

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124958.5 + phase: 0
(938 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
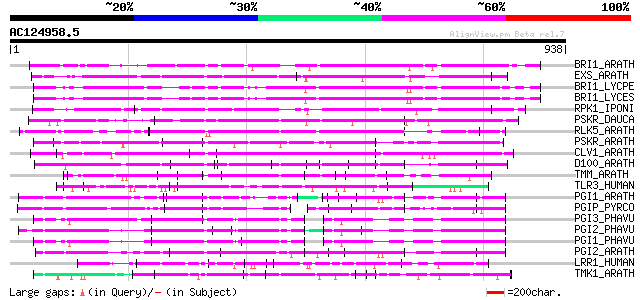
Score E
Sequences producing significant alignments: (bits) Value
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 275 5e-73
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 245 5e-64
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 234 6e-61
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 232 3e-60
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 220 2e-56
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 214 9e-55
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 206 3e-52
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 202 4e-51
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 196 2e-49
D100_ARATH (Q00874) DNA-damage-repair/toleration protein DRT100 ... 132 6e-30
TMM_ARATH (Q9SSD1) TOO MANY MOUTHS protein precursor (TMM) 114 2e-24
TLR3_HUMAN (O15455) Toll-like receptor 3 precursor 108 5e-23
PGI1_ARATH (Q9M5J9) Polygalacturonase inhibitor 1 precursor (Pol... 102 4e-21
PGIP_PYRCO (Q05091) Polygalacturonase inhibitor precursor (Polyg... 98 9e-20
PGI3_PHAVU (P58823) Polygalacturonase inhibitor 3 precursor (Pol... 98 9e-20
PGI2_PHAVU (P58822) Polygalacturonase inhibitor 2 precursor (Pol... 98 1e-19
PGI1_PHAVU (P35334) Polygalacturonase inhibitor 1 precursor (Pol... 97 2e-19
PGI2_ARATH (Q9M5J8) Polygalacturonase inhibitor 2 precursor (Pol... 97 3e-19
LRR1_HUMAN (Q9BTT6) Leucine-rich repeat-containing protein 1 (LA... 96 3e-19
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 95 8e-19
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 275 bits (702), Expect = 5e-73
Identities = 281/904 (31%), Positives = 393/904 (43%), Gaps = 159/904 (17%)
Query: 34 SQFIASEAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKGVGCNTTTGHVISLNLHCSNSL 93
SQ + E L+ FK+ L D NLL W K+ C + GV C I L+ S L
Sbjct: 29 SQSLYREIHQLISFKDVLPD-KNLLPDWSSNKNPCTFDGVTCRDDKVTSIDLS---SKPL 84
Query: 94 DKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNL--LD 151
+ ++SSLL L L L LS N + +V F + +L LDLS + G + L
Sbjct: 85 NVGFSAVSSSLLSLTGLESLFLS-NSHINGSVSGF-KCSASLTSLDLSRNSLSGPVTTLT 142
Query: 152 NLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSL 211
+LG+ S L+ L++S N+ LD G
Sbjct: 143 SLGSCSGLKFLNVSSNT-----------------LDFPG--------------------- 164
Query: 212 DTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFN-MTIPDW-LFENCHHLQNLNLSN 269
++SG + +SL LDLS N+ + + W L + C L++L +S
Sbjct: 165 ---KVSG------------GLKLNSLEVLDLSANSISGANVVGWVLSDGCGELKHLAISG 209
Query: 270 NNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLG 329
N + G + S R L LD+S N+ + IP F L LD+S N LSG +
Sbjct: 210 NKISGDVDVS--RCVNLEFLDVSSNNFSTGIP-FLGDCSALQHLDISGNKLSGDFSRAIS 266
Query: 330 QDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVL 389
LK L +S NQ G + L +L L+LA N G I D L L
Sbjct: 267 TC---TELKLLNISSNQFVGPIPP--LPLKSLQYLSLAENKFTGEIPDFLSGACDTLTGL 321
Query: 390 DLSFNHVTLNMSKNWVPPF-----QLETIGLANCHLGPQFPK-WIQTQKNFSHIDISNAG 443
DLS NH VPPF LE++ L++ + + P + + +D+S
Sbjct: 322 DLSGNHFY-----GAVPPFFGSCSLLESLALSSNNFSGELPMDTLLKMRGLKVLDLSFNE 376
Query: 444 VSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCP-LPRLPPN 502
S +P +LS + L TLDLS+N+FS P LP L N
Sbjct: 377 FSGELPESLTNLSAS-----------------------LLTLDLSSNNFSGPILPNLCQN 413
Query: 503 LRN----LDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILN 558
+N L L +N F G I L + L +L LSFN LSG IP+ + + + L
Sbjct: 414 PKNTLQELYLQNNGFTGKIPPT---LSNCSELVSLHLSFNYLSGTIPSSLGSLSKLRDLK 470
Query: 559 LAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPY 618
L +N G IP +K L LI+ N+L+G+IP L NC L ++L +NRL G IP
Sbjct: 471 LWLNMLEGEIPQELMYVKTLETLILDFNDLTGEIPSGLSNCTNLNWISLSNNRLTGEIPK 530
Query: 619 WIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLAL----- 673
WIG ++ L +L L NNSF NIP L +SL LDL+ N G IP +F
Sbjct: 531 WIGR-LENLAILKLSNNSFSGNIPAELGDCRSLIWLDLNTNLFNGTIPAAMFKQSGKIAA 589
Query: 674 ---------------TTEESINEKSYMEFMTIEESLPIYLSRTKHPLLIPWKGVN----- 713
+E + +EF I LS T++P I +
Sbjct: 590 NFIAGKRYVYIKNDGMKKECHGAGNLLEFQGIRSEQLNRLS-TRNPCNITSRVYGGHTSP 648
Query: 714 VFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELES 773
F N G + F +D+S N L+ IP EIG + L LNL N + GSIP +G+L
Sbjct: 649 TFDNNGSMMF-----LDMSYNMLSGYIPKEIGSMPYLFILNLGHNDISGSIPDEVGDLRG 703
Query: 774 LNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVFYKGNPHLC 833
LN+LDLS N L IP +M+ + L+ +DLS N LSG IP Q ++F + NP LC
Sbjct: 704 LNILDLSSNKLDGRIPQAMSALTMLTEIDLSNNNLSGPIPEMGQFETFPPAKFLNNPGLC 763
Query: 834 GPPLRKACPRNSSFEDTHCSHSEEHENDGNHGDKVLGMEINPLYISMAMGFSTGFWVFWG 893
G PL + P N+ D + H H G L S+AMG F +G
Sbjct: 764 GYPLPRCDPSNA---DGYAHHQRSH-----------GRRPASLAGSVAMGLLFSFVCIFG 809
Query: 894 SLIL 897
+++
Sbjct: 810 LILV 813
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells
protein) (EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 245 bits (625), Expect = 5e-64
Identities = 250/853 (29%), Positives = 373/853 (43%), Gaps = 106/853 (12%)
Query: 37 IASEAEALLEFKEGLKDPSNLLSSWKHGKDC--CQWKGVGCNTTTGHVISLNLHCSNSLD 94
++SE +L+ FK L++PS LLSSW C W GV C
Sbjct: 23 LSSETTSLISFKRSLENPS-LLSSWNVSSSASHCDWVGVTC------------------- 62
Query: 95 KLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLG 154
L G +NS L LP LS ++ +P +S+ KNL+ L L+ F G + +
Sbjct: 63 -LLGRVNS--LSLPSLS---------LRGQIPKEISSLKNLRELCLAGNQFSGKIPPEIW 110
Query: 155 NLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTL 214
NL L++LDLS NS + L L L LDLS S F L SLD
Sbjct: 111 NLKHLQTLDLSGNSLTGLLPRLLSELPQLLYLDLSDNHFSGSLPPSFFISLPALSSLDVS 170
Query: 215 RLSGCQLHKLPTSPPPEM-NFDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQ 273
+ L PPE+ +L L + N+F+ IP + N L+N +
Sbjct: 171 N------NSLSGEIPPEIGKLSNLSNLYMGLNSFSGQIPSEI-GNISLLKNFAAPSCFFN 223
Query: 274 GQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHG 333
G + I ++ LA LDLS N L IP F +L NL L+L L G IP LG
Sbjct: 224 GPLPKEISKLKHLAKLDLSYNPLKCSIPKSFGELHNLSILNLVSAELIGLIPPELG---N 280
Query: 334 QNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSF 393
SLK L LS N L+G L + ++ L+ + N + G + + + L L L+
Sbjct: 281 CKSLKSLMLSFNSLSGPLPLELSEIP-LLTFSAERNQLSGSLPS-WMGKWKVLDSLLLAN 338
Query: 394 NHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFW 453
N + + L+ + LA+ L P+ + + ID+S +S + F
Sbjct: 339 NRFSGEIPHEIEDCPMLKHLSLASNLLSGSIPRELCGSGSLEAIDLSGNLLSGTIEEVF- 397
Query: 454 DLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPR--------------- 498
D ++ + L++N++ + K L LDL +N+F+ +P+
Sbjct: 398 DGCSSLGELLLTNNQINGSIPEDLWKLPLMALDLDSNNFTGEIPKSLWKSTNLMEFTASY 457
Query: 499 ------LP------PNLRNLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPN 546
LP +L+ L LS N G I L SL L+L+ N G IP
Sbjct: 458 NRLEGYLPAEIGNAASLKRLVLSDNQLTGEIPREIGKL---TSLSVLNLNANMFQGKIPV 514
Query: 547 CWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPE------------ 594
+ T++ L+L NN G IPD +L L L++ NNLSG IP
Sbjct: 515 ELGDCTSLTTLDLGSNNLQGQIPDKITALAQLQCLVLSYNNLSGSIPSKPSAYFHQIEMP 574
Query: 595 TLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHIL 654
L Q + +L NRL GPIP +G + +L+ + L NN IP +L +L +L IL
Sbjct: 575 DLSFLQHHGIFDLSYNRLSGPIPEELG-ECLVLVEISLSNNHLSGEIPASLSRLTNLTIL 633
Query: 655 DLSENQLTGAIPRCVFLALTTE------ESINEKSYMEFMTIEESLPIYLSRTKHPLLIP 708
DLS N LTG+IP+ + +L + +N F + + + L++ K +P
Sbjct: 634 DLSGNALTGSIPKEMGNSLKLQGLNLANNQLNGHIPESFGLLGSLVKLNLTKNKLDGPVP 693
Query: 709 WKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSI 768
N+ + L +DLS N L+ E+ E+ + +L L + +N+ G IPS +
Sbjct: 694 ASLGNL---------KELTHMDLSFNNLSGELSSELSTMEKLVGLYIEQNKFTGEIPSEL 744
Query: 769 GELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVFYKG 828
G L L LD+S N LS EIPT + + L +L+L+ N L G++P Q + G
Sbjct: 745 GNLTQLEYLDVSENLLSGEIPTKICGLPNLEFLNLAKNNLRGEVPSDGVCQDPSKALLSG 804
Query: 829 NPHLCGPPLRKAC 841
N LCG + C
Sbjct: 805 NKELCGRVVGSDC 817
Score = 106 bits (265), Expect = 3e-22
Identities = 97/332 (29%), Positives = 145/332 (43%), Gaps = 33/332 (9%)
Query: 485 LDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCE---ILCFNNSLENLDLSFNNLS 541
+DLS+ + S L +L N L S+ + + C+ + C + +L L +L
Sbjct: 21 VDLSSETTS--LISFKRSLENPSLLSSWNVSSSASHCDWVGVTCLLGRVNSLSLPSLSLR 78
Query: 542 GVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQV 601
G IP ++ N+ L LA N F G IP +LK+L L + N+L+G +P L
Sbjct: 79 GQIPKEISSLKNLRELCLAGNQFSGKIPPEIWNLKHLQTLDLSGNSLTGLLPRLLSELPQ 138
Query: 602 LTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQL 661
L L+L N G +P + L L + NNS IP + +L +L L + N
Sbjct: 139 LLYLDLSDNHFSGSLPPSFFISLPALSSLDVSNNSLSGEIPPEIGKLSNLSNLYMGLNSF 198
Query: 662 TGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRL 721
+G IP + S+ + LP +S+ KH
Sbjct: 199 SGQIPSEI-----GNISLLKNFAAPSCFFNGPLPKEISKLKH------------------ 235
Query: 722 FFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSR 781
L +DLS N L IP G+L LS LNL +L+G IP +G +SL L LS
Sbjct: 236 ----LAKLDLSYNPLKCSIPKSFGELHNLSILNLVSAELIGLIPPELGNCKSLKSLMLSF 291
Query: 782 NNLSCEIPTSMANIDRLSWLDLSYNALSGKIP 813
N+LS +P ++ I L++ N LSG +P
Sbjct: 292 NSLSGPLPLELSEIPLLTF-SAERNQLSGSLP 322
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 234 bits (598), Expect = 6e-61
Identities = 260/894 (29%), Positives = 391/894 (43%), Gaps = 151/894 (16%)
Query: 40 EAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKGVGCNTTTGHVISLNLHCSNSLDKLQGH 99
+++ LL FK L LL +W D C + GV C + +S+D
Sbjct: 43 DSQQLLSFKAALPPTPTLLQNWLSSTDPCSFTGVSCKNSR----------VSSID----- 87
Query: 100 LNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLS-- 157
L+++ L + + S V +L NL+ L L +AN G+L +
Sbjct: 88 LSNTFLSVDF-------------SLVTSYLLPLSNLESLVLKNANLSGSLTSAAKSQCGV 134
Query: 158 LLESLDLSDNSFY--VNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTL- 214
L+S+DL++N+ ++++ S+LK L+LS L + L LD
Sbjct: 135 TLDSIDLAENTISGPISDISSFGVCSNLKSLNLSKNFLDPPGKEMLKGATFSLQVLDLSY 194
Query: 215 -RLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQ 273
+SG L +S M F L + GN +IP+ F+N
Sbjct: 195 NNISGFNLFPWVSS----MGFVELEFFSIKGNKLAGSIPELDFKN--------------- 235
Query: 274 GQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHG 333
L+ LDLS N+ + + P+F D NL LDLS N G I S+L
Sbjct: 236 ------------LSYLDLSANNFSTVFPSFKD-CSNLQHLDLSSNKFYGDIGSSLSSC-- 280
Query: 334 QNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKV-LDLS 392
L L L+ NQ G + + +L L L N+ +G+ + LA+ V LDLS
Sbjct: 281 -GKLSFLNLTNNQFVGLVPK--LPSESLQYLYLRGNDFQGVYPN-QLADLCKTVVELDLS 336
Query: 393 FNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWF 452
+N+ + VP L C + +DISN S +P
Sbjct: 337 YNNFS-----GMVPE------SLGEC-------------SSLELVDISNNNFSGKLPVDT 372
Query: 453 WDLSPNVEYMNLSSNE-LRRCGQDFSQKFKLKTLDLSNNSFSCPLPR-----LPPNLRNL 506
N++ M LS N+ + FS KL+TLD+S+N+ + +P NL+ L
Sbjct: 373 LLKLSNIKTMVLSFNKFVGGLPDSFSNLPKLETLDMSSNNLTGIIPSGICKDPMNNLKVL 432
Query: 507 DLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIG 566
L +NLF G I + L + L +LDLSFN L+G IP+ + + + L L +N G
Sbjct: 433 YLQNNLFKGPIP---DSLSNCSQLVSLDLSFNYLTGSIPSSLGSLSKLKDLILWLNQLSG 489
Query: 567 SIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQI 626
IP L+ L LI+ N+L+G IP +L NC L ++L +N+L G IP +G +
Sbjct: 490 EIPQELMYLQALENLILDFNDLTGPIPASLSNCTKLNWISLSNNQLSGEIPASLGR-LSN 548
Query: 627 LMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVF-------LALTT---- 675
L +L LGNNS NIP L +SL LDL+ N L G+IP +F +AL T
Sbjct: 549 LAILKLGNNSISGNIPAELGNCQSLIWLDLNTNFLNGSIPPPLFKQSGNIAVALLTGKRY 608
Query: 676 --------EESINEKSYMEFMTIEESLPIYLSRTKHPLLIP--WKGVN--VFFNEGRLFF 723
+E + +EF I + +S T+HP ++G+ F + G + F
Sbjct: 609 VYIKNDGSKECHGAGNLLEFGGIRQEQLDRIS-TRHPCNFTRVYRGITQPTFNHNGSMIF 667
Query: 724 EILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNN 783
+DLS N L IP E+G + LS LNL N L G IP +G L+++ +LDLS N
Sbjct: 668 -----LDLSYNKLEGSIPKELGAMYYLSILNLGHNDLSGMIPQQLGGLKNVAILDLSYNR 722
Query: 784 LSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVFYKGNPHLCGPPLRKACPR 843
+ IP S+ ++ L +DLS N LSG IP +F + + N LCG PL C
Sbjct: 723 FNGTIPNSLTSLTLLGEIDLSNNNLSGMIPESAPFDTFPDYRFANN-SLCGYPLPLPCSS 781
Query: 844 NSSFEDTHCSHSEEHENDGNHGDKVLGMEINPLYISMAMGFSTGFWVFWGSLIL 897
S + +H+ + G S+AMG + +G +I+
Sbjct: 782 GPK------SDANQHQKSHRRQASLAG--------SVAMGLLFSLFCIFGLIIV 821
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor
(EC 2.7.1.37) (tBRI1) (Altered brassinolide sensitivity
1) (Systemin receptor SR160)
Length = 1207
Score = 232 bits (592), Expect = 3e-60
Identities = 259/894 (28%), Positives = 389/894 (42%), Gaps = 151/894 (16%)
Query: 40 EAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKGVGCNTTTGHVISLNLHCSNSLDKLQGH 99
+++ LL FK L LL +W C + GV C + +S+D
Sbjct: 43 DSQQLLSFKAALPPTPTLLQNWLSSTGPCSFTGVSCKNSR----------VSSID----- 87
Query: 100 LNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLS-- 157
L+++ L + + S V +L NL+ L L +AN G+L +
Sbjct: 88 LSNTFLSVDF-------------SLVTSYLLPLSNLESLVLKNANLSGSLTSAAKSQCGV 134
