

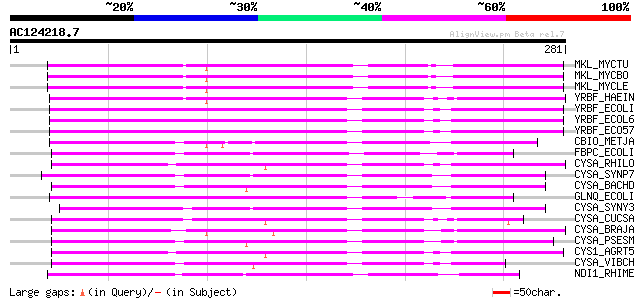
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124218.7 + phase: 0 /partial
(281 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
MKL_MYCTU (P63357) Possible ribonucleotide transport ATP-binding... 160 4e-39
MKL_MYCBO (P63358) Possible ribonucleotide transport ATP-binding... 160 4e-39
MKL_MYCLE (P30769) Possible ribonucleotide transport ATP-binding... 159 7e-39
YRBF_HAEIN (P45031) Probable ABC transporter ATP-binding protein... 155 1e-37
YRBF_ECOLI (P63386) Hypothetical ABC transporter ATP-binding pro... 152 1e-36
YRBF_ECOL6 (P63387) Hypothetical ABC transporter ATP-binding pro... 152 1e-36
YRBF_ECO57 (P63388) Hypothetical ABC transporter ATP-binding pro... 152 1e-36
CBIO_METJA (Q58488) Probable cobalt transport ATP-binding protei... 139 6e-33
FBPC_ECOLI (P37009) Ferric cations import ATP-binding protein fb... 137 3e-32
CYSA_RHILO (Q98K23) Sulfate/thiosulfate import ATP-binding prote... 137 4e-32
CYSA_SYNP7 (P14788) Sulfate/thiosulfate import ATP-binding prote... 135 9e-32
CYSA_BACHD (Q9K876) Sulfate/thiosulfate import ATP-binding prote... 135 1e-31
GLNQ_ECOLI (P10346) Glutamine transport ATP-binding protein glnQ 135 1e-31
CYSA_SYNY3 (P74548) Sulfate/thiosulfate import ATP-binding prote... 135 1e-31
CYSA_CUCSA (Q9G4F5) Sulfate/thiosulfate import ATP-binding prote... 135 1e-31
CYSA_BRAJA (Q89UD2) Sulfate/thiosulfate import ATP-binding prote... 135 1e-31
CYSA_PSESM (Q88AS5) Sulfate/thiosulfate import ATP-binding prote... 134 2e-31
CYS1_AGRT5 (Q8UH62) Sulfate/thiosulfate import ATP-binding prote... 134 3e-31
CYSA_VIBCH (Q9KUI0) Sulfate/thiosulfate import ATP-binding prote... 132 7e-31
NDI1_RHIME (O52618) Nod factor export ATP-binding protein I (Nod... 132 1e-30
>MKL_MYCTU (P63357) Possible ribonucleotide transport ATP-binding
protein mkl
Length = 359
Score = 160 bits (404), Expect = 4e-39
Identities = 99/263 (37%), Positives = 144/263 (54%), Gaps = 19/263 (7%)
Query: 20 VLIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVY 79
V IE + KSFG +I V+ I GE ++GPSGTGKS LK + GLL P++G +
Sbjct: 26 VSIEVNGLTKSFGSSRIWEDVTLTIPAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGSII 85
Query: 80 IRGRKRDGLISDDEISGLR--IGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELV 137
I G S E+ +R G++FQ ALF S+ + +N F L EH+ E EI ++V
Sbjct: 86 IDGTDIIEC-SAKELYEIRTLFGVLFQDGALFGSMNLYDNTAFPLREHTKKKESEIRDIV 144
Query: 138 KETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPI 197
E LA VGL G E + P E+SGGM+KR LAR+++ D P+++L DEP +GLDP+
Sbjct: 145 MEKLALVGLGGDEKKFPGEISGGMRKRAGLARALVLD-------PQIILCDEPDSGLDPV 197
Query: 198 ASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMT 257
+ + LI ++ + DA + ++VTH + + D + L + LV G
Sbjct: 198 RTAYLSQLIMDINAQ-IDA--------TILIVTHNINIARTVPDNMGMLFRKHLVMFGPR 248
Query: 258 HEFTTSTNPIVQQFASGSLDGPI 280
TS P+V+QF +G GPI
Sbjct: 249 EVLLTSDEPVVRQFLNGRRIGPI 271
>MKL_MYCBO (P63358) Possible ribonucleotide transport ATP-binding
protein mkl
Length = 359
Score = 160 bits (404), Expect = 4e-39
Identities = 99/263 (37%), Positives = 144/263 (54%), Gaps = 19/263 (7%)
Query: 20 VLIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVY 79
