbjct: 525 RKENYRPISLMNIDAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQGWFNIRKSINVIQH 584
Query: 131 AHRCK-KELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVNG 189
++ K K+ ++ +D E+A+D + ++ + K+G K I+ T ++++NG
Sbjct: 585 INKLKNKDHMILSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKLIEAIYSKPTANIILNG 644
Query: 190 SPTDEFPLKRGCK 202
FPL+ G +
Sbjct: 645 VKLKSFPLRSGTR 657
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 52.0 bits (123), Expect = 6e-06
Identities = 37/133 (27%), Positives = 65/133 (48%), Gaps = 1/133 (0%)
Query: 71 RLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVVDD 130
++ +F ISL+ KIL K+LANR++ I +I Q F+ G Q I + V+
Sbjct: 552 KIENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQVGFIPGMQGWFNIRKSINVIHY 611
Query: 131 AHRCK-KELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVNG 189
++ K K ++ +D E+A+D + ++ +V+ + G IK ++ VNG
Sbjct: 612 INKLKDKNHMIISLDAEKAFDKIQHPFMIKVLERSGIQGPYLNMIKAIYSKPVANIKVNG 671
Query: 190 SPTDEFPLKRGCK 202
+ PLK G +
Sbjct: 672 EKLEAIPLKSGTR 684
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 52.0 bits (123), Expect = 6e-06
Identities = 30/89 (33%), Positives = 48/89 (53%), Gaps = 3/89 (3%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEV-GGYGVRRIREFNSALLGKWCWQLLIEKESLWYR 451
F+ +N +K++WV W +C SKE GG G R + FN ALL K ++++ + +L R
Sbjct: 28 FWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRDLGWFNQALLAKQSFRIIHQPHTLLSR 87
Query: 452 VLSARYNEEGGRL--LDGGRSTSAWWRVI 478
+L +RY + G R + AW +I
Sbjct: 88 LLRSRYFPHSSMMECSVGTRPSYAWRSII 116
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 48.5 bits (114), Expect = 6e-05
Identities = 39/156 (25%), Positives = 73/156 (46%), Gaps = 3/156 (1%)
Query: 50 GGLPKESIALSLLLFLR--LRARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDS 107
G LP S++L + + +F ISL+ KIL K+LAN+++ I +I
Sbjct: 502 GILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQQHIKKLIHHD 561
Query: 108 QSTFVKGRQILDGILVANEVVDDAHRCKK-ELLLFKVDFEEAYDIVDWSYLDEVMYKMGF 166
Q F+ Q I + ++ +R K ++ +D E+A+D + ++ + + K+G
Sbjct: 562 QVGFIPAMQGWFNIRKSINIIQHINRTKDTNHMIISIDAEKAFDKIQQPFMLKPLNKLGI 621
Query: 167 PVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
K I+ T ++++NG + PLK G +
Sbjct: 622 DGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTR 657
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 35.8 bits (81), Expect = 0.42
Identities = 37/155 (23%), Positives = 68/155 (43%), Gaps = 5/155 (3%)
Query: 50 GGLPKESIALSLLLFL--RLRARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDS 107
G LP SI L+ +F+ + A+R DF IS+ + + L +LA RL I
Sbjct: 389 GNLP-HSIRLARTVFIPKTVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSINW--DPR 445
Query: 108 QSTFVKGRQILDGILVANEVVDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFP 167
Q F+ D + + V+ +H+ + + +D +A+D + + + + + G P
Sbjct: 446 QRGFLPTDGCADNATIVDLVLRHSHKHFRSCYIANLDVSKAFDSLSHASIYDTLRAYGAP 505
Query: 168 VL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
++ TS+ +G ++EF RG K
Sbjct: 506 KGFVDYVQNTYEGGGTSLNGDGWSSEEFVPARGVK 540
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 34.3 bits (77), Expect = 1.2
Identities = 30/126 (23%), Positives = 61/126 (47%), Gaps = 12/126 (9%)
Query: 33 IFCVIMLCVLLVNFIGMGGLPKESIALSLLLFLRLRARRLND---FCLISLVESMYKILA 89
+FC+ C+L F P++ SL++ L+L R +D + I L++++ K+L
Sbjct: 444 MFCLYKQCLLESYF------PQKWKIASLVILLKLLDRIRSDPGSYRPICLLDNLGKVLE 497
Query: 90 KVLANRL-RYVIGMVISDSQSTFVKGRQILDGILVANEVVDDAHRCKKELLLFKVDFEEA 148
++ RL + ++ + +S Q F G+ D V+ + K ++ +DF+ A
Sbjct: 498 GIMVKRLDQKLMDVEVSPYQFAFTYGKSTEDAWRCVQRHVECSEM--KYVIGLNIDFQGA 555
Query: 149 YDIVDW 154
+D + W
Sbjct: 556 FDNLGW 561
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 33.9 bits (76), Expect = 1.6
Identities = 27/124 (21%), Positives = 56/124 (44%), Gaps = 1/124 (0%)
Query: 78 ISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVVDDAHRCKKE 137
ISL + +I+ +++ +R+R ++S Q F+ R ++ + + + +K
Sbjct: 688 ISLTDPFARIMERIICSRIRSEYSHLLSPHQHGFLNFRSCPSSLVRSISLYHSILKNEKS 747
Query: 138 LLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVNG-SPTDEFP 196
L + DF +A+D V L + + G L KE + T S+ +N ++ +P
Sbjct: 748 LDILFFDFAKAFDKVSHPILLKKLALFGLDKLTCSWFKEFLHLRTFSVKINKFVSSNAYP 807
Query: 197 LKRG 200
+ G
Sbjct: 808 ISSG 811
>COBS_CLOPE (Q8XLK5) Cobalamin synthase (EC 2.-.-.-)
Length = 251
Score = 32.0 bits (71), Expect = 6.0
Identities = 16/60 (26%), Positives = 37/60 (61%), Gaps = 2/60 (3%)
Query: 20 LMVLFLASLKIFGIFCVIMLCVLLVNFIGMGGLPKESIALSLLLFLRLRARRLNDFCLIS 79
L +L +++ F + V++L LLV F GM + +++ + +L+F+ + +R +N + ++S
Sbjct: 102 LRILKDSTVGAFSVISVVLL--LLVEFAGMFTVLNKNLDMRVLIFIPIASRAINGYFIVS 159
>FTSW_BUCAP (Q8K9T3) Cell division protein ftsW
Length = 353
Score = 31.6 bits (70), Expect = 7.9
Identities = 20/66 (30%), Positives = 32/66 (48%), Gaps = 14/66 (21%)
Query: 10 GTVIIINIRALMVLFLASLKIFGIFCVIMLCVLLVNFIGMGGLPKESIALSLLLFLRLRA 69
GTVI++ + L VLFL+ +KI F +I L++ +L+LF R
Sbjct: 139 GTVIVLFLTTLSVLFLSGVKIKQFFIIIFFVTLIIT--------------ALVLFEPYRI 184
Query: 70 RRLNDF 75
+R+ F
Sbjct: 185 KRILSF 190
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.350 0.157 0.564
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 76,797,433
Number of Sequences: 164201
Number of extensions: 2983993
Number of successful extensions: 9998
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 9984
Number of HSP's gapped (non-prelim): 15
length of query: 739
length of database: 59,974,054
effective HSP length: 118
effective length of query: 621
effective length of database: 40,598,336
effective search space: 25211566656
effective search space used: 25211566656
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC124963.21