Query: 158 LLESLDLSDNSFY--VNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTL- 214
L+S+DL++N+ ++++ S+LK L+LS L + L LD
Sbjct: 135 TLDSIDLAENTISGPISDISSFGVCSNLKSLNLSKNFLDPPGKEMLKAATFSLQVLDLSY 194
Query: 215 -RLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQ 273
+SG L +S M F L L GN +IP+ F+N
Sbjct: 195 NNISGFNLFPWVSS----MGFVELEFFSLKGNKLAGSIPELDFKN--------------- 235
Query: 274 GQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHG 333
L+ LDLS N+ + + P+F D NL LDLS N G I S+L
Sbjct: 236 ------------LSYLDLSANNFSTVFPSFKD-CSNLQHLDLSSNKFYGDIGSSLSSC-- 280
Query: 334 QNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKV-LDLS 392
L L L+ NQ G + + +L L L N+ +G+ + LA+ V LDLS
Sbjct: 281 -GKLSFLNLTNNQFVGLVPK--LPSESLQYLYLRGNDFQGVYPN-QLADLCKTVVELDLS 336
Query: 393 FNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWF 452
+N+ + VP L C + +DIS S +P
Sbjct: 337 YNNFS-----GMVPE------SLGEC-------------SSLELVDISYNNFSGKLPVDT 372
Query: 453 WDLSPNVEYMNLSSNE-LRRCGQDFSQKFKLKTLDLSNNSFSCPLPR-----LPPNLRNL 506
N++ M LS N+ + FS KL+TLD+S+N+ + +P NL+ L
Sbjct: 373 LSKLSNIKTMVLSFNKFVGGLPDSFSNLLKLETLDMSSNNLTGVIPSGICKDPMNNLKVL 432
Query: 507 DLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIG 566
L +NLF G I + L + L +LDLSFN L+G IP+ + + + L L +N G
Sbjct: 433 YLQNNLFKGPIP---DSLSNCSQLVSLDLSFNYLTGSIPSSLGSLSKLKDLILWLNQLSG 489
Query: 567 SIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQI 626
IP L+ L LI+ N+L+G IP +L NC L ++L +N+L G IP +G +
Sbjct: 490 EIPQELMYLQALENLILDFNDLTGPIPASLSNCTKLNWISLSNNQLSGEIPASLGR-LSN 548
Query: 627 LMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVF-------LALTT---- 675
L +L LGNNS NIP L +SL LDL+ N L G+IP +F +AL T
Sbjct: 549 LAILKLGNNSISGNIPAELGNCQSLIWLDLNTNFLNGSIPPPLFKQSGNIAVALLTGKRY 608
Query: 676 --------EESINEKSYMEFMTIEESLPIYLSRTKHPLLIP--WKGVN--VFFNEGRLFF 723
+E + +EF I + +S T+HP ++G+ F + G + F
Sbjct: 609 VYIKNDGSKECHGAGNLLEFGGIRQEQLDRIS-TRHPCNFTRVYRGITQPTFNHNGSMIF 667
Query: 724 EILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNN 783
+DLS N L IP E+G + LS LNL N L G IP +G L+++ +LDLS N
Sbjct: 668 -----LDLSYNKLEGSIPKELGAMYYLSILNLGHNDLSGMIPQQLGGLKNVAILDLSYNR 722
Query: 784 LSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVFYKGNPHLCGPPLRKACPR 843
+ IP S+ ++ L +DLS N LSG IP +F + + N LCG PL C
Sbjct: 723 FNGTIPNSLTSLTLLGEIDLSNNNLSGMIPESAPFDTFPDYRFANN-SLCGYPLPIPCSS 781
Query: 844 NSSFEDTHCSHSEEHENDGNHGDKVLGMEINPLYISMAMGFSTGFWVFWGSLIL 897
S + +H+ + G S+AMG + +G +I+
Sbjct: 782 GPK------SDANQHQKSHRRQASLAG--------SVAMGLLFSLFCIFGLIIV 821
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 220 bits (560), Expect = 2e-56
Identities = 236/844 (27%), Positives = 354/844 (40%), Gaps = 118/844 (13%)
Query: 39 SEAEALLEF-KEGLKDPSNLLSSWKHGKDC-CQWKGVGCNTTTGHVISLNLHCSNSLDKL 96
S+ ALL + PS++ SW C W GV C D+
Sbjct: 26 SDGAALLSLTRHWTSIPSDITQSWNASDSTPCSWLGVEC------------------DRR 67
Query: 97 QGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNL 156
Q ++ LNLS P+ +S K+LK + LS F G++ LGN
Sbjct: 68 Q-----------FVDTLNLSSYGISGEFGPE-ISHLKHLKKVVLSGNGFFGSIPSQLGNC 115
Query: 157 SLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTLRL 216
SLLE +DLS NSF N L L +L+ L L F + + + L+T+
Sbjct: 116 SLLEHIDLSSNSFTGNIPDTLGALQNLRNLSL----FFNSLIGPFPESLLSIPHLETVYF 171
Query: 217 SGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQGQI 276
+G L+ + P N L TL L N F+ +P L N LQ L L++NNL G +
Sbjct: 172 TGNGLN--GSIPSNIGNMSELTTLWLDDNQFSGPVPSSL-GNITTLQELYLNDNNLVGTL 228
Query: 277 SYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNS 336
++ + L LD+ NSL G IP F + + LS N +G +P LG S
Sbjct: 229 PVTLNNLENLVYLDVRNNSLVGAIPLDFVSCKQIDTISLSNNQFTGGLPPGLG---NCTS 285
Query: 337 LKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHV 396
L+E L+G + QL+ L L LA N+ G I L ++ L L N +
Sbjct: 286 LREFGAFSCALSGPIPSCFGQLTKLDTLYLAGNHFSGRIPP-ELGKCKSMIDLQLQQNQL 344
Query: 397 TLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLS 456
+ QL+ + L +L + P I ++ + + +S +P +L
Sbjct: 345 EGEIPGELGMLSQLQYLHLYTNNLSGEVPLSIWKIQSLQSLQLYQNNLSGELPVDMTELK 404
Query: 457 PNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPN------LRNLDLSS 510
V ++ QD L+ LDL+ N F+ +PPN L+ L L
Sbjct: 405 QLVSLALYENHFTGVIPQDLGANSSLEVLDLTRNMFT---GHIPPNLCSQKKLKRLLLGY 461
Query: 511 NLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPD 570
N G++ L ++LE L L NNL G +P+ + N++ +L+ NNF G IP
Sbjct: 462 NYLEGSVP---SDLGGCSTLERLILEENNLRGGLPD-FVEKQNLLFFDLSGNNFTGPIPP 517
Query: 571 SFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVL 630
S G+LKN+ + + +N LSG IP L + L LNL N L+G +P + ++ L L
Sbjct: 518 SLGNLKNVTAIYLSSNQLSGSIPPELGSLVKLEHLNLSHNILKGILPSEL-SNCHKLSEL 576
Query: 631 ILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTI 690
+N + +IP TL L L L L EN +G IP +F +
Sbjct: 577 DASHNLLNGSIPSTLGSLTELTKLSLGENSFSGGIPTSLFQS------------------ 618
Query: 691 EESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVEL 750
N N + L N L +IP +G L L
Sbjct: 619 ----------------------NKLLN-----------LQLGGNLLAGDIP-PVGALQAL 644
Query: 751 SALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSG 810
+LNLS N+L G +P +G+L+ L LD+S NNLS + ++ I L+++++S+N SG
Sbjct: 645 RSLNLSSNKLNGQLPIDLGKLKMLEELDVSHNNLSGTLRV-LSTIQSLTFINISHNLFSG 703
Query: 811 KIPIG-NQMQSFDEVFYKGNPHLC--GPPLRKACPRNSSFEDTHCSHSEEHENDGNHGDK 867
+P + + + GN LC P ACP +S + N G G
Sbjct: 704 PVPPSLTKFLNSSPTSFSGNSDLCINCPADGLACPESSILRPCNM-----QSNTGKGGLS 758
Query: 868 VLGM 871
LG+
Sbjct: 759 TLGI 762
Score = 194 bits (492), Expect = 1e-48
Identities = 184/609 (30%), Positives = 274/609 (44%), Gaps = 72/609 (11%)
Query: 211 LDTLRLSGCQLHKLPTSPPPEMN-FDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSN 269
+DTL LS + + PE++ L + LSGN F +IP L NC L++++LS+
Sbjct: 70 VDTLNLSS---YGISGEFGPEISHLKHLKKVVLSGNGFFGSIPSQL-GNCSLLEHIDLSS 125
Query: 270 NNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLG 329
N+ G I ++ + L L L NSL G P + +L + + N L+GSIPS +G
Sbjct: 126 NSFTGNIPDTLGALQNLRNLSLFFNSLIGPFPESLLSIPHLETVYFTGNGLNGSIPSNIG 185
Query: 330 QDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVL 389
+ L L L NQ +G + S+ ++ L L L NN+ G + V L N NL L
Sbjct: 186 N---MSELTTLWLDDNQFSGPVPSSLGNITTLQELYLNDNNLVGTLP-VTLNNLENLVYL 241
Query: 390 DLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVP 449
D+ N + + ++V Q++TI L+N P + + + +S +P
Sbjct: 242 DVRNNSLVGAIPLDFVSCKQIDTISLSNNQFTGGLPPGLGNCTSLREFGAFSCALSGPIP 301
Query: 450 NWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLS 509
+ F L+ KL TL L+ N FS R+PP L
Sbjct: 302 SCFGQLT------------------------KLDTLYLAGNHFS---GRIPPELGKC--- 331
Query: 510 SNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIP 569
S+ +L L N L G IP + + L+L NN G +P
Sbjct: 332 ------------------KSMIDLQLQQNQLEGEIPGELGMLSQLQYLHLYTNNLSGEVP 373
Query: 570 DSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMV 629
S +++L L +Y NNLSG++P + + L L L N G IP +G + L V
Sbjct: 374 LSIWKIQSLQSLQLYQNNLSGELPVDMTELKQLVSLALYENHFTGVIPQDLGANSS-LEV 432
Query: 630 LILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYM---- 685
L L N F +IP LC K L L L N L G++P + T E I E++ +
Sbjct: 433 LDLTRNMFTGHIPPNLCSQKKLKRLLLGYNYLEGSVPSDLGGCSTLERLILEENNLRGGL 492
Query: 686 -EFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEI 744
+F+ + L LS IP N+ + + I LSSN L+ IP E+
Sbjct: 493 PDFVEKQNLLFFDLSGNNFTGPIPPSLGNL---------KNVTAIYLSSNQLSGSIPPEL 543
Query: 745 GKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLS 804
G LV+L LNLS N L G +PS + L+ LD S N L+ IP+++ ++ L+ L L
Sbjct: 544 GSLVKLEHLNLSHNILKGILPSELSNCHKLSELDASHNLLNGSIPSTLGSLTELTKLSLG 603
Query: 805 YNALSGKIP 813
N+ SG IP
Sbjct: 604 ENSFSGGIP 612
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 214 bits (545), Expect = 9e-55
Identities = 193/634 (30%), Positives = 302/634 (47%), Gaps = 42/634 (6%)
Query: 245 NNFNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFF 304
++F+ DW+ +C +L L + N G++ L+L + L+G +
Sbjct: 57 SSFSSNCCDWVGISCKSSVSLGLDDVNESGRV----------VELELGRRKLSGKLSESV 106
Query: 305 DKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVL 364
KL L L+L++N LSGSI ++L ++L+ L LS N +G L S+ L +L VL
Sbjct: 107 AKLDQLKVLNLTHNSLSGSIAASL---LNLSNLEVLDLSSNDFSG-LFPSLINLPSLRVL 162
Query: 365 NLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQF 424
N+ N+ G+I N ++ +DL+ N+ ++ +E +GLA+ +L
Sbjct: 163 NVYENSFHGLIPASLCNNLPRIREIDLAMNYFDGSIPVGIGNCSSVEYLGLASNNLSGSI 222
Query: 425 PKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQD-FSQKFKLK 483
P+ + N S + + N +S + + LS N+ +++SSN+ D F + KL
Sbjct: 223 PQELFQLSNLSVLALQNNRLSGALSSKLGKLS-NLGRLDISSNKFSGKIPDVFLELNKLW 281
Query: 484 TLDLSNNSFSCPLPRLPPNLRNLDLSS---NLFYGTISHVCEILCFNNSLENLDLSFNNL 540
+N F+ +PR N R++ L S N G I C + +L +LDL+ N+
Sbjct: 282 YFSAQSNLFNGEMPRSLSNSRSISLLSLRNNTLSGQIYLNCSAM---TNLTSLDLASNSF 338
Query: 541 SGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNN---NLSGKIPETLK 597
SG IP+ N + +N A FI IP+SF + ++L L N+ N+S + E L+
Sbjct: 339 SGSIPSNLPNCLRLKTINFAKIKFIAQIPESFKNFQSLTSLSFSNSSIQNISSAL-EILQ 397
Query: 598 NCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLS 657
+CQ L L L N + +P + L VLI+ + +P+ L SL +LDLS
Sbjct: 398 HCQNLKTLVLTLNFQKEELPSVPSLQFKNLKVLIIASCQLRGTVPQWLSNSPSLQLLDLS 457
Query: 658 ENQLTGAIPRCV-------FLALTTEESINEKSYMEFMTIEESLPIYLSRTKHPL----L 706
NQL+G IP + +L L+ I E + +T +SL + + P
Sbjct: 458 WNQLSGTIPPWLGSLNSLFYLDLSNNTFIGEIPHS--LTSLQSLVSKENAVEEPSPDFPF 515
Query: 707 IPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPS 766
K N + MIDLS N L I E G L +L LNL N L G+IP+
Sbjct: 516 FKKKNTNAGGLQYNQPSSFPPMIDLSYNSLNGSIWPEFGDLRQLHVLNLKNNNLSGNIPA 575
Query: 767 SIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVFY 826
++ + SL VLDLS NNLS IP S+ + LS ++YN LSG IP G Q Q+F +
Sbjct: 576 NLSGMTSLEVLDLSHNNLSGNIPPSLVKLSFLSTFSVAYNKLSGPIPTGVQFQTFPNSSF 635
Query: 827 KGNPHLCGPPLRKACPRNSSFEDTHCSHSEEHEN 860
+GN LCG A P + + + H S + +N
Sbjct: 636 EGNQGLCG---EHASPCHITDQSPHGSAVKSKKN 666
Score = 181 bits (459), Expect = 8e-45
Identities = 187/691 (27%), Positives = 301/691 (43%), Gaps = 146/691 (21%)
Query: 32 LNSQFIASEAEALLEFKEGLKDPSNLLSSWKHGK------DCCQWKGVGCNTT------- 78
+NSQ + + L + ++ + + WK + +CC W G+ C ++
Sbjct: 22 VNSQNLTCNSNDLKALEGFMRGLESSIDGWKWNESSSFSSNCCDWVGISCKSSVSLGLDD 81
Query: 79 ---TGHVISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNL 135
+G V+ L L KL G L+ S+ +L L LNL+ N S L+ + NL
Sbjct: 82 VNESGRVVELEL----GRRKLSGKLSESVAKLDQLKVLNLTHNSLSGSIAASLLNLS-NL 136
Query: 136 KHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVLSR 195
+ LDLS +F G L +L NL L L++ +NSF HGL + S
Sbjct: 137 EVLDLSSNDFSG-LFPSLINLPSLRVLNVYENSF--------HGL----------IPASL 177
Query: 196 CQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDWL 255
C N LP + +DL+ N F+ +IP +
Sbjct: 178 CNN-------------------------LP----------RIREIDLAMNYFDGSIPVGI 202
Query: 256 FENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDL 315
NC ++ L L++NNL G I + +++ L++L L N L+G + + KL NL LD+
Sbjct: 203 -GNCSSVEYLGLASNNLSGSIPQELFQLSNLSVLALQNNRLSGALSSKLGKLSNLGRLDI 261
Query: 316 SYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGII 375
S N SG IP + N L N NG + RS+ ++ +L+L N + G I
Sbjct: 262 SSNKFSGKIPDVFLE---LNKLWYFSAQSNLFNGEMPRSLSNSRSISLLSLRNNTLSGQI 318
Query: 376 SDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFS 435
++ + +NL LDL+ N + ++ N +L+TI A Q P+ + ++ +
Sbjct: 319 Y-LNCSAMTNLTSLDLASNSFSGSIPSNLPNCLRLKTINFAKIKFIAQIPESFKNFQSLT 377
Query: 436 HIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCP 495
+ SN+ + ++S +E + N LKTL L+ N
Sbjct: 378 SLSFSNSSIQ--------NISSALEILQHCQN--------------LKTLVLTLNFQKEE 415
Query: 496 LPRLP----PNLRNLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNG 551
LP +P NL+ L ++S GT+ + L + SL+ LDLS+N LSG IP +
Sbjct: 416 LPSVPSLQFKNLKVLIIASCQLRGTVP---QWLSNSPSLQLLDLSWNQLSGTIPPWLGSL 472
Query: 552 TNMIILNLAMNNFIGSIPDSFGSLKNL--------------------------------- 578
++ L+L+ N FIG IP S SL++L
Sbjct: 473 NSLFYLDLSNNTFIGEIPHSLTSLQSLVSKENAVEEPSPDFPFFKKKNTNAGGLQYNQPS 532
Query: 579 ---HMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNN 635
M+ + N+L+G I + + L +LNLK+N L G IP + + + L VL L +N
Sbjct: 533 SFPPMIDLSYNSLNGSIWPEFGDLRQLHVLNLKNNNLSGNIPANL-SGMTSLEVLDLSHN 591
Query: 636 SFDENIPKTLCQLKSLHILDLSENQLTGAIP 666
+ NIP +L +L L ++ N+L+G IP
Sbjct: 592 NLSGNIPPSLVKLSFLSTFSVAYNKLSGPIP 622
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 206 bits (523), Expect = 3e-52
Identities = 209/675 (30%), Positives = 304/675 (44%), Gaps = 114/675 (16%)
Query: 17 ILCLLMHGHVLCNGG--LNSQFIASEAEALLEFKEGLKDPSNLLSSWKHGKDC--CQWKG 72
+ CL++ +LC L S + +A L + K GL DP+ LSSW D C+W G
Sbjct: 2 LYCLIL---LLCLSSTYLPSLSLNQDATILRQAKLGLSDPAQSLSSWSDNNDVTPCKWLG 58
Query: 73 VGCNTTTGHVISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTT 132
V C+ T+ +V+S++L S L G S L LP L L+L N S D T
Sbjct: 59 VSCDATS-NVVSVDL----SSFMLVGPFPSILCHLPSLHSLSLYNNSINGSLSADDFDTC 113
Query: 133 KNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVV 192
NL LDLS +NL S+ +SL + L +LK L++SG
Sbjct: 114 HNLISLDLS---------ENLLVGSIPKSLPFN--------------LPNLKFLEISGNN 150