V IE + KSFG +I V+ I GE ++GPSGTGKS LK + GLL P++G +
Sbjct: 26 VSIEVNGLTKSFGSSRIWEDVTLTIPAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGSII 85
Query: 80 IRGRKRDGLISDDEISGLR--IGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELV 137
I G S E+ +R G++FQ ALF S+ + +N F L EH+ E EI ++V
Sbjct: 86 IDGTDIIEC-SAKELYEIRTLFGVLFQDGALFGSMNLYDNTAFPLREHTKKKESEIRDIV 144
Query: 138 KETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPI 197
E LA VGL G E + P E+SGGM+KR LAR+++ D P+++L DEP +GLDP+
Sbjct: 145 MEKLALVGLGGDEKKFPGEISGGMRKRAGLARALVLD-------PQIILCDEPDSGLDPV 197
Query: 198 ASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMT 257
+ + LI ++ + DA + ++VTH + + D + L + LV G
Sbjct: 198 RTAYLSQLIMDINAQ-IDA--------TILIVTHNINIARTVPDNMGMLFRKHLVMFGPR 248
Query: 258 HEFTTSTNPIVQQFASGSLDGPI 280
TS P+V+QF +G GPI
Sbjct: 249 EVLLTSDEPVVRQFLNGRRIGPI 271
>MKL_MYCLE (P30769) Possible ribonucleotide transport ATP-binding
protein mkl
Length = 347
Score = 159 bits (402), Expect = 7e-39
Identities = 98/263 (37%), Positives = 144/263 (54%), Gaps = 19/263 (7%)
Query: 20 VLIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVY 79
V IE + + KSFG +I V+ I GE ++GPSGTGKS LK + GLL P++G +
Sbjct: 14 VAIEVKGLTKSFGSSRIWEDVTLDIPAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGSIL 73
Query: 80 IRGRKRDGLISDDEISGLR--IGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELV 137
I G S E+ +R G++FQ ALF S+ + +N F L EH+ E EI ++V
Sbjct: 74 IDGTDIIEC-SAKELYEIRTLFGVLFQDGALFGSMNLYDNTAFPLREHTKKKESEIRDIV 132
Query: 138 KETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPI 197
E L VGL G E + P E+SGGM+KR LAR+++ D P+++L DEP +GLDP+
Sbjct: 133 MEKLQLVGLGGDEKKFPGEISGGMRKRAGLARALVLD-------PQIILCDEPDSGLDPV 185
Query: 198 ASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMT 257
+ + LI ++ + DA + ++VTH + + D + L + LV G
Sbjct: 186 RTAYLSQLIMDINAQ-IDA--------TILIVTHNVNIARTVPDNMGMLFRKHLVMFGPR 236
Query: 258 HEFTTSTNPIVQQFASGSLDGPI 280
TS P+V+QF +G GPI
Sbjct: 237 EVLLTSDEPVVRQFLNGRRIGPI 259
>YRBF_HAEIN (P45031) Probable ABC transporter ATP-binding protein
HI1087
Length = 264
Score = 155 bits (392), Expect = 1e-37
Identities = 93/263 (35%), Positives = 158/263 (59%), Gaps = 19/263 (7%)
Query: 21 LIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYI 80
LIE +++ G++ I + ++ +++ G+ I+GPSG GK+T+LK+I G L P++GE+
Sbjct: 5 LIEVKNLTFKRGDRVIYDNLNLQVKKGKITAIMGPSGIGKTTLLKLIGGQLMPEQGEILF 64
Query: 81 RGRKRDGLISDDEISGLR--IGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVK 138
G+ L S+ E+ +R +G++FQS ALF ++ +NV F + EH+ +PE I ++V
Sbjct: 65 DGQDICRL-SNRELYEVRKRMGMLFQSGALFTDISTFDNVAFPIREHTHLPENLIRQIVL 123
Query: 139 ETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIA 198
L AVGL+G +PSELSGGM +R ALAR+I +++P+++++DEP G DPI+
Sbjct: 124 MKLEAVGLRGAAALMPSELSGGMARRAALARAI-------ALDPDLIMFDEPFTGQDPIS 176
Query: 199 STVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTH 258
V+ LI+ ++ +AL N++S +VV+H + D + K++ EG +
Sbjct: 177 MGVILSLIKRLN----EAL----NLTS-IVVSHDVEEVLSIADYAYIIADQKVIAEGTSE 227
Query: 259 EFTTSTNPIVQQFASGSLDGPIK 281
+ S + V QF G DGP++
Sbjct: 228 QLLQSQDLRVVQFLKGESDGPVR 250
>YRBF_ECOLI (P63386) Hypothetical ABC transporter ATP-binding