Query: 193 LSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMT-I 251
LS F + R L++L L+G L T P N +L L L+ N F+ + I
Sbjct: 151 LSDTIPSSFGEFR----KLESLNLAGNFLSG--TIPASLGNVTTLKELKLAYNLFSPSQI 204
Query: 252 PDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLV 311
P L N LQ L L+ NL G I S+ R+T+L LDL+ N L G IP++ +L +
Sbjct: 205 PSQL-GNLTELQVLWLAGCNLVGPIPPSLSRLTSLVNLDLTFNQLTGSIPSWITQLKTVE 263
Query: 312 ALDLSYNMLSGSIPSTLGQ-------DHGQN-------------SLKELRLSINQLNGSL 351
++L N SG +P ++G D N +L+ L L N L G L
Sbjct: 264 QIELFNNSFSGELPESMGNMTTLKRFDASMNKLTGKIPDNLNLLNLESLNLFENMLEGPL 323
Query: 352 ERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLE 411
SI + L L L N + G++ AN S L+ +DLS+N + + N +LE
Sbjct: 324 PESITRSKTLSELKLFNNRLTGVLPSQLGAN-SPLQYVDLSYNRFSGEIPANVCGEGKLE 382
Query: 412 TIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRR 471
+ L + + + K+ + + +SN +S +P+ FW L
Sbjct: 383 YLILIDNSFSGEISNNLGKCKSLTRVRLSNNKLSGQIPHGFWGLP--------------- 427
Query: 472 CGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCEILCFNNSLE 531
+L L+LS+NSF+ +P+ +N L
Sbjct: 428 ---------RLSLLELSDNSFTGSIPKTIIGAKN------------------------LS 454
Query: 532 NLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGK 591
NL +S N SG IPN + +I ++ A N+F G IP+S LK L L + N LSG+
Sbjct: 455 NLRISKNRFSGSIPNEIGSLNGIIEISGAENDFSGEIPESLVKLKQLSRLDLSKNQLSGE 514
Query: 592 IPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSL 651
IP L+ + L LNL +N L G IP +G + +L L L +N F IP L LK L
Sbjct: 515 IPRELRGWKNLNELNLANNHLSGEIPKEVGI-LPVLNYLDLSSNQFSGEIPLELQNLK-L 572
Query: 652 HILDLSENQLTGAIP 666
++L+LS N L+G IP
Sbjct: 573 NVLNLSYNHLSGKIP 587
Score = 201 bits (510), Expect = 1e-50
Identities = 182/566 (32%), Positives = 279/566 (49%), Gaps = 70/566 (12%)
Query: 236 SLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSIE-RVTTLAILDLSKN 294
SL +L L N+ N ++ F+ CH+L +L+LS N L G I S+ + L L++S N
Sbjct: 90 SLHSLSLYNNSINGSLSADDFDTCHNLISLDLSENLLVGSIPKSLPFNLPNLKFLEISGN 149
Query: 295 SLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGS-LER 353
+L+ IP+ F + L +L+L+ N LSG+IP++LG +LKEL+L+ N + S +
Sbjct: 150 NLSDTIPSSFGEFRKLESLNLAGNFLSGTIPASLGN---VTTLKELKLAYNLFSPSQIPS 206
Query: 354 SIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQ-LET 412
+ L+ L VL LA N+ G I L+ ++L LDL+FN +T ++ +W+ + +E
Sbjct: 207 QLGNLTELQVLWLAGCNLVGPIPP-SLSRLTSLVNLDLTFNQLTGSIP-SWITQLKTVEQ 264
Query: 413 IGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRR- 471
I L N + P+ + D S ++ +P+ L N+E +NL N L
Sbjct: 265 IELFNNSFSGELPESMGNMTTLKRFDASMNKLTGKIPDNLNLL--NLESLNLFENMLEGP 322
Query: 472 CGQDFSQKFKLKTLDLSNNSFSCPLP-RLPPN--LRNLDLSSNLFYGTISHVCEILCFNN 528
+ ++ L L L NN + LP +L N L+ +DLS N F G I +C
Sbjct: 323 LPESITRSKTLSELKLFNNRLTGVLPSQLGANSPLQYVDLSYNRFSGEIP---ANVCGEG 379
Query: 529 SLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNL 588
LE L L N+ SG I N ++ + L+ N G IP F L L +L + +N+
Sbjct: 380 KLEYLILIDNSFSGEISNNLGKCKSLTRVRLSNNKLSGQIPHGFWGLPRLSLLELSDNSF 439
Query: 589 SGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQL 648
+G IP+T+ + L+ L + NR G IP IG+ + ++ + N F IP++L +L
Sbjct: 440 TGSIPKTIIGAKNLSNLRISKNRFSGSIPNEIGS-LNGIIEISGAENDFSGEIPESLVKL 498
Query: 649 KSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHPLLIP 708
K L LDLS+NQL+G IPR L
Sbjct: 499 KQLSRLDLSKNQLSGEIPR-------------------------------------ELRG 521
Query: 709 WKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSI 768
WK +N ++L++N L+ EIP E+G L L+ L+LS NQ G IP +
Sbjct: 522 WKNLNE--------------LNLANNHLSGEIPKEVGILPVLNYLDLSSNQFSGEIPLEL 567
Query: 769 GELESLNVLDLSRNNLSCEIPTSMAN 794
L+ LNVL+LS N+LS +IP AN
Sbjct: 568 QNLK-LNVLNLSYNHLSGKIPPLYAN 592
Score = 191 bits (486), Expect = 6e-48
Identities = 180/615 (29%), Positives = 289/615 (46%), Gaps = 74/615 (12%)
Query: 235 DSLVTLDLSGNNFNMTIPDWLFENCHHLQNL---NLSNNNLQGQISYSIERVTTLAILDL 291
D +L +N ++T WL +C N+ +LS+ L G + + +L L L
Sbjct: 37 DPAQSLSSWSDNNDVTPCKWLGVSCDATSNVVSVDLSSFMLVGPFPSILCHLPSLHSLSL 96
Query: 292 SKNSLNG-LIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGS 350
NS+NG L + FD NL++LDLS N+L GSIP +L +LK L +S N L+ +
Sbjct: 97 YNNSINGSLSADDFDTCHNLISLDLSENLLVGSIPKSL--PFNLPNLKFLEISGNNLSDT 154
Query: 351 LERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPF-Q 409
+ S + L LNLA N + G I L N + LK L L++N + + + + +
Sbjct: 155 IPSSFGEFRKLESLNLAGNFLSGTIP-ASLGNVTTLKELKLAYNLFSPSQIPSQLGNLTE 213
Query: 410 LETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNEL 469
L+ + LA C+L P + + ++D++ ++ +P+W L VE + L +N
Sbjct: 214 LQVLWLAGCNLVGPIPPSLSRLTSLVNLDLTFNQLTGSIPSWITQLK-TVEQIELFNNSF 272
Query: 470 R-RCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSS-NLFYGTISH-VCEILCF 526
+ LK D S N + ++P NL L+L S NLF + + E +
Sbjct: 273 SGELPESMGNMTTLKRFDASMNKLT---GKIPDNLNLLNLESLNLFENMLEGPLPESITR 329
Query: 527 NNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNN 586
+ +L L L N L+GV+P+ + + ++L+ N F G IP + L LI+ +N
Sbjct: 330 SKTLSELKLFNNRLTGVLPSQLGANSPLQYVDLSYNRFSGEIPANVCGEGKLEYLILIDN 389
Query: 587 NLSGKIPETLKNCQVLTLLNLKSNRLRGPIP--YWIGTDIQILMVLILGNNSFDENIPKT 644
+ SG+I L C+ LT + L +N+L G IP +W
Sbjct: 390 SFSGEISNNLGKCKSLTRVRLSNNKLSGQIPHGFW------------------------- 424
Query: 645 LCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHP 704
L L +L+LS+N TG+IP+ + A + +S+ +
Sbjct: 425 --GLPRLSLLELSDNSFTGSIPKTIIGAKNLSN------------------LRISKNRFS 464
Query: 705 LLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSI 764
IP NE I+++ ++F + EIP + KL +LS L+LS+NQL G I
Sbjct: 465 GSIP--------NEIGSLNGIIEISGAENDF-SGEIPESLVKLKQLSRLDLSKNQLSGEI 515
Query: 765 PSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEV 824
P + ++LN L+L+ N+LS EIP + + L++LDLS N SG+IP+ Q + +
Sbjct: 516 PRELRGWKNLNELNLANNHLSGEIPKEVGILPVLNYLDLSSNQFSGEIPLELQNLKLNVL 575
Query: 825 FYKGNPHLCG--PPL 837
N HL G PPL
Sbjct: 576 NLSYN-HLSGKIPPL 589
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 202 bits (513), Expect = 4e-51
Identities = 183/617 (29%), Positives = 280/617 (44%), Gaps = 98/617 (15%)
Query: 258 NCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSY 317
N + L L N L G++S S+ ++ + +L+LS+N + IP L NL LDLS
Sbjct: 74 NTGRVIRLELGNKKLSGKLSESLGKLDEIRVLNLSRNFIKDSIPLSIFNLKNLQTLDLSS 133
Query: 318 NMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLV-VLNLAVNNMEGIIS 376
N LSG IP+++ +L+ LS N+ NGSL I S + V+ LAVN G +
Sbjct: 134 NDLSGGIPTSINLP----ALQSFDLSSNKFNGSLPSHICHNSTQIRVVKLAVNYFAGNFT 189
Query: 377 DVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSH 436
L+ L L N +T N+ ++ +L +G+ L + I+ +
Sbjct: 190 S-GFGKCVLLEHLCLGMNDLTGNIPEDLFHLKRLNLLGIQENRLSGSLSREIRNLSSLVR 248
Query: 437 IDISNAGVSDYVPNWFWDL-----------------------SPNVEYMNLSSNELR-RC 472
+D+S S +P+ F +L SP++ +NL +N L R
Sbjct: 249 LDVSWNLFSGEIPDVFDELPQLKFFLGQTNGFIGGIPKSLANSPSLNLLNLRNNSLSGRL 308
Query: 473 GQDFSQKFKLKTLDLSNNSFSCPLPRLPPN---LRNLDLSSNLFYGTISHVCEILCFNN- 528
+ + L +LDL N F+ LP P+ L+N++L+ N F+G + F N
Sbjct: 309 MLNCTAMIALNSLDLGTNRFNGRLPENLPDCKRLKNVNLARNTFHGQVPE-----SFKNF 363
Query: 529 -SLENLDLSFNNLS------GVIPNCWTNGTNMIILNLAMNNFIGSIPDSFG-SLKNLHM 580
SL LS ++L+ G++ +C N+ L L +N ++PD + L +
Sbjct: 364 ESLSYFSLSNSSLANISSALGILQHC----KNLTTLVLTLNFHGEALPDDSSLHFEKLKV 419
Query: 581 LIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDEN 640
L++ N L+G +P L + L LL+L NRL G IP WIG
Sbjct: 420 LVVANCRLTGSMPRWLSSSNELQLLDLSWNRLTGAIPSWIG------------------- 460
Query: 641 IPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEE---SLPIY 697
K+L LDLS N TG IP+ +LT ES+ ++ +++ E P +
Sbjct: 461 ------DFKALFYLDLSNNSFTGEIPK----SLTKLESLTSRN----ISVNEPSPDFPFF 506
Query: 698 LSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSR 757
+ R + + + + F I+L N L+ I E G L +L +L
Sbjct: 507 MKRNESARALQYNQI----------FGFPPTIELGHNNLSGPIWEEFGNLKKLHVFDLKW 556
Query: 758 NQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQ 817
N L GSIPSS+ + SL LDLS N LS IP S+ + LS ++YN LSG IP G Q
Sbjct: 557 NALSGSIPSSLSGMTSLEALDLSNNRLSGSIPVSLQQLSFLSKFSVAYNNLSGVIPSGGQ 616
Query: 818 MQSFDEVFYKGNPHLCG 834
Q+F ++ N HLCG
Sbjct: 617 FQTFPNSSFESN-HLCG 632
Score = 189 bits (481), Expect = 2e-47
Identities = 179/602 (29%), Positives = 284/602 (46%), Gaps = 48/602 (7%)
Query: 40 EAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKGVGCNT-TTGHVISLNLHCSNSLDKLQG 98
+ EAL +F L+ + + DCC W G+ CN+ TG VI L L KL G
Sbjct: 35 DLEALRDFIAHLEPKPDGWINSSSSTDCCNWTGITCNSNNTGRVIRLEL----GNKKLSG 90
Query: 99 HLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLSL 158
L+ SL +L + LNLS N F++ ++P + KNL+ LDLS + G + ++ NL
Sbjct: 91 KLSESLGKLDEIRVLNLSRN-FIKDSIPLSIFNLKNLQTLDLSSNDLSGGIPTSI-NLPA 148
Query: 159 LESLDLSDNSFYVNNLKWL-HGLSSLKILDL----------SG----VVLSRC---QNDW 200
L+S DLS N F + + H + ++++ L SG V+L ND
Sbjct: 149 LQSFDLSSNKFNGSLPSHICHNSTQIRVVKLAVNYFAGNFTSGFGKCVLLEHLCLGMNDL 208
Query: 201 FHDIRVILHSLDTLRLSGCQLHKLPTSPPPEM-NFDSLVTLDLSGNNFNMTIPDWLFENC 259
+I L L L L G Q ++L S E+ N SLV LD+S N F+ IPD +F+
Sbjct: 209 TGNIPEDLFHLKRLNLLGIQENRLSGSLSREIRNLSSLVRLDVSWNLFSGEIPD-VFDEL 267
Query: 260 HHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNM 319
L+ N G I S+ +L +L+L NSL+G + ++ L +LDL N
Sbjct: 268 PQLKFFLGQTNGFIGGIPKSLANSPSLNLLNLRNNSLSGRLMLNCTAMIALNSLDLGTNR 327
Query: 320 LSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDV- 378
+G +P L LK + L+ N +G + S +L +L+ +++ I S +
Sbjct: 328 FNGRLPENLPD---CKRLKNVNLARNTFHGQVPESFKNFESLSYFSLSNSSLANISSALG 384
Query: 379 ---HLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFS 435
H N + L VL L+F+ L + + +L+ + +ANC L P+W+ +
Sbjct: 385 ILQHCKNLTTL-VLTLNFHGEALPDDSS-LHFEKLKVLVVANCRLTGSMPRWLSSSNELQ 442
Query: 436 HIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCP 495
+D+S ++ +P+W D + Y++LS+N G+ KL++L N S + P
Sbjct: 443 LLDLSWNRLTGAIPSWIGDFKA-LFYLDLSNNSF--TGEIPKSLTKLESLTSRNISVNEP 499
Query: 496 LPRLPPNLRNLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMI 555
P P ++ + + L Y +I F ++E L NNLSG I + N +
Sbjct: 500 SPDFPFFMKRNESARALQYN------QIFGFPPTIE---LGHNNLSGPIWEEFGNLKKLH 550
Query: 556 ILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGP 615
+ +L N GSIP S + +L L + NN LSG IP +L+ L+ ++ N L G
Sbjct: 551 VFDLKWNALSGSIPSSLSGMTSLEALDLSNNRLSGSIPVSLQQLSFLSKFSVAYNNLSGV 610
Query: 616 IP 617
IP
Sbjct: 611 IP 612
Score = 75.1 bits (183), Expect = 8e-13
Identities = 77/253 (30%), Positives = 115/253 (45%), Gaps = 29/253 (11%)
Query: 75 CNTTTGHVISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKN 134
C T V++LN H D H L++ ++ L+G+ +P +LS++
Sbjct: 389 CKNLTTLVLTLNFHGEALPDDSSLHFEK--LKVLVVANCRLTGS------MPRWLSSSNE 440
Query: 135 LKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVLS 194
L+ LDLS G + +G+ L LDLS+NSF K L L SL ++S
Sbjct: 441 LQLLDLSWNRLTGAIPSWIGDFKALFYLDLSNNSFTGEIPKSLTKLESLTSRNIS----- 495
Query: 195 RCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDW 254
N+ D + ++ R Q +++ PP T++L NN ++ P W
Sbjct: 496 --VNEPSPDFPFFMKRNESAR--ALQYNQIFGFPP---------TIELGHNN--LSGPIW 540
Query: 255 L-FENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVAL 313
F N L +L N L G I S+ +T+L LDLS N L+G IP +L L
Sbjct: 541 EEFGNLKKLHVFDLKWNALSGSIPSSLSGMTSLEALDLSNNRLSGSIPVSLQQLSFLSKF 600
Query: 314 DLSYNMLSGSIPS 326
++YN LSG IPS
Sbjct: 601 SVAYNNLSGVIPS 613
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 196 bits (498), Expect = 2e-49
Identities = 163/569 (28%), Positives = 270/569 (46%), Gaps = 27/569 (4%)
Query: 305 DKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVL 364
D +++L++S+ L G+I +G L L L+ N G L + L++L VL
Sbjct: 67 DDDARVISLNVSFTPLFGTISPEIGM---LTHLVNLTLAANNFTGELPLEMKSLTSLKVL 123
Query: 365 NLAVN-NMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQ 423
N++ N N+ G L +L+VLD N+ + +L+ + +
Sbjct: 124 NISNNGNLTGTFPGEILKAMVDLEVLDTYNNNFNGKLPPEMSELKKLKYLSFGGNFFSGE 183
Query: 424 FPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVE-YMNLSSNELRRCGQDFSQKFKL 482
P+ ++ ++ ++ AG+S P + L E Y+ ++ +F KL
Sbjct: 184 IPESYGDIQSLEYLGLNGAGLSGKSPAFLSRLKNLREMYIGYYNSYTGGVPPEFGGLTKL 243
Query: 483 KTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSG 542
+ LD+++ + + +P NL++L H+ L SL++LDLS N L+G
Sbjct: 244 EILDMASCTLTGEIPTSLSNLKHLHTLFLHINNLTGHIPPELSGLVSLKSLDLSINQLTG 303
Query: 543 VIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVL 602
IP + N N+ ++NL NN G IP++ G L L + ++ NN + ++P L L
Sbjct: 304 EIPQSFINLGNITLINLFRNNLYGQIPEAIGELPKLEVFEVWENNFTLQLPANLGRNGNL 363
Query: 603 TLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLT 662
L++ N L G IP + ++ M LIL NN F IP+ L + KSL + + +N L
Sbjct: 364 IKLDVSDNHLTGLIPKDLCRGEKLEM-LILSNNFFFGPIPEELGKCKSLTKIRIVKNLLN 422
Query: 663 GAIPRCVF-LALTTEESINEKSYMEFMTIEESLP----IYLSRTKHPLLIP-----WKGV 712
G +P +F L L T + + + + + S IYLS IP + +
Sbjct: 423 GTVPAGLFNLPLVTIIELTDNFFSGELPVTMSGDVLDQIYLSNNWFSGEIPPAIGNFPNL 482
Query: 713 NVFF--------NEGRLFFEI--LKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLG 762
F N R FE+ L I+ S+N +T IP I + L +++LSRN++ G
Sbjct: 483 QTLFLDRNRFRGNIPREIFELKHLSRINTSANNITGGIPDSISRCSTLISVDLSRNRING 542
Query: 763 SIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFD 822
IP I +++L L++S N L+ IPT + N+ L+ LDLS+N LSG++P+G Q F+
Sbjct: 543 EIPKGINNVKNLGTLNISGNQLTGSIPTGIGNMTSLTTLDLSFNDLSGRVPLGGQFLVFN 602
Query: 823 EVFYKGNPHLCGPPLRKACPRNSSFEDTH 851
E + GN +LC P R +CP H
Sbjct: 603 ETSFAGNTYLC-LPHRVSCPTRPGQTSDH 630
Score = 151 bits (382), Expect = 7e-36
Identities = 173/612 (28%), Positives = 247/612 (40%), Gaps = 71/612 (11%)
Query: 36 FIASEAEALLEFKEGLKDPSNL-LSSWKHGKDC---CQWKGVGCNTTTGHVISLNL---- 87
F ++ E LL K + P L W H C + GV C+ VISLN+
Sbjct: 23 FAYTDMEVLLNLKSSMIGPKGHGLHDWIHSSSPDAHCSFSGVSCDDDA-RVISLNVSFTP 81
Query: 88 -------------HCSN---SLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVP-DFLS 130
H N + + G L + L L LN+S N + T P + L
Sbjct: 82 LFGTISPEIGMLTHLVNLTLAANNFTGELPLEMKSLTSLKVLNISNNGNLTGTFPGEILK 141
Query: 131 TTKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSG 190
+L+ LD + NF G L + L L+ L N F + + SL+ L L+G
Sbjct: 142 AMVDLEVLDTYNNNFNGKLPPEMSELKKLKYLSFGGNFFSGEIPESYGDIQSLEYLGLNG 201
Query: 191 VVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTS--PPPEMNFDSLVTLDLSGNNFN 248
LS L L LR + T PP L LD++
Sbjct: 202 AGLSGKSP-------AFLSRLKNLREMYIGYYNSYTGGVPPEFGGLTKLEILDMASCTLT 254
Query: 249 MTIPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLV 308
IP L N HL L L NNL G I + + +L LDLS N L G IP F L
Sbjct: 255 GEIPTSL-SNLKHLHTLFLHINNLTGHIPPELSGLVSLKSLDLSINQLTGEIPQSFINLG 313
Query: 309 NLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAV 368
N+ ++L N L G IP +G +L L V +
Sbjct: 314 NITLINLFRNNLYGQIPEAIG---------------------------ELPKLEVFEVWE 346
Query: 369 NNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWI 428
NN + +L NL LD+S NH+T + K+ +LE + L+N P+ +
Sbjct: 347 NNFT-LQLPANLGRNGNLIKLDVSDNHLTGLIPKDLCRGEKLEMLILSNNFFFGPIPEEL 405
Query: 429 QTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLS 488
K+ + I I ++ VP ++L P V + L+ N L + LS
Sbjct: 406 GKCKSLTKIRIVKNLLNGTVPAGLFNL-PLVTIIELTDNFFSGELPVTMSGDVLDQIYLS 464
Query: 489 NNSFSCPLPRLP---PNLRNLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIP 545
NN FS +P PNL+ L L N F G I EI + L ++ S NN++G IP
Sbjct: 465 NNWFSGEIPPAIGNFPNLQTLFLDRNRFRGNIPR--EIFELKH-LSRINTSANNITGGIP 521
Query: 546 NCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLL 605
+ + + +I ++L+ N G IP ++KNL L + N L+G IP + N LT L
Sbjct: 522 DSISRCSTLISVDLSRNRINGEIPKGINNVKNLGTLNISGNQLTGSIPTGIGNMTSLTTL 581
Query: 606 NLKSNRLRGPIP 617
+L N L G +P
Sbjct: 582 DLSFNDLSGRVP 593
Score = 101 bits (251), Expect = 1e-20
Identities = 92/289 (31%), Positives = 135/289 (45%), Gaps = 30/289 (10%)
Query: 80 GHVISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLD 139
G++ +NL +N L G + ++ +LP L + N+F +P L NL LD
Sbjct: 313 GNITLINLFRNN----LYGQIPEAIGELPKLEVFEVWENNFTLQ-LPANLGRNGNLIKLD 367
Query: 140 LSHANFKGNLLDNLGNLSLLESLDLSDNSFY------------------VNNLKWLHGLS 181
+S + G + +L LE L LS+N F+ V NL L+G
Sbjct: 368 VSDNHLTGLIPKDLCRGEKLEMLILSNNFFFGPIPEELGKCKSLTKIRIVKNL--LNGTV 425
Query: 182 SLKILDLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEM-NFDSLVTL 240
+ +L V + +++F + S D L + PP + NF +L TL
Sbjct: 426 PAGLFNLPLVTIIELTDNFFSGELPVTMSGDVLDQIYLSNNWFSGEIPPAIGNFPNLQTL 485
Query: 241 DLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLI 300
L N F IP +FE HL +N S NN+ G I SI R +TL +DLS+N +NG I
Sbjct: 486 FLDRNRFRGNIPREIFE-LKHLSRINTSANNITGGIPDSISRCSTLISVDLSRNRINGEI 544
Query: 301 PNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNG 349
P + + NL L++S N L+GSIP+ +G SL L LS N L+G
Sbjct: 545 PKGINNVKNLGTLNISGNQLTGSIPTGIG---NMTSLTTLDLSFNDLSG 590
>D100_ARATH (Q00874) DNA-damage-repair/toleration protein DRT100
precursor
Length = 372
Score = 132 bits (331), Expect = 6e-30
Identities = 97/319 (30%), Positives = 147/319 (45%), Gaps = 53/319 (16%)
Query: 524 LCFNNSLENLDLS-FNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLI 582
+C +L +L L+ + ++G IP C T+ ++ IL+LA N G IP G L L +L
Sbjct: 106 VCDLTALTSLVLADWKGITGEIPPCITSLASLRILDLAGNKITGEIPAEIGKLSKLAVLN 165
Query: 583 MYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIP 642
+ N +SG+IP +L + L L L N + G IP G+ +++L ++LG N +IP
Sbjct: 166 LAENQMSGEIPASLTSLIELKHLELTENGITGVIPADFGS-LKMLSRVLLGRNELTGSIP 224
Query: 643 KTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTK 702
+++ ++ L LDLS+N + G IP
Sbjct: 225 ESISGMERLADLDLSKNHIEGPIPE----------------------------------- 249
Query: 703 HPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLG 762
W G ++L +++L N LT IP + L NLSRN L G
Sbjct: 250 ------WMGN----------MKVLSLLNLDCNSLTGPIPGSLLSNSGLDVANLSRNALEG 293
Query: 763 SIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFD 822
+IP G L LDLS N+LS IP S+++ + LD+S+N L G+IP G +
Sbjct: 294 TIPDVFGSKTYLVSLDLSHNSLSGRIPDSLSSAKFVGHLDISHNKLCGRIPTGFPFDHLE 353
Query: 823 EVFYKGNPHLCGPPLRKAC 841
+ N LCG PL +C
Sbjct: 354 ATSFSDNQCLCGGPLTTSC 372
Score = 111 bits (277), Expect = 1e-23
Identities = 94/321 (29%), Positives = 152/321 (47%), Gaps = 23/321 (7%)
Query: 43 ALLEFKEGLKDPS-NLLSSWKHGKDCC-QWKGVGCNTTTGHVISLNLHCSNS-------- 92
AL FK L +P+ + ++W DCC +W G+ C+ +G V ++L +
Sbjct: 34 ALNAFKSSLSEPNLGIFNTWSENTDCCKEWYGISCDPDSGRVTDISLRGESEDAIFQKAG 93
Query: 93 -LDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLD 151
+ G ++ ++ L L+ L L+ + +P +++ +L+ LDL+ G +
Sbjct: 94 RSGYMSGSIDPAVCDLTALTSLVLADWKGITGEIPPCITSLASLRILDLAGNKITGEIPA 153
Query: 152 NLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSL 211
+G LS L L+L++N L L LK L+L+ +N I SL
Sbjct: 154 EIGKLSKLAVLNLAENQMSGEIPASLTSLIELKHLELT-------ENGITGVIPADFGSL 206
Query: 212 DTLRLSGCQLHKLPTSPPPEMN-FDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNN 270
L ++L S P ++ + L LDLS N+ IP+W+ N L LNL N
Sbjct: 207 KMLSRVLLGRNELTGSIPESISGMERLADLDLSKNHIEGPIPEWM-GNMKVLSLLNLDCN 265
Query: 271 NLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQ 330
+L G I S+ + L + +LS+N+L G IP+ F LV+LDLS+N LSG IP +L
Sbjct: 266 SLTGPIPGSLLSNSGLDVANLSRNALEGTIPDVFGSKTYLVSLDLSHNSLSGRIPDSLS- 324
Query: 331 DHGQNSLKELRLSINQLNGSL 351
+ L +S N+L G +
Sbjct: 325 --SAKFVGHLDISHNKLCGRI 343
Score = 98.6 bits (244), Expect = 7e-20
Identities = 84/288 (29%), Positives = 121/288 (41%), Gaps = 56/288 (19%)
Query: 502 NLRNLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAM 561
+LR LDL+ N G I L + L L+L+ N +SG IP T+ + L L
Sbjct: 136 SLRILDLAGNKITGEIPAEIGKL---SKLAVLNLAENQMSGEIPASLTSLIELKHLELTE 192
Query: 562 NNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIG 621
N G IP FGSLK L +++ N L+G IPE++ + L L+L N + GPIP W+G
Sbjct: 193 NGITGVIPADFGSLKMLSRVLLGRNELTGSIPESISGMERLADLDLSKNHIEGPIPEWMG 252
Query: 622 TDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINE 681
++++L +L L NS IP +L L + +LS N L G IP +
Sbjct: 253 -NMKVLSLLNLDCNSLTGPIPGSLLSNSGLDVANLSRNALEGTIP----------DVFGS 301
Query: 682 KSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIP 741
K+Y L +DLS N L+ IP
Sbjct: 302 KTY-----------------------------------------LVSLDLSHNSLSGRIP 320
Query: 742 VEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIP 789
+ + L++S N+L G IP+ + L S N C P
Sbjct: 321 DSLSSAKFVGHLDISHNKLCGRIPTGF-PFDHLEATSFSDNQCLCGGP 367
Score = 93.6 bits (231), Expect = 2e-18
Identities = 72/274 (26%), Positives = 133/274 (48%), Gaps = 30/274 (10%)
Query: 347 LNGSLERSIYQLSNLVVLNLAVNNMEGIISDVH--LANFSNLKVLDLSFNHVTLNMSKNW 404
++GS++ ++ L+ L L LA + +GI ++ + + ++L++LDL+ N +T +
Sbjct: 98 MSGSIDPAVCDLTALTSLVLA--DWKGITGEIPPCITSLASLRILDLAGNKITGEIPAEI 155
Query: 405 VPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNL 464
+L + LA + + P + + H++++ G++ +P F L + + L
Sbjct: 156 GKLSKLAVLNLAENQMSGEIPASLTSLIELKHLELTENGITGVIPADFGSLKM-LSRVLL 214
Query: 465 SSNELR-RCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCEI 523
NEL + S +L LDLS N P+P N++ L L
Sbjct: 215 GRNELTGSIPESISGMERLADLDLSKNHIEGPIPEWMGNMKVLSL--------------- 259
Query: 524 LCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIM 583
L+L N+L+G IP + + + + NL+ N G+IPD FGS L L +
Sbjct: 260 ---------LNLDCNSLTGPIPGSLLSNSGLDVANLSRNALEGTIPDVFGSKTYLVSLDL 310
Query: 584 YNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIP 617
+N+LSG+IP++L + + + L++ N+L G IP
Sbjct: 311 SHNSLSGRIPDSLSSAKFVGHLDISHNKLCGRIP 344
Score = 81.6 bits (200), Expect = 9e-15
Identities = 68/228 (29%), Positives = 113/228 (48%), Gaps = 9/228 (3%)
Query: 443 GVSDYVPNWFWDLSPNVEYMNLSSNELR-RCGQDFSQKFKLKTLDLSNNSFSCPLPRLPP 501
G++ +P L+ ++ ++L+ N++ + + KL L+L+ N S +P
Sbjct: 122 GITGEIPPCITSLA-SLRILDLAGNKITGEIPAEIGKLSKLAVLNLAENQMSGEIPASLT 180
Query: 502 NL---RNLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILN 558
+L ++L+L+ N G I L L + L N L+G IP + + L+
Sbjct: 181 SLIELKHLELTENGITGVIPADFGSL---KMLSRVLLGRNELTGSIPESISGMERLADLD 237
Query: 559 LAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPY 618
L+ N+ G IP+ G++K L +L + N+L+G IP +L + L + NL N L G IP
Sbjct: 238 LSKNHIEGPIPEWMGNMKVLSLLNLDCNSLTGPIPGSLLSNSGLDVANLSRNALEGTIPD 297
Query: 619 WIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIP 666
G+ L+ L L +NS IP +L K + LD+S N+L G IP
Sbjct: 298 VFGSK-TYLVSLDLSHNSLSGRIPDSLSSAKFVGHLDISHNKLCGRIP 344
Score = 74.3 bits (181), Expect = 1e-12
Identities = 79/305 (25%), Positives = 124/305 (39%), Gaps = 59/305 (19%)
Query: 272 LQGQISYSIERVTTLAILDLSK-NSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQ 330
+ G I ++ +T L L L+ + G IP L +L LDL+ N ++G IP+ +G
Sbjct: 98 MSGSIDPAVCDLTALTSLVLADWKGITGEIPPCITSLASLRILDLAGNKITGEIPAEIG- 156
Query: 331 DHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLD 390
+LS L VLNLA N M G I L + LK L+
Sbjct: 157 --------------------------KLSKLAVLNLAENQMSGEIP-ASLTSLIELKHLE 189
Query: 391 LSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPN 450
L+ N +T + ++ L + L L P+ I + + +D+S + +P
Sbjct: 190 LTENGITGVIPADFGSLKMLSRVLLGRNELTGSIPESISGMERLADLDLSKNHIEGPIPE 249
Query: 451 WFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLD--- 507
W ++ L L+L NS + P+P + LD
Sbjct: 250 WMGNMKV------------------------LSLLNLDCNSLTGPIPGSLLSNSGLDVAN 285
Query: 508 LSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGS 567
LS N GTI ++ L +LDLS N+LSG IP+ ++ + L+++ N G
Sbjct: 286 LSRNALEGTIP---DVFGSKTYLVSLDLSHNSLSGRIPDSLSSAKFVGHLDISHNKLCGR 342
Query: 568 IPDSF 572
IP F
Sbjct: 343 IPTGF 347
>TMM_ARATH (Q9SSD1) TOO MANY MOUTHS protein precursor (TMM)
Length = 496
Score = 114 bits (284), Expect = 2e-24
Identities = 92/265 (34%), Positives = 127/265 (47%), Gaps = 32/265 (12%)
Query: 551 GTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSN 610
G+++ L L N F+G IPD G+L NL +L ++ N+L+G IP + L L+L N
Sbjct: 158 GSSLQTLVLRENGFLGPIPDELGNLTNLKVLDLHKNHLNGSIPLSFNRFSGLRSLDLSGN 217
Query: 611 RLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVF 670
RL G IP G + L VL L N +P TL SL +DLS N++TG IP
Sbjct: 218 RLTGSIP---GFVLPALSVLDLNQNLLTGPVPPTLTSCGSLIKIDLSRNRVTGPIP---- 270
Query: 671 LALTTEESINEKSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMID 730
ESIN + + L + +R P +G+N L+ +
Sbjct: 271 ------ESINRLNQLVL------LDLSYNRLSGPFPSSLQGLN-----------SLQALM 307
Query: 731 LSSNF-LTHEIPVEIGK-LVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEI 788
L N + IP K L L L LS + GSIP S+ L SL VL L NNL+ EI
Sbjct: 308 LKGNTKFSTTIPENAFKGLKNLMILVLSNTNIQGSIPKSLTRLNSLRVLHLEGNNLTGEI 367
Query: 789 PTSMANIDRLSWLDLSYNALSGKIP 813
P ++ LS L L+ N+L+G +P
Sbjct: 368 PLEFRDVKHLSELRLNDNSLTGPVP 392
Score = 81.6 bits (200), Expect = 9e-15
Identities = 87/283 (30%), Positives = 127/283 (44%), Gaps = 35/283 (12%)
Query: 359 SNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANC 418
S+L L L N G I D L N +NLKVLDL NH+ ++ ++ L ++ L+
Sbjct: 159 SSLQTLVLRENGFLGPIPD-ELGNLTNLKVLDLHKNHLNGSIPLSFNRFSGLRSLDLSGN 217
Query: 419 HLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQ 478
L P ++ S +D++ ++ VP L CG
Sbjct: 218 RLTGSIPGFVLPA--LSVLDLNQNLLTGPVPP-----------------TLTSCGS---- 254
Query: 479 KFKLKTLDLSNNSFSCPLPRLPPNLRNL---DLSSNLFYGTISHVCEILCFNNSLENLDL 535
L +DLS N + P+P L L DLS N G + L NSL+ L L
Sbjct: 255 ---LIKIDLSRNRVTGPIPESINRLNQLVLLDLSYNRLSGPFPSSLQGL---NSLQALML 308
Query: 536 SFNN-LSGVIP-NCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIP 593
N S IP N + N++IL L+ N GSIP S L +L +L + NNL+G+IP
Sbjct: 309 KGNTKFSTTIPENAFKGLKNLMILVLSNTNIQGSIPKSLTRLNSLRVLHLEGNNLTGEIP 368
Query: 594 ETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNS 636
++ + L+ L L N L GP+P+ T ++ L L NN+
Sbjct: 369 LEFRDVKHLSELRLNDNSLTGPVPFERDTVWRMRRKLRLYNNA 411
Score = 80.5 bits (197), Expect = 2e-14
Identities = 72/250 (28%), Positives = 118/250 (46%), Gaps = 10/250 (4%)
Query: 251 IPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNL 310
IP +L LQ L L N G I + +T L +LDL KN LNG IP F++ L
Sbjct: 150 IPAFLGRLGSSLQTLVLRENGFLGPIPDELGNLTNLKVLDLHKNHLNGSIPLSFNRFSGL 209
Query: 311 VALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNN 370
+LDLS N L+GSIP + +L L L+ N L G + ++ +L+ ++L+ N
Sbjct: 210 RSLDLSGNRLTGSIPGFV-----LPALSVLDLNQNLLTGPVPPTLTSCGSLIKIDLSRNR 264