protein yrbF
Length = 269
Score = 152 bits (383), Expect = 1e-36
Identities = 92/261 (35%), Positives = 140/261 (53%), Gaps = 17/261 (6%)
Query: 21 LIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYI 80
L++ RDV + G + I + +S + G+ I+GPSG GK+T+L++I G +APD GE+
Sbjct: 8 LVDMRDVSFTRGNRCIFDNISLTVPRGKITAIMGPSGIGKTTLLRLIGGQIAPDHGEILF 67
Query: 81 RGRKRDGLISDDEIS-GLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKE 139
G + + R+ ++FQS ALF + V +NV + L EH+ +P + V
Sbjct: 68 DGENIPAMSRSRLYTVRKRMSMLFQSGALFTDMNVFDNVAYPLREHTQLPAPLLHSTVMM 127
Query: 140 TLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIAS 199
L AVGL+G +PSELSGGM +R ALAR+I ++EP+++++DEP G DPI
Sbjct: 128 KLEAVGLRGAAKLMPSELSGGMARRAALARAI-------ALEPDLIMFDEPFVGQDPITM 180
Query: 200 TVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHE 259
V+ LI ++ ALG + VVV+H + D L K+V G
Sbjct: 181 GVLVKLISELN----SALG-----VTCVVVSHDVPEVLSIADHAWILADKKIVAHGSAQA 231
Query: 260 FTTSTNPIVQQFASGSLDGPI 280
+ +P V+QF G DGP+
Sbjct: 232 LQANPDPRVRQFLDGIADGPV 252
>YRBF_ECOL6 (P63387) Hypothetical ABC transporter ATP-binding
protein yrbF
Length = 269
Score = 152 bits (383), Expect = 1e-36
Identities = 92/261 (35%), Positives = 140/261 (53%), Gaps = 17/261 (6%)
Query: 21 LIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYI 80
L++ RDV + G + I + +S + G+ I+GPSG GK+T+L++I G +APD GE+
Sbjct: 8 LVDMRDVSFTRGNRCIFDNISLTVPRGKITAIMGPSGIGKTTLLRLIGGQIAPDHGEILF 67
Query: 81 RGRKRDGLISDDEIS-GLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKE 139
G + + R+ ++FQS ALF + V +NV + L EH+ +P + V
Sbjct: 68 DGENIPAMSRSRLYTVRKRMSMLFQSGALFTDMNVFDNVAYPLREHTQLPAPLLHSTVMM 127
Query: 140 TLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIAS 199
L AVGL+G +PSELSGGM +R ALAR+I ++EP+++++DEP G DPI
Sbjct: 128 KLEAVGLRGAAKLMPSELSGGMARRAALARAI-------ALEPDLIMFDEPFVGQDPITM 180
Query: 200 TVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHE 259
V+ LI ++ ALG + VVV+H + D L K+V G
Sbjct: 181 GVLVKLISELN----SALG-----VTCVVVSHDVPEVLSIADHAWILADKKIVAHGSAQA 231
Query: 260 FTTSTNPIVQQFASGSLDGPI 280
+ +P V+QF G DGP+
Sbjct: 232 LQANPDPRVRQFLDGIADGPV 252
>YRBF_ECO57 (P63388) Hypothetical ABC transporter ATP-binding
protein yrbF
Length = 269
Score = 152 bits (383), Expect = 1e-36
Identities = 92/261 (35%), Positives = 140/261 (53%), Gaps = 17/261 (6%)
Query: 21 LIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYI 80
L++ RDV + G + I + +S + G+ I+GPSG GK+T+L++I G +APD GE+
Sbjct: 8 LVDMRDVSFTRGNRCIFDNISLTVPRGKITAIMGPSGIGKTTLLRLIGGQIAPDHGEILF 67
Query: 81 RGRKRDGLISDDEIS-GLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKE 139
G + + R+ ++FQS ALF + V +NV + L EH+ +P + V
Sbjct: 68 DGENIPAMSRSRLYTVRKRMSMLFQSGALFTDMNVFDNVAYPLREHTQLPAPLLHSTVMM 127
Query: 140 TLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIAS 199
L AVGL+G +PSELSGGM +R ALAR+I ++EP+++++DEP G DPI
Sbjct: 128 KLEAVGLRGAAKLMPSELSGGMARRAALARAI-------ALEPDLIMFDEPFVGQDPITM 180
Query: 200 TVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHE 259
V+ LI ++ ALG + VVV+H + D L K+V G
Sbjct: 181 GVLVKLISELN----SALG-----VTCVVVSHDVPEVLSIADHAWILADKKIVAHGSAQA 231
Query: 260 FTTSTNPIVQQFASGSLDGPI 280
+ +P V+QF G DGP+