Query: 371 MEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGL-ANCHLGPQFPK-WI 428
+ G I + + + L +LDLS+N ++ + L+ + L N P+
Sbjct: 265 VTGPIPE-SINRLNQLVLLDLSYNRLSGPFPSSLQGLNSLQALMLKGNTKFSTTIPENAF 323
Query: 429 QTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELR-RCGQDFSQKFKLKTLDL 487
+ KN + +SN + +P L+ ++ ++L N L +F L L L
Sbjct: 324 KGLKNLMILVLSNTNIQGSIPKSLTRLN-SLRVLHLEGNNLTGEIPLEFRDVKHLSELRL 382
Query: 488 SNNSFSCPLP 497
++NS + P+P
Sbjct: 383 NDNSLTGPVP 392
Score = 79.3 bits (194), Expect = 4e-14
Identities = 81/247 (32%), Positives = 117/247 (46%), Gaps = 28/247 (11%)
Query: 98 GHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLS 157
G L SSL L L N F+ +PD L NLK LDL + G++ + S
Sbjct: 155 GRLGSSLQTLV------LRENGFL-GPIPDELGNLTNLKVLDLHKNHLNGSIPLSFNRFS 207
Query: 158 LLESLDLSDNSFYVNNLKWLHGLSSLKILDLS-----GVV---LSRC---------QNDW 200
L SLDLS N + ++ L +L +LDL+ G V L+ C +N
Sbjct: 208 GLRSLDLSGNRLTGSIPGFV--LPALSVLDLNQNLLTGPVPPTLTSCGSLIKIDLSRNRV 265
Query: 201 FHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMN-FDSLVTLDLSGNN-FNMTIPDWLFEN 258
I ++ L+ L L ++L P + +SL L L GN F+ TIP+ F+
Sbjct: 266 TGPIPESINRLNQLVLLDLSYNRLSGPFPSSLQGLNSLQALMLKGNTKFSTTIPENAFKG 325
Query: 259 CHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYN 318
+L L LSN N+QG I S+ R+ +L +L L N+L G IP F + +L L L+ N
Sbjct: 326 LKNLMILVLSNTNIQGSIPKSLTRLNSLRVLHLEGNNLTGEIPLEFRDVKHLSELRLNDN 385
Query: 319 MLSGSIP 325
L+G +P
Sbjct: 386 SLTGPVP 392
Score = 76.3 bits (186), Expect = 4e-13
Identities = 82/276 (29%), Positives = 116/276 (41%), Gaps = 59/276 (21%)
Query: 568 IPDSFGSLKNLHMLIMYN--NNLSGKIPETLKNC-QVLTLLNLKSNRLRGPIPYWIGTDI 624
+ +S LK+L L Y +IP L L L L+ N GPIP +G ++
Sbjct: 124 VSESLTRLKHLKALFFYRCLGRAPQRIPAFLGRLGSSLQTLVLRENGFLGPIPDELG-NL 182
Query: 625 QILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSY 684
L VL L N + +IP + + L LDLS N+LTG+IP V AL+
Sbjct: 183 TNLKVLDLHKNHLNGSIPLSFNRFSGLRSLDLSGNRLTGSIPGFVLPALSV--------- 233
Query: 685 MEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEI 744
+ + ++L L+ P L L IDLS N +T IP I
Sbjct: 234 ---LDLNQNL---LTGPVPPTLTSCGS--------------LIKIDLSRNRVTGPIPESI 273
Query: 745 GKLVELSALNLSRNQLLGSIPSSIGELESLN--------------------------VLD 778
+L +L L+LS N+L G PSS+ L SL +L
Sbjct: 274 NRLNQLVLLDLSYNRLSGPFPSSLQGLNSLQALMLKGNTKFSTTIPENAFKGLKNLMILV 333
Query: 779 LSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPI 814
LS N+ IP S+ ++ L L L N L+G+IP+
Sbjct: 334 LSNTNIQGSIPKSLTRLNSLRVLHLEGNNLTGEIPL 369
Score = 55.1 bits (131), Expect = 9e-07
Identities = 59/196 (30%), Positives = 89/196 (45%), Gaps = 12/196 (6%)
Query: 92 SLDKLQGHLNSSL--LQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNL 149
SLD L S+ LP LS L+L+ N + VP L++ +L +DLS G +
Sbjct: 211 SLDLSGNRLTGSIPGFVLPALSVLDLNQN-LLTGPVPPTLTSCGSLIKIDLSRNRVTGPI 269
Query: 150 LDNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVL--SRCQNDWFHDIRVI 207
+++ L+ L LDLS N L GL+SL+ L L G + + F ++
Sbjct: 270 PESINRLNQLVLLDLSYNRLSGPFPSSLQGLNSLQALMLKGNTKFSTTIPENAFKGLK-- 327
Query: 208 LHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNL 267
+L L LS + + P +SL L L GNN IP F + HL L L
Sbjct: 328 --NLMILVLSNTNIQG--SIPKSLTRLNSLRVLHLEGNNLTGEIP-LEFRDVKHLSELRL 382
Query: 268 SNNNLQGQISYSIERV 283
++N+L G + + + V
Sbjct: 383 NDNSLTGPVPFERDTV 398
Score = 36.6 bits (83), Expect = 0.32
Identities = 24/68 (35%), Positives = 32/68 (46%), Gaps = 2/68 (2%)
Query: 764 IPSSIGEL-ESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIG-NQMQSF 821
IP+ +G L SL L L N IP + N+ L LDL N L+G IP+ N+
Sbjct: 150 IPAFLGRLGSSLQTLVLRENGFLGPIPDELGNLTNLKVLDLHKNHLNGSIPLSFNRFSGL 209
Query: 822 DEVFYKGN 829
+ GN
Sbjct: 210 RSLDLSGN 217
>TLR3_HUMAN (O15455) Toll-like receptor 3 precursor
Length = 904
Score = 108 bits (271), Expect = 5e-23
Identities = 154/607 (25%), Positives = 254/607 (41%), Gaps = 86/607 (14%)
Query: 125 VPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLK 184
VPD L T N+ L+L+H + N S L SLD+ N+ + L LK
Sbjct: 45 VPDDLPT--NITVLNLTHNQLRRLPAANFTRYSQLTSLDVGFNTISKLEPELCQKLPMLK 102
Query: 185 ILDLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSG 244
+L+L LS+ + F +L L L + K+ +P + +L+TLDLS
Sbjct: 103 VLNLQHNELSQLSDKTF----AFCTNLTELHLMSNSIQKIKNNPFVKQK--NLITLDLSH 156
Query: 245 NNFNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSIE--RVTTLAILDLSKNSLNGLIPN 302
N + T + +LQ L LSNN +Q S ++ ++L L+LS N + P
Sbjct: 157 NGLSSTKLGTQVQ-LENLQELLLSNNKIQALKSEELDIFANSSLKKLELSSNQIKEFSPG 215
Query: 303 FFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQL--SN 360
F + L L L+ L S+ L + S++ L LS +QL+ + + L +N
Sbjct: 216 CFHAIGRLFGLFLNNVQLGPSLTEKLCLELANTSIRNLSLSNSQLSTTSNTTFLGLKWTN 275
Query: 361 LVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHL 420
L +L+L+ NN+ ++ + A L+ L +N++ F GL N
Sbjct: 276 LTMLDLSYNNL-NVVGNDSFAWLPQLEYFFLEYNNIQ--------HLFSHSLHGLFNV-- 324
Query: 421 GPQFPKWIQTQKNFSHIDISNAG---VSDYVPNWFWDLSPNVEYMNLSSNELRRCGQD-F 476
+++ +++F+ IS A + D+ W L E++N+ N++ + F
Sbjct: 325 -----RYLNLKRSFTKQSISLASLPKIDDFSFQWLKCL----EHLNMEDNDIPGIKSNMF 375
Query: 477 SQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSS------NLFYGTISHV-CEILCFNNS 529
+ LK L LSN+ S R N + L+ NL IS + + +
Sbjct: 376 TGLINLKYLSLSNSFTSL---RTLTNETFVSLAHSPLHILNLTKNKISKIESDAFSWLGH 432
Query: 530 LENLDLSFNNLSGVIPNC-WTNGTNMIILNLAMNNF------------------------ 564
LE LDL N + + W N+ + L+ N +
Sbjct: 433 LEVLDLGLNEIGQELTGQEWRGLENIFEIYLSYNKYLQLTRNSFALVPSLQRLMLRRVAL 492
Query: 565 --IGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLR--------- 613
+ S P F L+NL +L + NNN++ + L+ + L +L+L+ N L
Sbjct: 493 KNVDSSPSPFQPLRNLTILDLSNNNIANINDDMLEGLEKLEILDLQHNNLARLWKHANPG 552
Query: 614 GPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLAL 673
GPI + G + L +L L +N FDE + L L I+DL N L +P VF
Sbjct: 553 GPIYFLKG--LSHLHILNLESNGFDEIPVEVFKDLFELKIIDLGLNNL-NTLPASVFNNQ 609
Query: 674 TTEESIN 680
+ +S+N
Sbjct: 610 VSLKSLN 616
Score = 79.0 bits (193), Expect = 6e-14
Identities = 162/638 (25%), Positives = 235/638 (36%), Gaps = 164/638 (25%)
Query: 91 NSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTT------------------ 132
N++ KL+ L +LP L LNL N+ Q + F T
Sbjct: 85 NTISKLEPELCQ---KLPMLKVLNLQHNELSQLSDKTFAFCTNLTELHLMSNSIQKIKNN 141
Query: 133 -----KNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLH--GLSSLKI 185
KNL LDLSH L L L+ L LS+N + L SSLK
Sbjct: 142 PFVKQKNLITLDLSHNGLSSTKLGTQVQLENLQELLLSNNKIQALKSEELDIFANSSLKK 201
Query: 186 LDLSGVVLSRCQNDWFHDIRVILH-----------------------SLDTLRLSGCQLH 222
L+LS + FH I + S+ L LS QL
Sbjct: 202 LELSSNQIKEFSPGCFHAIGRLFGLFLNNVQLGPSLTEKLCLELANTSIRNLSLSNSQLS 261
Query: 223 KLPTSPPPEMNFDSLVTLDLSGNNFNMTIPD---WL---------FENCHHL-------- 262
+ + + +L LDLS NN N+ D WL + N HL
Sbjct: 262 TTSNTTFLGLKWTNLTMLDLSYNNLNVVGNDSFAWLPQLEYFFLEYNNIQHLFSHSLHGL 321
Query: 263 ---QNLNLSNNNLQGQIS---------YSIERVTTLAILDLSKNSLNGLIPNFFDKLVNL 310
+ LNL + + IS +S + + L L++ N + G+ N F L+NL
Sbjct: 322 FNVRYLNLKRSFTKQSISLASLPKIDDFSFQWLKCLEHLNMEDNDIPGIKSNMFTGLINL 381
Query: 311 VALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNN 370
L LS NS LR N+ SL S L +LNL N
Sbjct: 382 KYLSLS------------------NSFTSLRTLTNETFVSLAHS-----PLHILNLTKNK 418
Query: 371 MEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANC-HLGPQFPKWIQ 429
+ I SD + +L+VLDL N + ++ E GL N + + K++Q
Sbjct: 419 ISKIESDA-FSWLGHLEVLDLGLNEIGQELTGQ-------EWRGLENIFEIYLSYNKYLQ 470
Query: 430 TQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSN 489
+N + L P+++ + L ++ LK +D S
Sbjct: 471 LTRNS------------------FALVPSLQRLML-------------RRVALKNVDSSP 499
Query: 490 NSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWT 549
+ F PL NL LDLS+N + E L LE LDL NNL+ + +
Sbjct: 500 SPFQ-PLR----NLTILDLSNNNIANINDDMLEGL---EKLEILDLQHNNLARLWKHANP 551
Query: 550 NG--------TNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQV 601
G +++ ILNL N F + F L L ++ + NNL+ +P ++ N QV
Sbjct: 552 GGPIYFLKGLSHLHILNLESNGFDEIPVEVFKDLFELKIIDLGLNNLN-TLPASVFNNQV 610
Query: 602 -LTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFD 638
L LNL+ N + G + L L + N FD
Sbjct: 611 SLKSLNLQKNLITSVEKKVFGPAFRNLTELDMRFNPFD 648
Score = 70.9 bits (172), Expect = 2e-11
Identities = 128/563 (22%), Positives = 220/563 (38%), Gaps = 107/563 (19%)
Query: 284 TTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLS 343
T + +L+L+ N L L F + L +LD+ +N +S P + LK L L
Sbjct: 51 TNITVLNLTHNQLRRLPAANFTRYSQLTSLDVGFNTISKLEPELCQK---LPMLKVLNLQ 107
Query: 344 INQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKN 403
N+L+ +++ +NL L+L N+++ I ++ + NL LDLS N
Sbjct: 108 HNELSQLSDKTFAFCTNLTELHLMSNSIQKIKNNPFVKQ-KNLITLDLSHN--------- 157
Query: 404 WVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMN 463
GL++ LG Q +Q + N + +SN +
Sbjct: 158 ----------GLSSTKLGTQ----VQLE-NLQELLLSNNKIQA----------------- 185
Query: 464 LSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTIS---HV 520
L S EL D LK L+LS+N P + LF + +
Sbjct: 186 LKSEEL-----DIFANSSLKKLELSSNQIK---EFSPGCFHAIGRLFGLFLNNVQLGPSL 237
Query: 521 CEILCF---NNSLENLDLSFNNLSGVIPNCWTN--GTNMIILNLAMNNFIGSIPDSFGSL 575
E LC N S+ NL LS + LS + TN+ +L+L+ NN DSF L
Sbjct: 238 TEKLCLELANTSIRNLSLSNSQLSTTSNTTFLGLKWTNLTMLDLSYNNLNVVGNDSFAWL 297
Query: 576 KNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNN 635
L + NN+ +L + LNLK + + I + + + +
Sbjct: 298 PQLEYFFLEYNNIQHLFSHSLHGLFNVRYLNLKRSFTK--------QSISLASLPKIDDF 349
Query: 636 SFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESIN-EKSYMEFMTIEESL 694
SF LK L L++ +N + G I +F L + ++ S+ T+
Sbjct: 350 SFQ--------WLKCLEHLNMEDNDIPG-IKSNMFTGLINLKYLSLSNSFTSLRTLTNET 400
Query: 695 PIYLSRTK-HPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVE----IGKLVE 749
+ L+ + H L + ++ ++ + L+++DL N + E+ + + + E
Sbjct: 401 FVSLAHSPLHILNLTKNKISKIESDAFSWLGHLEVLDLGLNEIGQELTGQEWRGLENIFE 460
Query: 750 L-----SALNLSRNQL------------------LGSIPSSIGELESLNVLDLSRNNLSC 786
+ L L+RN + S PS L +L +LDLS NN++
Sbjct: 461 IYLSYNKYLQLTRNSFALVPSLQRLMLRRVALKNVDSSPSPFQPLRNLTILDLSNNNIAN 520
Query: 787 EIPTSMANIDRLSWLDLSYNALS 809
+ +++L LDL +N L+
Sbjct: 521 INDDMLEGLEKLEILDLQHNNLA 543
Score = 45.4 bits (106), Expect = 7e-04
Identities = 60/205 (29%), Positives = 92/205 (44%), Gaps = 29/205 (14%)
Query: 80 GHVISLNLHCSNSLDKLQGHLNSSL-----LQLPYLSYLNLSGNDF-MQSTVPDFLSTTK 133
GH+ L+L + +L G L + L Y YL L+ N F + ++ +
Sbjct: 431 GHLEVLDLGLNEIGQELTGQEWRGLENIFEIYLSYNKYLQLTRNSFALVPSLQRLMLRRV 490
Query: 134 NLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVL 193
LK++D S + F+ L NL++L DLS+N+ N L GL L+ILDL L
Sbjct: 491 ALKNVDSSPSPFQP-----LRNLTIL---DLSNNNIANINDDMLEGLEKLEILDLQHNNL 542
Query: 194 SRCQNDWFHD--------IRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGN 245
+R W H ++ + H L L L ++P ++ L +DL N
Sbjct: 543 ARL---WKHANPGGPIYFLKGLSH-LHILNLESNGFDEIPVEVFKDL--FELKIIDLGLN 596
Query: 246 NFNMTIPDWLFENCHHLQNLNLSNN 270
N N T+P +F N L++LNL N
Sbjct: 597 NLN-TLPASVFNNQVSLKSLNLQKN 620
>PGI1_ARATH (Q9M5J9) Polygalacturonase inhibitor 1 precursor
(Polygalacturonase-inhibiting protein) (PGIP-1)
Length = 330
Score = 102 bits (255), Expect = 4e-21
Identities = 87/282 (30%), Positives = 128/282 (44%), Gaps = 31/282 (10%)
Query: 557 LNLAMNNFIGSIPDSFGSLKNLHMLIMYN-NNLSGKIPETLKNCQVLTLLNLKSNRLRGP 615
L + G IP G L L L+ +NL+G I T+ + L +L L L GP
Sbjct: 75 LTIFSGQISGQIPAEVGDLPYLETLVFRKLSNLTGTIQPTIAKLKNLRMLRLSWTNLTGP 134
Query: 616 IPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTT 675
IP +I + ++ L L L N +IP +L L + L+LS N+LTG+IP
Sbjct: 135 IPDFI-SQLKNLEFLELSFNDLSGSIPSSLSTLPKILALELSRNKLTGSIP--------- 184
Query: 676 EESINEKSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNF 735
+S+ F L + ++ P IP N+ FN IDLS N
Sbjct: 185 ------ESFGSFPGTVPDLRLSHNQLSGP--IPKSLGNIDFNR----------IDLSRNK 226
Query: 736 LTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANI 795
L + + G +++LSRN I S + ++L +LDL+ N ++ IP
Sbjct: 227 LQGDASMLFGSNKTTWSIDLSRNMFQFDI-SKVDIPKTLGILDLNHNGITGNIPVQWTEA 285
Query: 796 DRLSWLDLSYNALSGKIPIGNQMQSFDEVFYKGNPHLCGPPL 837
L + ++SYN L G IP G ++Q+FD Y N LCG PL
Sbjct: 286 P-LQFFNVSYNKLCGHIPTGGKLQTFDSYSYFHNKCLCGAPL 326
Score = 82.0 bits (201), Expect = 7e-15
Identities = 92/315 (29%), Positives = 138/315 (43%), Gaps = 20/315 (6%)
Query: 16 AILCLLMHGHVLCNGGLNSQFIASEAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKGVGC 75
A LCLL L ++ LL+ K+ L +P +L +SW DCC W + C
Sbjct: 5 ATLCLLFLFTFLTTCLSKDLCNQNDKNTLLKIKKSLNNPYHL-ASWDPQTDCCSWYCLEC 63
Query: 76 N--TTTGHVISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTK 133
T V +L + ++ G + + + LPYL L + T+ ++ K
Sbjct: 64 GDATVNHRVTALTIFSG----QISGQIPAEVGDLPYLETLVFRKLSNLTGTIQPTIAKLK 119
Query: 134 NLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVL 193
NL+ L LS N G + D + L LE L+LS N + L L + L+LS L
Sbjct: 120 NLRMLRLSWTNLTGPIPDFISQLKNLEFLELSFNDLSGSIPSSLSTLPKILALELSRNKL 179
Query: 194 SRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEM--NFDSLVTLDLSGNNFNMTI 251
+ + F ++ LRLS QL + P P+ N D +DLS N
Sbjct: 180 TGSIPESFGSFP---GTVPDLRLSHNQL----SGPIPKSLGNID-FNRIDLSRNKLQGD- 230
Query: 252 PDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLV 311
LF + +++LS N Q IS ++ TL ILDL+ N + G IP + + L
Sbjct: 231 ASMLFGSNKTTWSIDLSRNMFQFDIS-KVDIPKTLGILDLNHNGITGNIPVQWTE-APLQ 288
Query: 312 ALDLSYNMLSGSIPS 326
++SYN L G IP+
Sbjct: 289 FFNVSYNKLCGHIPT 303
Score = 80.9 bits (198), Expect = 1e-14
Identities = 70/252 (27%), Positives = 107/252 (41%), Gaps = 31/252 (12%)
Query: 538 NNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLK 597
+NL+G I N+ +L L+ N G IPD LKNL L + N+LSG IP +L
Sbjct: 105 SNLTGTIQPTIAKLKNLRMLRLSWTNLTGPIPDFISQLKNLEFLELSFNDLSGSIPSSLS 164
Query: 598 NCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLS 657
+ L L N+L G IP G+ + L L +N IPK+L + + +DLS
Sbjct: 165 TLPKILALELSRNKLTGSIPESFGSFPGTVPDLRLSHNQLSGPIPKSLGNI-DFNRIDLS 223
Query: 658 ENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFN 717
N+L G TT ++ +F + +P
Sbjct: 224 RNKLQGDASMLFGSNKTTWSIDLSRNMFQFDISKVDIP---------------------- 261
Query: 718 EGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVL 777
+ L ++DL+ N +T IPV+ + L N+S N+L G IP+ G+L++ +
Sbjct: 262 ------KTLGILDLNHNGITGNIPVQWTE-APLQFFNVSYNKLCGHIPTG-GKLQTFDSY 313
Query: 778 DLSRNNLSCEIP 789
N C P
Sbjct: 314 SYFHNKCLCGAP 325
Score = 77.8 bits (190), Expect = 1e-13
Identities = 69/226 (30%), Positives = 119/226 (52%), Gaps = 34/226 (15%)
Query: 261 HLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNML 320
+L+ L LS NL G I I ++ L L+LS N L+G IP+ L ++AL+LS N L
Sbjct: 120 NLRMLRLSWTNLTGPIPDFISQLKNLEFLELSFNDLSGSIPSSLSTLPKILALELSRNKL 179
Query: 321 SGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHL 380
+GSIP + G G ++ +LRLS NQL+G + +S+ + + ++L+ N ++G S +
Sbjct: 180 TGSIPESFGSFPG--TVPDLRLSHNQLSGPIPKSLGNI-DFNRIDLSRNKLQGDASMLFG 236
Query: 381 ANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDIS 440
+N + + DLS N ++SK +P +T+G+ +D++
Sbjct: 237 SNKTTWSI-DLSRNMFQFDISKVDIP----KTLGI---------------------LDLN 270
Query: 441 NAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLD 486
+ G++ +P W +P +++ N+S N+L CG KL+T D
Sbjct: 271 HNGITGNIPVQ-WTEAP-LQFFNVSYNKL--CGH-IPTGGKLQTFD 311
Score = 68.2 bits (165), Expect = 1e-10
Identities = 81/326 (24%), Positives = 131/326 (39%), Gaps = 38/326 (11%)
Query: 266 NLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVN--LVALDLSYNMLSGS 323
N ++ N +I S+ LA D + + D VN + AL + +SG
Sbjct: 26 NQNDKNTLLKIKKSLNNPYHLASWDPQTDCCSWYCLECGDATVNHRVTALTIFSGQISGQ 85
Query: 324 IPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANF 383
IP+ +G +L +LS L G+++ +I +L NL +L L+ N+ G I D ++
Sbjct: 86 IPAEVGDLPYLETLVFRKLS--NLTGTIQPTIAKLKNLRMLRLSWTNLTGPIPDF-ISQL 142
Query: 384 SNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAG 443
NL+ L+LSFN L P + T +++S
Sbjct: 143 KNLEFLELSFND------------------------LSGSIPSSLSTLPKILALELSRNK 178
Query: 444 VSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNL 503
++ +P F V + LS N+L +DLS N L +
Sbjct: 179 LTGSIPESFGSFPGTVPDLRLSHNQLSGPIPKSLGNIDFNRIDLSRNKLQGDASMLFGSN 238
Query: 504 R---NLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLA 560
+ ++DLS N+F IS V +L LDL+ N ++G IP WT + N++
Sbjct: 239 KTTWSIDLSRNMFQFDISKVD----IPKTLGILDLNHNGITGNIPVQWTEAP-LQFFNVS 293
Query: 561 MNNFIGSIPDSFGSLKNLHMLIMYNN 586
N G IP G L+ ++N
Sbjct: 294 YNKLCGHIPTG-GKLQTFDSYSYFHN 318
Score = 64.7 bits (156), Expect = 1e-09
Identities = 53/161 (32%), Positives = 76/161 (46%), Gaps = 24/161 (14%)
Query: 529 SLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNL 588
+L L LS+ NL+G IP+ + N+ L L+ N+ GSIP S +L + L + N L
Sbjct: 120 NLRMLRLSWTNLTGPIPDFISQLKNLEFLELSFNDLSGSIPSSLSTLPKILALELSRNKL 179
Query: 589 SGKIPETLKNCQ-VLTLLNLKSNRLRGPIPYWIGT---------------DIQILM---- 628
+G IPE+ + + L L N+L GPIP +G D +L
Sbjct: 180 TGSIPESFGSFPGTVPDLRLSHNQLSGPIPKSLGNIDFNRIDLSRNKLQGDASMLFGSNK 239
Query: 629 ---VLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIP 666
+ L N F +I K K+L ILDL+ N +TG IP
Sbjct: 240 TTWSIDLSRNMFQFDISKVDIP-KTLGILDLNHNGITGNIP 279
Score = 40.8 bits (94), Expect = 0.017
Identities = 26/89 (29%), Positives = 43/89 (48%), Gaps = 25/89 (28%)
Query: 750 LSALNLSRNQLLGSIPSSIGE-------------------------LESLNVLDLSRNNL 784
++AL + Q+ G IP+ +G+ L++L +L LS NL
Sbjct: 72 VTALTIFSGQISGQIPAEVGDLPYLETLVFRKLSNLTGTIQPTIAKLKNLRMLRLSWTNL 131
Query: 785 SCEIPTSMANIDRLSWLDLSYNALSGKIP 813
+ IP ++ + L +L+LS+N LSG IP
Sbjct: 132 TGPIPDFISQLKNLEFLELSFNDLSGSIP 160
>PGIP_PYRCO (Q05091) Polygalacturonase inhibitor precursor
(Polygalacturonase-inhibiting protein)
Length = 330
Score = 98.2 bits (243), Expect = 9e-20
Identities = 97/316 (30%), Positives = 146/316 (45%), Gaps = 20/316 (6%)
Query: 13 KFIAILCLLMHGHVLCNGGLNSQFIASEAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKG 72
KF L L + + N L+ + + LL+ K+ DP +L+SWK DCC W
Sbjct: 4 KFSTFLSLTLLFSSVLNPALSDLCNPDDKKVLLQIKKAFGDPY-VLASWKSDTDCCDWYC 62
Query: 73 VGCNTTTGHVISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTT 132
V C++TT + SL + ++ G + + + LPYL L + + ++
Sbjct: 63 VTCDSTTNRINSLTIFAG----QVSGQIPALVGDLPYLETLEFHKQPNLTGPIQPAIAKL 118
Query: 133 KNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVV 192
K LK L LS N G++ D L L L LDLS NNL S ++ +L +
Sbjct: 119 KGLKSLRLSWTNLSGSVPDFLSQLKNLTFLDLS-----FNNLTGAIPSSLSELPNLGALR 173
Query: 193 LSRCQNDWFHDIRV--ILHSLDTLRLSGCQLH-KLPTSPPPEMNFDSLVTLDLSGNNFNM 249
L R + I + ++ L LS QL +PTS +M+F S+ DLS N
Sbjct: 174 LDRNKLTGHIPISFGQFIGNVPDLYLSHNQLSGNIPTSFA-QMDFTSI---DLSRNKLEG 229
Query: 250 TIPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVN 309
+F Q ++LS N L+ +S +E T+L LD++ N + G IP F +L N
Sbjct: 230 DA-SVIFGLNKTTQIVDLSRNLLEFNLS-KVEFPTSLTSLDINHNKIYGSIPVEFTQL-N 286
Query: 310 LVALDLSYNMLSGSIP 325
L++SYN L G IP
Sbjct: 287 FQFLNVSYNRLCGQIP 302
Score = 95.5 bits (236), Expect = 6e-19
Identities = 86/283 (30%), Positives = 135/283 (47%), Gaps = 51/283 (18%)
Query: 578 LHMLIMYNNNLSGKIPETLKNCQVLTLLNL-KSNRLRGPIPYWIGTDIQILMVLILGNNS 636
++ L ++ +SG+IP + + L L K L GPI I ++ L L L +
Sbjct: 72 INSLTIFAGQVSGQIPALVGDLPYLETLEFHKQPNLTGPIQPAIAK-LKGLKSLRLSWTN 130
Query: 637 FDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPI 696
++P L QLK+L LDLS N LTGAIP S++E + + ++ +
Sbjct: 131 LSGSVPDFLSQLKNLTFLDLSFNNLTGAIP----------SSLSELPNLGALRLDRN--- 177
Query: 697 YLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLS 756
T H + + F + F + + LS N L+ IP ++ + ++++LS
Sbjct: 178 --KLTGH--------IPISFGQ---FIGNVPDLYLSHNQLSGNIPTSFAQM-DFTSIDLS 223
Query: 757 RNQLLGSIPSSIGELESLNVLDLSRN----NLS-CEIPTSMANID--------------- 796
RN+L G G ++ ++DLSRN NLS E PTS+ ++D
Sbjct: 224 RNKLEGDASVIFGLNKTTQIVDLSRNLLEFNLSKVEFPTSLTSLDINHNKIYGSIPVEFT 283
Query: 797 --RLSWLDLSYNALSGKIPIGNQMQSFDEVFYKGNPHLCGPPL 837
+L++SYN L G+IP+G ++QSFDE Y N LCG PL
Sbjct: 284 QLNFQFLNVSYNRLCGQIPVGGKLQSFDEYSYFHNRCLCGAPL 326
Score = 85.5 bits (210), Expect = 6e-16
Identities = 77/251 (30%), Positives = 106/251 (41%), Gaps = 31/251 (12%)
Query: 539 NLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKN 598
NL+G I + L L+ N GS+PD LKNL L + NNL+G IP +L
Sbjct: 106 NLTGPIQPAIAKLKGLKSLRLSWTNLSGSVPDFLSQLKNLTFLDLSFNNLTGAIPSSLSE 165
Query: 599 CQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSE 658
L L L N+L G IP G I + L L +N NIP + Q+ I DLS
Sbjct: 166 LPNLGALRLDRNKLTGHIPISFGQFIGNVPDLYLSHNQLSGNIPTSFAQMDFTSI-DLSR 224
Query: 659 NQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNE 718
N+L G L TT+ ++ +EF LS+ + P
Sbjct: 225 NKLEGDASVIFGLNKTTQIVDLSRNLLEF---------NLSKVEFP-------------- 261
Query: 719 GRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLD 778
L +D++ N + IPVE +L LN+S N+L G IP G+L+S +
Sbjct: 262 -----TSLTSLDINHNKIYGSIPVEFTQL-NFQFLNVSYNRLCGQIPVG-GKLQSFDEYS 314
Query: 779 LSRNNLSCEIP 789
N C P
Sbjct: 315 YFHNRCLCGAP 325
Score = 82.8 bits (203), Expect = 4e-15
Identities = 66/213 (30%), Positives = 108/213 (49%), Gaps = 33/213 (15%)
Query: 262 LQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLS 321
L++L LS NL G + + ++ L LDLS N+L G IP+ +L NL AL L N L+
Sbjct: 121 LKSLRLSWTNLSGSVPDFLSQLKNLTFLDLSFNNLTGAIPSSLSELPNLGALRLDRNKLT 180
Query: 322 GSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLA 381
G IP + GQ G ++ +L LS NQL+G++ S Q+ + ++L+ N +EG S +
Sbjct: 181 GHIPISFGQFIG--NVPDLYLSHNQLSGNIPTSFAQM-DFTSIDLSRNKLEGDASVIFGL 237
Query: 382 NFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISN 441
N +++DLS N + N+SK ++ + + +DI++
Sbjct: 238 N-KTTQIVDLSRNLLEFNLSK-------------------------VEFPTSLTSLDINH 271
Query: 442 AGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQ 474
+ +P F L N +++N+S N L CGQ
Sbjct: 272 NKIYGSIPVEFTQL--NFQFLNVSYNRL--CGQ 300
Score = 68.2 bits (165), Expect = 1e-10
Identities = 54/165 (32%), Positives = 83/165 (49%), Gaps = 7/165 (4%)
Query: 503 LRNLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMN 562
L++L LS G++ + L +L LDLSFNNL+G IP+ + N+ L L N
Sbjct: 121 LKSLRLSWTNLSGSVP---DFLSQLKNLTFLDLSFNNLTGAIPSSLSELPNLGALRLDRN 177
Query: 563 NFIGSIPDSFGS-LKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIG 621
G IP SFG + N+ L + +N LSG IP + T ++L N+L G G
Sbjct: 178 KLTGHIPISFGQFIGNVPDLYLSHNQLSGNIPTSFAQMD-FTSIDLSRNKLEGDASVIFG 236
Query: 622 TDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIP 666
+ + ++ L N + N+ K SL LD++ N++ G+IP
Sbjct: 237 LN-KTTQIVDLSRNLLEFNLSKVEFP-TSLTSLDINHNKIYGSIP 279
>PGI3_PHAVU (P58823) Polygalacturonase inhibitor 3 precursor
(Polygalacturonase-inhibiting protein) (PGIP-2) (PGIP-3)
Length = 342
Score = 98.2 bits (243), Expect = 9e-20
Identities = 84/312 (26%), Positives = 132/312 (41%), Gaps = 58/312 (18%)
Query: 530 LENLDLSFNNLSGVIPNCWTNGTNMIILNL----AMNNFIGSIPDSFGSLKNLHMLIMYN 585
+ NLDLS +NL P ++ N+ LN +NN +G IP + L LH L + +
Sbjct: 81 VNNLDLSGHNLPKPYP-IPSSLANLPYLNFLYIGGINNLVGPIPPAIAKLTQLHYLYITH 139
Query: 586 NNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTL 645
N+SG IP+ L + L L+ N L G +P ++
Sbjct: 140 TNVSGAIPDFLSQIKTLVTLDFSYNALSG-------------------------TLPPSI 174
Query: 646 CQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHPL 705
L +L + N+++GAIP SY F + S+ I +R +
Sbjct: 175 SSLPNLVGITFDGNRISGAIP---------------DSYGSFSKLFTSMTISRNRLTGKI 219
Query: 706 LIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIP 765
+ +N+ F +DLS N L + V G ++L++N L +
Sbjct: 220 PPTFANLNLAF------------VDLSRNMLQGDASVLFGSDKNTQKIHLAKNSLDFDL- 266
Query: 766 SSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVF 825
+G ++LN LDL N + +P + + L L++S+N L G+IP G +Q FD
Sbjct: 267 EKVGLSKNLNGLDLRNNRIYGTLPQGLTQLKFLHSLNVSFNNLCGEIPQGGNLQRFDVSA 326
Query: 826 YKGNPHLCGPPL 837
Y N LCG PL
Sbjct: 327 YANNKCLCGSPL 338
Score = 94.0 bits (232), Expect = 2e-18
Identities = 86/298 (28%), Positives = 138/298 (45%), Gaps = 31/298 (10%)
Query: 40 EAEALLEFKEGLKDPSNLLSSWKHGKDCCQ--WKGVGCNTTTGHVISLNLHCSNSLDKLQ 97
+ +ALL+ K+ L +P+ L SSW DCC W GV C+T T + N+LD L
Sbjct: 36 DKQALLQIKKDLGNPTTL-SSWLPTTDCCNRTWLGVLCDTDT------QTYRVNNLD-LS 87
Query: 98 GH-------LNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLL 150
GH + SSL LPYL++L + G + + +P ++ L +L ++H N G +
Sbjct: 88 GHNLPKPYPIPSSLANLPYLNFLYIGGINNLVGPIPPAIAKLTQLHYLYITHTNVSGAIP 147
Query: 151 DNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHS 210
D L + L +LD S N+ + L +L + G +S D + + S
Sbjct: 148 DFLSQIKTLVTLDFSYNALSGTLPPSISSLPNLVGITFDGNRISGAIPDSYGSFSKLFTS 207
Query: 211 LDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNN 270
+ R ++L PP +L +DLS N LF + + Q ++L+ N
Sbjct: 208 MTISR------NRLTGKIPPTFANLNLAFVDLS-RNMLQGDASVLFGSDKNTQKIHLAKN 260
Query: 271 NLQGQISYSIERV---TTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIP 325
+L + +E+V L LDL N + G +P +L L +L++S+N L G IP
Sbjct: 261 SL----DFDLEKVGLSKNLNGLDLRNNRIYGTLPQGLTQLKFLHSLNVSFNNLCGEIP 314
Score = 73.6 bits (179), Expect = 2e-12
Identities = 79/266 (29%), Positives = 125/266 (46%), Gaps = 18/266 (6%)
Query: 240 LDLSGNNFNMTIP-DWLFENCHHLQNLNLSN-NNLQGQISYSIERVTTLAILDLSKNSLN 297
LDLSG+N P N +L L + NNL G I +I ++T L L ++ +++
Sbjct: 84 LDLSGHNLPKPYPIPSSLANLPYLNFLYIGGINNLVGPIPPAIAKLTQLHYLYITHTNVS 143
Query: 298 GLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQ 357
G IP+F ++ LV LD SYN LSG++P ++ +L + N+++G++ S
Sbjct: 144 GAIPDFLSQIKTLVTLDFSYNALSGTLPPSIS---SLPNLVGITFDGNRISGAIPDSYGS 200
Query: 358 LSNLVV-LNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLA 416
S L + ++ N + G I AN NL +DLS N + + S + + I LA
Sbjct: 201 FSKLFTSMTISRNRLTGKIPPT-FANL-NLAFVDLSRNMLQGDASVLFGSDKNTQKIHLA 258
Query: 417 NCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDF 476
L K + KN + +D+ N + +P L + +N+S N L CG +
Sbjct: 259 KNSLDFDLEK-VGLSKNLNGLDLRNNRIYGTLPQGLTQLK-FLHSLNVSFNNL--CG-EI 313
Query: 477 SQKFKLKTLDLS---NNSFSC--PLP 497
Q L+ D+S NN C PLP
Sbjct: 314 PQGGNLQRFDVSAYANNKCLCGSPLP 339
>PGI2_PHAVU (P58822) Polygalacturonase inhibitor 2 precursor
(Polygalacturonase-inhibiting protein) (PGIP-2)
Length = 342
Score = 97.8 bits (242), Expect = 1e-19
Identities = 84/312 (26%), Positives = 131/312 (41%), Gaps = 58/312 (18%)
Query: 530 LENLDLSFNNLSGVIPNCWTNGTNMIILNL----AMNNFIGSIPDSFGSLKNLHMLIMYN 585
+ NLDLS NL P ++ N+ LN +NN +G IP + L LH L + +
Sbjct: 81 VNNLDLSGLNLPKPYP-IPSSLANLPYLNFLYIGGINNLVGPIPPAIAKLTQLHYLYITH 139
Query: 586 NNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTL 645
N+SG IP+ L + L L+ N L G +P ++
Sbjct: 140 TNVSGAIPDFLSQIKTLVTLDFSYNALSG-------------------------TLPPSI 174
Query: 646 CQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHPL 705
L +L + N+++GAIP SY F + S+ I +R +
Sbjct: 175 SSLPNLVGITFDGNRISGAIP---------------DSYGSFSKLFTSMTISRNRLTGKI 219
Query: 706 LIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIP 765
+ +N+ F +DLS N L + V G ++L++N L +
Sbjct: 220 PPTFANLNLAF------------VDLSRNMLEGDASVLFGSDKNTQKIHLAKNSLAFDL- 266
Query: 766 SSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVF 825
+G ++LN LDL N + +P + + L L++S+N L G+IP G +Q FD
Sbjct: 267 GKVGLSKNLNGLDLRNNRIYGTLPQGLTQLKFLHSLNVSFNNLCGEIPQGGNLQRFDVSA 326
Query: 826 YKGNPHLCGPPL 837
Y N LCG PL
Sbjct: 327 YANNKCLCGSPL 338
Score = 95.5 bits (236), Expect = 6e-19
Identities = 99/364 (27%), Positives = 148/364 (40%), Gaps = 71/364 (19%)
Query: 15 IAILCLLMHGHV-LCNGGLNSQFIASEAEALLEFKEGLKDPSNLLSSWKHGKDCCQ--WK 71
+ IL L H LCN + +ALL+ K+ L +P+ L SSW DCC W
Sbjct: 18 LVILVSLSTAHSELCN--------PQDKQALLQIKKDLGNPTTL-SSWLPTTDCCNRTWL 68
Query: 72 GVGCNTTTGHVISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLST 131
GV C+T T NL S + SSL LPYL++L + G + + +P ++
Sbjct: 69 GVLCDTDTQTYRVNNLDLSGLNLPKPYPIPSSLANLPYLNFLYIGGINNLVGPIPPAIAK 128
Query: 132 TKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGV 191
L +L ++H N G + D L + L +LD S N+ LSG
Sbjct: 129 LTQLHYLYITHTNVSGAIPDFLSQIKTLVTLDFSYNA-------------------LSG- 168
Query: 192 VLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTI 251
T PP + +LV + GN + I
Sbjct: 169 ----------------------------------TLPPSISSLPNLVGITFDGNRISGAI 194
Query: 252 PDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLV 311
PD ++ +S N L G+I + + LA +DLS+N L G F N
Sbjct: 195 PDSYGSFSKLFTSMTISRNRLTGKIPPTFANL-NLAFVDLSRNMLEGDASVLFGSDKNTQ 253
Query: 312 ALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNM 371
+ L+ N S+ LG+ +L L L N++ G+L + + QL L LN++ NN+
Sbjct: 254 KIHLAKN----SLAFDLGKVGLSKNLNGLDLRNNRIYGTLPQGLTQLKFLHSLNVSFNNL 309
Query: 372 EGII 375
G I
Sbjct: 310 CGEI 313
Score = 72.0 bits (175), Expect = 7e-12
Identities = 79/266 (29%), Positives = 124/266 (45%), Gaps = 18/266 (6%)
Query: 240 LDLSGNNFNMTIP-DWLFENCHHLQNLNLSN-NNLQGQISYSIERVTTLAILDLSKNSLN 297
LDLSG N P N +L L + NNL G I +I ++T L L ++ +++
Sbjct: 84 LDLSGLNLPKPYPIPSSLANLPYLNFLYIGGINNLVGPIPPAIAKLTQLHYLYITHTNVS 143
Query: 298 GLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQ 357
G IP+F ++ LV LD SYN LSG++P ++ +L + N+++G++ S
Sbjct: 144 GAIPDFLSQIKTLVTLDFSYNALSGTLPPSIS---SLPNLVGITFDGNRISGAIPDSYGS 200
Query: 358 LSNLVV-LNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLA 416
S L + ++ N + G I AN NL +DLS N + + S + + I LA
Sbjct: 201 FSKLFTSMTISRNRLTGKIPPT-FANL-NLAFVDLSRNMLEGDASVLFGSDKNTQKIHLA 258
Query: 417 NCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDF 476
L K + KN + +D+ N + +P L + +N+S N L CG +
Sbjct: 259 KNSLAFDLGK-VGLSKNLNGLDLRNNRIYGTLPQGLTQLK-FLHSLNVSFNNL--CG-EI 313
Query: 477 SQKFKLKTLDLS---NNSFSC--PLP 497
Q L+ D+S NN C PLP
Sbjct: 314 PQGGNLQRFDVSAYANNKCLCGSPLP 339
Score = 49.3 bits (116), Expect = 5e-05
Identities = 62/251 (24%), Positives = 92/251 (35%), Gaps = 50/251 (19%)
Query: 344 INQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKN 403
IN L G + +I +L+ L L + N+ G I D L+ L LD S+N
Sbjct: 115 INNLVGPIPPAIAKLTQLHYLYITHTNVSGAIPDF-LSQIKTLVTLDFSYNA-------- 165
Query: 404 WVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMN 463
L P I + N I +S +P+ + S M
Sbjct: 166 ----------------LSGTLPPSISSLPNLVGITFDGNRISGAIPDSYGSFSKLFTSMT 209
Query: 464 LSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCEI 523
+S N L L +DLS N L+ +++ +G+ + +I
Sbjct: 210 ISRNRLTGKIPPTFANLNLAFVDLSRNM--------------LEGDASVLFGSDKNTQKI 255
Query: 524 LCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIM 583
NSL DL LS N+ L+L N G++P LK LH L +
Sbjct: 256 HLAKNSLA-FDLGKVGLS----------KNLNGLDLRNNRIYGTLPQGLTQLKFLHSLNV 304
Query: 584 YNNNLSGKIPE 594
NNL G+IP+
Sbjct: 305 SFNNLCGEIPQ 315
>PGI1_PHAVU (P35334) Polygalacturonase inhibitor 1 precursor
(Polygalacturonase-inhibiting protein) (PGIP-1)
Length = 342
Score = 97.4 bits (241), Expect = 2e-19
Identities = 82/312 (26%), Positives = 132/312 (42%), Gaps = 58/312 (18%)
Query: 530 LENLDLSFNNLSGVIPNCWTNGTNMIILNL----AMNNFIGSIPDSFGSLKNLHMLIMYN 585
+ NLDLS +NL P ++ N+ LN +NN +G IP + L LH L + +
Sbjct: 81 VNNLDLSGHNLPKPYP-IPSSLANLPYLNFLYIGGINNLVGPIPPAIAKLTQLHYLYITH 139
Query: 586 NNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTL 645
N+SG IP+ L + L L+ N L G +P ++
Sbjct: 140 TNVSGAIPDFLSQIKTLVTLDFSYNALSG-------------------------TLPPSI 174
Query: 646 CQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHPL 705
L +L + N+++GAIP SY F + ++ I +R +
Sbjct: 175 SSLPNLGGITFDGNRISGAIP---------------DSYGSFSKLFTAMTISRNRLTGKI 219
Query: 706 LIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIP 765
+ +N+ F +DLS N L + V G ++L++N L +
Sbjct: 220 PPTFANLNLAF------------VDLSRNMLEGDASVLFGSDKNTKKIHLAKNSLAFDL- 266
Query: 766 SSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVF 825
+G ++LN LDL N + +P + + L L++S+N L G+IP G ++ FD
Sbjct: 267 GKVGLSKNLNGLDLRNNRIYGTLPQGLTQLKFLQSLNVSFNNLCGEIPQGGNLKRFDVSS 326
Query: 826 YKGNPHLCGPPL 837
Y N LCG PL
Sbjct: 327 YANNKCLCGSPL 338
Score = 94.4 bits (233), Expect = 1e-18
Identities = 98/362 (27%), Positives = 149/362 (41%), Gaps = 81/362 (22%)
Query: 40 EAEALLEFKEGLKDPSNLLSSWKHGKDCCQ--WKGVGCNTTTGHVISLNLHCSNSLDKLQ 97
+ +ALL+ K+ L +P+ L SSW DCC W GV C+T T + N+LD L
Sbjct: 36 DKQALLQIKKDLGNPTTL-SSWLPTTDCCNRTWLGVLCDTDT------QTYRVNNLD-LS 87
Query: 98 GH-------LNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLL 150
GH + SSL LPYL++L + G + + +P ++ L +L ++H N G +
Sbjct: 88 GHNLPKPYPIPSSLANLPYLNFLYIGGINNLVGPIPPAIAKLTQLHYLYITHTNVSGAIP 147
Query: 151 DNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHS 210
D L + L +LD S N+ LSG
Sbjct: 148 DFLSQIKTLVTLDFSYNA-------------------LSG-------------------- 168
Query: 211 LDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNN 270
T PP + +L + GN + IPD + +S N
Sbjct: 169 ---------------TLPPSISSLPNLGGITFDGNRISGAIPDSYGSFSKLFTAMTISRN 213
Query: 271 NLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQ 330
L G+I + + LA +DLS+N L G F N + L+ N S+ LG+
Sbjct: 214 RLTGKIPPTFANL-NLAFVDLSRNMLEGDASVLFGSDKNTKKIHLAKN----SLAFDLGK 268
Query: 331 DHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLD 390
+L L L N++ G+L + + QL L LN++ NN+ G + NLK D
Sbjct: 269 VGLSKNLNGLDLRNNRIYGTLPQGLTQLKFLQSLNVSFNNLCG-----EIPQGGNLKRFD 323
Query: 391 LS 392
+S
Sbjct: 324 VS 325
Score = 75.9 bits (185), Expect = 5e-13
Identities = 80/266 (30%), Positives = 126/266 (47%), Gaps = 18/266 (6%)
Query: 240 LDLSGNNFNMTIP-DWLFENCHHLQNLNLSN-NNLQGQISYSIERVTTLAILDLSKNSLN 297
LDLSG+N P N +L L + NNL G I +I ++T L L ++ +++
Sbjct: 84 LDLSGHNLPKPYPIPSSLANLPYLNFLYIGGINNLVGPIPPAIAKLTQLHYLYITHTNVS 143
Query: 298 GLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQ 357
G IP+F ++ LV LD SYN LSG++P ++ +L + N+++G++ S
Sbjct: 144 GAIPDFLSQIKTLVTLDFSYNALSGTLPPSIS---SLPNLGGITFDGNRISGAIPDSYGS 200
Query: 358 LSNL-VVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLA 416
S L + ++ N + G I AN NL +DLS N + + S + + I LA
Sbjct: 201 FSKLFTAMTISRNRLTGKIPPT-FANL-NLAFVDLSRNMLEGDASVLFGSDKNTKKIHLA 258
Query: 417 NCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDF 476
L K + KN + +D+ N + +P L ++ +N+S N L CG +
Sbjct: 259 KNSLAFDLGK-VGLSKNLNGLDLRNNRIYGTLPQGLTQLK-FLQSLNVSFNNL--CG-EI 313
Query: 477 SQKFKLKTLDLS---NNSFSC--PLP 497
Q LK D+S NN C PLP
Sbjct: 314 PQGGNLKRFDVSSYANNKCLCGSPLP 339
>PGI2_ARATH (Q9M5J8) Polygalacturonase inhibitor 2 precursor
(Polygalacturonase-inhibiting protein) (PGIP-2)
Length = 330
Score = 96.7 bits (239), Expect = 3e-19
Identities = 83/274 (30%), Positives = 121/274 (43%), Gaps = 33/274 (12%)
Query: 566 GSIPDSFGSLKNLHMLIMYN-NNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDI 624
G IP G L L LI NL+G I T+ + LT L L L GP+P ++ + +
Sbjct: 84 GQIPPEVGDLPYLTSLIFRKLTNLTGHIQPTIAKLKNLTFLRLSWTNLTGPVPEFL-SQL 142
Query: 625 QILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSY 684
+ L + L N +IP +L L+ L L+LS N+LTG IP
Sbjct: 143 KNLEYIDLSFNDLSGSIPSSLSSLRKLEYLELSRNKLTGPIPE----------------- 185
Query: 685 MEFMTIEESLP-IYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVE 743
F T +P ++LS + IP N F IDLS N L + +
Sbjct: 186 -SFGTFSGKVPSLFLSHNQLSGTIPKSLGNPDFYR----------IDLSRNKLQGDASIL 234
Query: 744 IGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDL 803
G +++SRN + S + ++LN LD++ N ++ IP + L++
Sbjct: 235 FGAKKTTWIVDISRNMFQFDL-SKVKLAKTLNNLDMNHNGITGSIPAEWSKA-YFQLLNV 292
Query: 804 SYNALSGKIPIGNQMQSFDEVFYKGNPHLCGPPL 837
SYN L G+IP G +Q FD + N LCG PL
Sbjct: 293 SYNRLCGRIPKGEYIQRFDSYSFFHNKCLCGAPL 326
Score = 77.8 bits (190), Expect = 1e-13
Identities = 72/252 (28%), Positives = 104/252 (40%), Gaps = 33/252 (13%)
Query: 539 NLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKN 598
NL+G I N+ L L+ N G +P+ LKNL + + N+LSG IP +L +
Sbjct: 106 NLTGHIQPTIAKLKNLTFLRLSWTNLTGPVPEFLSQLKNLEYIDLSFNDLSGSIPSSLSS 165
Query: 599 CQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSE 658
+ L L L N+L GPIP GT + L L +N IPK+L + +DLS
Sbjct: 166 LRKLEYLELSRNKLTGPIPESFGTFSGKVPSLFLSHNQLSGTIPKSLGN-PDFYRIDLSR 224
Query: 659 NQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNE 718
N+L G TT ++ +F LS+ K
Sbjct: 225 NKLQGDASILFGAKKTTWIVDISRNMFQF---------DLSKVK---------------- 259
Query: 719 GRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGE-LESLNVL 777
+ L +D++ N +T IP E K LN+S N+L G IP GE ++ +
Sbjct: 260 ---LAKTLNNLDMNHNGITGSIPAEWSK-AYFQLLNVSYNRLCGRIPK--GEYIQRFDSY 313
Query: 778 DLSRNNLSCEIP 789
N C P
Sbjct: 314 SFFHNKCLCGAP 325
Score = 77.4 bits (189), Expect = 2e-13
Identities = 91/317 (28%), Positives = 136/317 (42%), Gaps = 47/317 (14%)
Query: 44 LLEFKEGLKDPSNLLSSWKHGKDCCQWKGVGCNTTTGHVISLNLHCSNSLDKLQGHLNSS 103
LL+ K+ L +P +L +SW DCC W + C T + +L + ++ G +
Sbjct: 33 LLKIKKSLNNPYHL-ASWDPKTDCCSWYCLECGDATVNHRVTSLIIQDG--EISGQIPPE 89
Query: 104 LLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLSLLESLD 163
+ LPYL+ L + + ++ KNL L LS N G + + L L LE +D
Sbjct: 90 VGDLPYLTSLIFRKLTNLTGHIQPTIAKLKNLTFLRLSWTNLTGPVPEFLSQLKNLEYID 149
Query: 164 LSDNSFYVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHK 223
LS N DLSG + S + L L+ L LS +L
Sbjct: 150 LSFN-------------------DLSGSIPSSLSS---------LRKLEYLELSRNKL-- 179
Query: 224 LPTSPPPEM--NFDSLV-TLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSI 280
T P PE F V +L LS N + TIP L ++LS N LQG S
Sbjct: 180 --TGPIPESFGTFSGKVPSLFLSHNQLSGTIPKSL--GNPDFYRIDLSRNKLQGDASILF 235
Query: 281 ERVTTLAILDLSKNSLNGLIPNFFDKLV-NLVALDLSYNMLSGSIPSTLGQDHGQNSLKE 339
T I+D+S+N + KL L LD+++N ++GSIP+ + + Q
Sbjct: 236 GAKKTTWIVDISRNMFQFDLSKV--KLAKTLNNLDMNHNGITGSIPAEWSKAYFQ----L 289
Query: 340 LRLSINQLNGSLERSIY 356
L +S N+L G + + Y
Sbjct: 290 LNVSYNRLCGRIPKGEY 306
Score = 68.2 bits (165), Expect = 1e-10
Identities = 54/196 (27%), Positives = 93/196 (46%), Gaps = 28/196 (14%)
Query: 498 RLPPNLRNLDLSSNLFYGTIS----HVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTN 553
++PP + +L ++L + ++ H+ + +L L LS+ NL+G +P + N
Sbjct: 85 QIPPEVGDLPYLTSLIFRKLTNLTGHIQPTIAKLKNLTFLRLSWTNLTGPVPEFLSQLKN 144
Query: 554 MIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQ-VLTLLNLKSNRL 612
+ ++L+ N+ GSIP S SL+ L L + N L+G IPE+ + L L N+L
Sbjct: 145 LEYIDLSFNDLSGSIPSSLSSLRKLEYLELSRNKLTGPIPESFGTFSGKVPSLFLSHNQL 204
Query: 613 RGPIPYWIGT---------------DIQILM-------VLILGNNSFDENIPKTLCQLKS 650
G IP +G D IL ++ + N F ++ K K+
Sbjct: 205 SGTIPKSLGNPDFYRIDLSRNKLQGDASILFGAKKTTWIVDISRNMFQFDLSKVKL-AKT 263
Query: 651 LHILDLSENQLTGAIP 666
L+ LD++ N +TG+IP
Sbjct: 264 LNNLDMNHNGITGSIP 279
Score = 64.7 bits (156), Expect = 1e-09
Identities = 67/233 (28%), Positives = 109/233 (46%), Gaps = 17/233 (7%)
Query: 271 NLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQ 330
NL G I +I ++ L L LS +L G +P F +L NL +DLS+N LSGSIPS+L
Sbjct: 106 NLTGHIQPTIAKLKNLTFLRLSWTNLTGPVPEFLSQLKNLEYIDLSFNDLSGSIPSSLS- 164
Query: 331 DHGQNSLKELRLSINQLNGSLERSIYQLSNLV-VLNLAVNNMEGIISDVHLANFSNLKVL 389
L+ L LS N+L G + S S V L L+ N + G I L N + +
Sbjct: 165 --SLRKLEYLELSRNKLTGPIPESFGTFSGKVPSLFLSHNQLSGTIPK-SLGN-PDFYRI 220
Query: 390 DLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVP 449
DLS N + + S + + ++ K ++ K +++D+++ G++ +P
Sbjct: 221 DLSRNKLQGDASILFGAKKTTWIVDISRNMFQFDLSK-VKLAKTLNNLDMNHNGITGSIP 279
Query: 450 -NWFWDLSPNVEYMNLSSNELRRCGQ----DFSQKFKLKTLDLSNNSFSCPLP 497
W + +N+S N L CG+ ++ Q+F + + PLP
Sbjct: 280 AEW---SKAYFQLLNVSYNRL--CGRIPKGEYIQRFDSYSFFHNKCLCGAPLP 327
>LRR1_HUMAN (Q9BTT6) Leucine-rich repeat-containing protein 1 (LAP
and no PDZ protein) (LANO adapter protein)
Length = 524
Score = 96.3 bits (238), Expect = 3e-19
Identities = 107/376 (28%), Positives = 180/376 (47%), Gaps = 45/376 (11%)
Query: 447 YVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRN- 505
YVP + + ++E + L +N+LR + F Q KL+ L LS+N + RLPP + N
Sbjct: 26 YVPEEIYRYARSLEELLLDANQLRELPEQFFQLVKLRKLGLSDNE----IQRLPPEIANF 81
Query: 506 -----LDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLA 560
LD+S N I + E + F +L+ D S N L+ +P + N+ L++
Sbjct: 82 MQLVELDVSRN----EIPEIPESISFCKALQVADFSGNPLTR-LPESFPELQNLTCLSV- 135
Query: 561 MNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWI 620
+ + S+P++ G+L NL L + NL +P++L + L L+L +N + +P I
Sbjct: 136 NDISLQSLPENIGNLYNLASLEL-RENLLTYLPDSLTQLRRLEELDLGNNEIYN-LPESI 193
Query: 621 GTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESIN 680
G + + + + GN +P+ + LK+L LD+SEN+L L T+ I+
Sbjct: 194 GALLHLKDLWLDGNQL--SELPQEIGNLKNLLCLDVSENRLERLPEEISGLTSLTDLVIS 251
Query: 681 EKSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLF--------FEILKMIDLS 732
+ + E++P + + K ++ ++ RL E L + L+
Sbjct: 252 Q-------NLLETIPDGIGKLKKLSILK-------VDQNRLTQLPEAVGECESLTELVLT 297
Query: 733 SNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSM 792
N L +P IGKL +LS LN RN+L+ S+P IG SL V + N L+ IP +
Sbjct: 298 ENQLL-TLPKSIGKLKKLSNLNADRNKLV-SLPKEIGGCCSLTVFCVRDNRLT-RIPAEV 354
Query: 793 ANIDRLSWLDLSYNAL 808
+ L LD++ N L
Sbjct: 355 SQATELHVLDVAGNRL 370
Score = 72.0 bits (175), Expect = 7e-12
Identities = 95/359 (26%), Positives = 154/359 (42%), Gaps = 58/359 (16%)
Query: 336 SLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNH 395
SL+EL L NQL L +QL L L L+ N ++ + ++ ANF L LD+S N
Sbjct: 37 SLEELLLDANQLR-ELPEQFFQLVKLRKLGLSDNEIQRLPPEI--ANFMQLVELDVSRNE 93
Query: 396 VT-LNMSKNWVPPFQLETI-GLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFW 453
+ + S ++ Q+ G L FP+ +Q S DIS + + + N +
Sbjct: 94 IPEIPESISFCKALQVADFSGNPLTRLPESFPE-LQNLTCLSVNDISLQSLPENIGNLY- 151
Query: 454 DLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPN------LRNLD 507
N+ + L N L +Q +L+ LDL NN + LP + L++L
Sbjct: 152 ----NLASLELRENLLTYLPDSLTQLRRLEELDLGNNE----IYNLPESIGALLHLKDLW 203
Query: 508 LSSNLFYGT---ISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNF 564
L N I ++ +LC + S L+ +SG+ T+ T+++I N
Sbjct: 204 LDGNQLSELPQEIGNLKNLLCLDVSENRLERLPEEISGL-----TSLTDLVI----SQNL 254
Query: 565 IGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRG---------- 614
+ +IPD G LK L +L + N L+ ++PE + C+ LT L L N+L
Sbjct: 255 LETIPDGIGKLKKLSILKVDQNRLT-QLPEAVGECESLTELVLTENQLLTLPKSIGKLKK 313
Query: 615 ------------PIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQL 661
+P IG + + + N IP + Q LH+LD++ N+L
Sbjct: 314 LSNLNADRNKLVSLPKEIGGCCSLTVFCVRDNRL--TRIPAEVSQATELHVLDVAGNRL 370
Score = 53.1 bits (126), Expect = 3e-06
Identities = 89/338 (26%), Positives = 138/338 (40%), Gaps = 61/338 (18%)
Query: 115 LSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNL 174
LS ND ++P+ + NL L+L N L D+L L LE LDL +N Y NL
Sbjct: 133 LSVNDISLQSLPENIGNLYNLASLELRE-NLLTYLPDSLTQLRRLEELDLGNNEIY--NL 189
Query: 175 KWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNF 234
I +LH L L L G QL +LP N
Sbjct: 190 P--------------------------ESIGALLH-LKDLWLDGNQLSELPQEIG---NL 219
Query: 235 DSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKN 294
+L+ LD+S N +P+ + L +L +S N L+ I I ++ L+IL + +N
Sbjct: 220 KNLLCLDVSENRLE-RLPEEI-SGLTSLTDLVISQNLLE-TIPDGIGKLKKLSILKVDQN 276
Query: 295 SLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERS 354
L L P + +L L L+ N L ++P ++G+ L L N+L SL +
Sbjct: 277 RLTQL-PEAVGECESLTELVLTENQLL-TLPKSIGK---LKKLSNLNADRNKL-VSLPKE 330
Query: 355 IYQLSNLVVLNLAVNNMEGIISDV------HLANFSNLKVLDLSFNHVTLNMSKNWVP-- 406
I +L V + N + I ++V H+ + + ++L L + L + W+
Sbjct: 331 IGGCCSLTVFCVRDNRLTRIPAEVSQATELHVLDVAGNRLLHLPLSLTALKLKALWLSDN 390
Query: 407 ------PFQLET-----IGLANCHLGPQFPKWIQTQKN 433
FQ +T + C L PQ P Q+N
Sbjct: 391 QSQPLLTFQTDTDYTTGEKILTCVLLPQLPSEPTCQEN 428
Score = 51.6 bits (122), Expect = 1e-05
Identities = 58/185 (31%), Positives = 89/185 (47%), Gaps = 15/185 (8%)
Query: 214 LRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQ 273
LR G +++ PP NF LV LD+S N IP+ + C LQ + S N L
Sbjct: 61 LRKLGLSDNEIQRLPPEIANFMQLVELDVSRNEI-PEIPESI-SFCKALQVADFSGNPLT 118
Query: 274 --GQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQD 331
+ ++ +T L++ D+S SL P L NL +L+L N+L+ +P +L Q
Sbjct: 119 RLPESFPELQNLTCLSVNDISLQSL----PENIGNLYNLASLELRENLLT-YLPDSLTQ- 172
Query: 332 HGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDL 391
L+EL L N++ +L SI L +L L L N + + ++ N NL LD+
Sbjct: 173 --LRRLEELDLGNNEIY-NLPESIGALLHLKDLWLDGNQLSELPQEI--GNLKNLLCLDV 227
Query: 392 SFNHV 396
S N +
Sbjct: 228 SENRL 232
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 95.1 bits (235), Expect = 8e-19
Identities = 116/420 (27%), Positives = 190/420 (44%), Gaps = 31/420 (7%)
Query: 432 KNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNS 491
K + I I ++G+ + +LS +E + L N + S L+ L LSNN+
Sbjct: 64 KRVTRIQIGHSGLQGTLSPDLRNLS-ELERLELQWNNISGPVPSLSGLASLQVLMLSNNN 122
Query: 492 F-SCPLPRLP--PNLRNLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCW 548
F S P +L+++++ +N F + E L ++L+N + N+SG +P
Sbjct: 123 FDSIPSDVFQGLTSLQSVEIDNNPFKSW--EIPESLRNASALQNFSANSANVSGSLPGFL 180
Query: 549 --TNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLN 606
+ IL+LA NN G +P S + L + L+G I L+N L +
Sbjct: 181 GPDEFPGLSILHLAFNNLEGELPMSLAG-SQVQSLWLNGQKLTGDIT-VLQNMTGLKEVW 238
Query: 607 LKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIP 666
L SN+ GP+P + G ++ L L L +NSF +P +L L+SL +++L+ N L G +P
Sbjct: 239 LHSNKFSGPLPDFSG--LKELESLSLRDNSFTGPVPASLLSLESLKVVNLTNNHLQGPVP 296
Query: 667 RCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEIL 726
VF + + + + + + E P R K LLI F+ E
Sbjct: 297 --VFKSSVSVDLDKDSNSFCLSSPGECDP----RVKSLLLIASS-----FDYPPRLAESW 345
Query: 727 KMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSC 786
K D +N++ I G + +S L + +L G+I G ++SL + L NNL+
Sbjct: 346 KGNDPCTNWIG--IACSNGNITVIS---LEKMELTGTISPEFGAIKSLQRIILGINNLTG 400
Query: 787 EIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVFYKGNPHLCGPPLRKACPRNSS 846
IP + + L LD+S N L GK+P +S V GNP + + P +SS
Sbjct: 401 MIPQELTTLPNLKTLDVSSNKLFGKVP---GFRSNVVVNTNGNPDIGKDKSSLSSPGSSS 457
Score = 77.8 bits (190), Expect = 1e-13
Identities = 100/399 (25%), Positives = 164/399 (41%), Gaps = 48/399 (12%)
Query: 208 LHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNL 267
L +L L Q + + P SL L LS NNF+ +IP +F+ LQ++ +
Sbjct: 84 LRNLSELERLELQWNNISGPVPSLSGLASLQVLMLSNNNFD-SIPSDVFQGLTSLQSVEI 142
Query: 268 SNNNLQG-QISYSIERVTTLAILDLSKNSLNGLIPNFF--DKLVNLVALDLSYNMLSGSI 324
NN + +I S+ + L + +++G +P F D+ L L L++N L G +
Sbjct: 143 DNNPFKSWEIPESLRNASALQNFSANSANVSGSLPGFLGPDEFPGLSILHLAFNNLEGEL 202
Query: 325 PSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFS 384
P +L Q+ L L+ +L G + + ++ L + L N G + D +
Sbjct: 203 PMSLAGSQVQS----LWLNGQKLTGDIT-VLQNMTGLKEVWLHSNKFSGPLPD--FSGLK 255
Query: 385 NLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGV 444
L+ L L N T + + + L+ + L N HL P + K+ +D+
Sbjct: 256 ELESLSLRDNSFTGPVPASLLSLESLKVVNLTNNHLQGPVPVF----KSSVSVDL----- 306
Query: 445 SDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLR 504
D N F SP C ++K+L L +SF P PRL + +
Sbjct: 307 -DKDSNSFCLSSPG------------ECDP------RVKSLLLIASSFDYP-PRLAESWK 346
Query: 505 NLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNF 564
D +N I C N ++ + L L+G I + ++ + L +NN
Sbjct: 347 GNDPCTNWI--------GIACSNGNITVISLEKMELTGTISPEFGAIKSLQRIILGINNL 398
Query: 565 IGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLT 603
G IP +L NL L + +N L GK+P N V T
Sbjct: 399 TGMIPQELTTLPNLKTLDVSSNKLFGKVPGFRSNVVVNT 437
Score = 72.0 bits (175), Expect = 7e-12
Identities = 96/396 (24%), Positives = 173/396 (43%), Gaps = 34/396 (8%)
Query: 245 NNFNMTIPD---WLFENC---HHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNG 298
++F + PD W C + + + ++ LQG +S + ++ L L+L N+++G
Sbjct: 43 SSFGWSDPDPCKWTHIVCTGTKRVTRIQIGHSGLQGTLSPDLRNLSELERLELQWNNISG 102
Query: 299 LIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNG-SLERSIYQ 357
+P+ L +L L LS N SIPS + Q G SL+ + + N + S+
Sbjct: 103 PVPS-LSGLASLQVLMLSNNNFD-SIPSDVFQ--GLTSLQSVEIDNNPFKSWEIPESLRN 158
Query: 358 LSNLVVLNLAVNNMEGIISD-VHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLA 416
S L + N+ G + + F L +L L+FN++ + + + Q++++ L
Sbjct: 159 ASALQNFSANSANVSGSLPGFLGPDEFPGLSILHLAFNNLEGELPMS-LAGSQVQSLWLN 217
Query: 417 NCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCG--- 473
L +Q + + + S +P D S E +LS + G
Sbjct: 218 GQKLTGDITV-LQNMTGLKEVWLHSNKFSGPLP----DFSGLKELESLSLRDNSFTGPVP 272
Query: 474 QDFSQKFKLKTLDLSNNSFSCPLPRLPPNLR-NLDLSSNLFYGTISHVCE-----ILCFN 527
LK ++L+NN P+P ++ +LD SN F + C+ +L
Sbjct: 273 ASLLSLESLKVVNLTNNHLQGPVPVFKSSVSVDLDKDSNSFCLSSPGECDPRVKSLLLIA 332
Query: 528 NSLENLDLSFNNLSGVIP-NCW-----TNGTNMIILNLAMNNFIGSIPDSFGSLKNLHML 581
+S + + G P W +NG N+ +++L G+I FG++K+L +
Sbjct: 333 SSFDYPPRLAESWKGNDPCTNWIGIACSNG-NITVISLEKMELTGTISPEFGAIKSLQRI 391
Query: 582 IMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIP 617
I+ NNL+G IP+ L L L++ SN+L G +P
Sbjct: 392 ILGINNLTGMIPQELTTLPNLKTLDVSSNKLFGKVP 427
Score = 71.2 bits (173), Expect = 1e-11
Identities = 77/283 (27%), Positives = 133/283 (46%), Gaps = 38/283 (13%)
Query: 585 NNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKT 644
++ L G + L+N L L L+ N + GP+P G + L VL+L NN+FD +IP
Sbjct: 73 HSGLQGTLSPDLRNLSELERLELQWNNISGPVPSLSG--LASLQVLMLSNNNFD-SIPSD 129
Query: 645 LCQ-LKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMT-----IEESLPIYL 698
+ Q L SL +++ N F + ES+ S ++ + + SLP +L
Sbjct: 130 VFQGLTSLQSVEIDNNP---------FKSWEIPESLRNASALQNFSANSANVSGSLPGFL 180
Query: 699 SRTKHPLLIPWKGVNVFFN--EGRLFFEI----LKMIDLSSNFLTHEIPVEIGKLVELSA 752
+ P L +++ FN EG L + ++ + L+ LT +I V + + L
Sbjct: 181 GPDEFPGL---SILHLAFNNLEGELPMSLAGSQVQSLWLNGQKLTGDITV-LQNMTGLKE 236
Query: 753 LNLSRNQLLGSIP--SSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSG 810
+ L N+ G +P S + ELESL++ D N+ + +P S+ +++ L ++L+ N L G
Sbjct: 237 VWLHSNKFSGPLPDFSGLKELESLSLRD---NSFTGPVPASLLSLESLKVVNLTNNHLQG 293
Query: 811 KIPIGNQMQSFD-----EVFYKGNPHLCGPPLRKACPRNSSFE 848
+P+ S D F +P C P ++ SSF+
Sbjct: 294 PVPVFKSSVSVDLDKDSNSFCLSSPGECDPRVKSLLLIASSFD 336
Score = 60.5 bits (145), Expect = 2e-08
Identities = 102/409 (24%), Positives = 163/409 (38%), Gaps = 70/409 (17%)
Query: 40 EAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKGVGCNTTT-------GH-----VISLNL 87
+ A+L K+ L PS+ W D C+W + C T GH +S +L
Sbjct: 28 DLSAMLSLKKSLNPPSSF--GWSD-PDPCKWTHIVCTGTKRVTRIQIGHSGLQGTLSPDL 84
Query: 88 HCSNSLDKLQGHLNS------SLLQLPYLSYLNLSGNDF-------------MQST---- 124
+ L++L+ N+ SL L L L LS N+F +QS
Sbjct: 85 RNLSELERLELQWNNISGPVPSLSGLASLQVLMLSNNNFDSIPSDVFQGLTSLQSVEIDN 144
Query: 125 -------VPDFLSTTKNLKHLDLSHANFKGNLLDNLG--NLSLLESLDLSDNSFYVNNLK 175
+P+ L L++ + AN G+L LG L L L+ N+
Sbjct: 145 NPFKSWEIPESLRNASALQNFSANSANVSGSLPGFLGPDEFPGLSILHLAFNNLEGELPM 204
Query: 176 WLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFD 235
L G S ++ L L+G L+ DI +L ++ L+ +K P
Sbjct: 205 SLAG-SQVQSLWLNGQKLT-------GDI-TVLQNMTGLKEVWLHSNKFSGPLPDFSGLK 255
Query: 236 SLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNS 295
L +L L N+F +P L + L+ +NL+NN+LQG + + + + LD NS
Sbjct: 256 ELESLSLRDNSFTGPVPASLL-SLESLKVVNLTNNHLQGPV--PVFKSSVSVDLDKDSNS 312
Query: 296 LNGLIPNFFD---KLVNLVALDLSY-----NMLSGSIPST--LGQDHGQNSLKELRLSIN 345
P D K + L+A Y G+ P T +G ++ + L
Sbjct: 313 FCLSSPGECDPRVKSLLLIASSFDYPPRLAESWKGNDPCTNWIGIACSNGNITVISLEKM 372
Query: 346 QLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFN 394
+L G++ + +L + L +NN+ G+I L NLK LD+S N
Sbjct: 373 ELTGTISPEFGAIKSLQRIILGINNLTGMIPQ-ELTTLPNLKTLDVSSN 420
Score = 42.7 bits (99), Expect = 0.004
Identities = 33/109 (30%), Positives = 47/109 (42%), Gaps = 9/109 (8%)
Query: 42 EALLEFKEGLKDPSNLLSSWKHGKDCCQWKGVGCNTTTGHVISL-NLHCSNSLDKLQGHL 100
++LL P L SWK C W G+ C+ VISL + + ++ G +
Sbjct: 326 KSLLLIASSFDYPPRLAESWKGNDPCTNWIGIACSNGNITVISLEKMELTGTISPEFGAI 385
Query: 101 NSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNL 149
S LQ L NL+G +P L+T NLK LD+S G +
Sbjct: 386 KS--LQRIILGINNLTG------MIPQELTTLPNLKTLDVSSNKLFGKV 426
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 111,864,004
Number of Sequences: 164201
Number of extensions: 4887462
Number of successful extensions: 17382
Number of sequences better than 10.0: 363
Number of HSP's better than 10.0 without gapping: 179
Number of HSP's successfully gapped in prelim test: 187
Number of HSP's that attempted gapping in prelim test: 11484
Number of HSP's gapped (non-prelim): 2370
length of query: 938
length of database: 59,974,054
effective HSP length: 120
effective length of query: 818
effective length of database: 40,269,934
effective search space: 32940806012
effective search space used: 32940806012
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 71 (32.0 bits)
Medicago: description of AC124958.5