Sbjct: 232 LQANPDPRVRQFLDGIADGPV 252
>CBIO_METJA (Q58488) Probable cobalt transport ATP-binding protein
cbiO
Length = 279
Score = 139 bits (351), Expect = 6e-33
Identities = 88/254 (34%), Positives = 147/254 (57%), Gaps = 30/254 (11%)
Query: 21 LIECRDVYKSFGE-KKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVY 79
++E +D+Y + + +L G++FK++ GE V ++GP+G GKST+ G+L P KGEV
Sbjct: 3 IVETKDLYFRYPDGTAVLKGINFKVKKGEMVSLLGPNGAGKSTLFLHFNGILRPTKGEVL 62
Query: 80 IRGRKRDGLISDDEISGLR----IGLVFQSA--ALFDSLTVRENVGFLLYEHSSMPEEEI 133
I+G+ I D+ S + +GLVFQ+ +F + TV+E+V F + +P+EE+
Sbjct: 63 IKGKP----IKYDKKSLVEVRKTVGLVFQNPDDQIF-APTVKEDVAFGPL-NLGLPKEEV 116
Query: 134 SELVKETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAG 193
+ VKE L AVG++G EN+ P LSGG KKRVA+A + +++PEV++ DEPTAG
Sbjct: 117 EKRVKEALKAVGMEGFENKPPHHLSGGQKKRVAIAGIL-------AMQPEVIVLDEPTAG 169
Query: 194 LDPIASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVW 253
LDP+ ++ + L+ ++ KG + ++ TH + D++ ++ GK++
Sbjct: 170 LDPVGASKIMKLLYDLNKKG----------MTIIISTHDVDLVPVYADKVYVMYDGKILK 219
Query: 254 EGMTHEFTTSTNPI 267
EG E + I
Sbjct: 220 EGTPKEVFSDVETI 233
>FBPC_ECOLI (P37009) Ferric cations import ATP-binding protein fbpC
(EC 3.6.3.30)
Length = 348
Score = 137 bits (345), Expect = 3e-32
Identities = 86/234 (36%), Positives = 136/234 (57%), Gaps = 21/234 (8%)
Query: 22 IECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIR 81
+E R+V K FG +++ ++ I G+ V ++GPSG GK+TIL+++AGL P +G+++I
Sbjct: 7 VELRNVTKRFGSNTVIDNINLTIPQGQMVTLLGPSGCGKTTILRLVAGLEKPSEGQIFID 66
Query: 82 GRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKETL 141
G ++ I I +VFQS ALF +++ ENVG+ L + +P E+ VKE L
Sbjct: 67 GED----VTHRSIQQRDICMVFQSYALFPHMSLGENVGYGL-KMLGVPRAELKARVKEAL 121
Query: 142 AAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIASTV 201
A V L+G E+R ++SGG ++RVALAR++I ++P+VLL+DEP + LD
Sbjct: 122 AMVDLEGFEDRFVDQISGGQQQRVALARALI-------LKPKVLLFDEPLSNLDANLRRS 174
Query: 202 VEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEG 255
+ D IR L K +I+S + VTH S D +L ++KG ++ G
Sbjct: 175 MRDKIRE--------LQKQFDITS-LYVTHDQSEAFAVSDTVLVMNKGHIMQIG 219
>CYSA_RHILO (Q98K23) Sulfate/thiosulfate import ATP-binding protein
cysA (EC 3.6.3.25) (Sulfate-transporting ATPase)
Length = 346
Score = 137 bits (344), Expect = 4e-32
Identities = 92/264 (34%), Positives = 136/264 (50%), Gaps = 24/264 (9%)
Query: 22 IECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIR 81
+ +V K F L+ VS I+ GE + ++GPSG+GK+T+L++IAGL P +G+++
Sbjct: 3 VRVANVRKEFERFPALHDVSLDIKSGELIALLGPSGSGKTTLLRLIAGLERPTRGKIFF- 61
Query: 82 GRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGF-LLYEHSSM--PEEEISELVK 138
D S I +G VFQ ALF +TV +N+GF L H S P +EI
Sbjct: 62 ---GDEDASQKSIQERNVGFVFQHYALFRHMTVADNIGFGLKVRHGSSRPPAQEIRRRAS 118
Query: 139 ETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIA 198
E L V L G+E R P++LSGG ++RVALAR++ +IEP+VLL DEP LD
Sbjct: 119 ELLDLVQLSGLEKRYPAQLSGGQRQRVALARAM-------AIEPKVLLLDEPFGALDAQV 171
Query: 199 STVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTH 258
+ +R +H D G + V VTH DR++ + +G++ G
Sbjct: 172 RRELRRWLREIH----DRTG-----HTTVFVTHDQEEALELADRVVVMSQGRIEQVGTAD 222
Query: 259 E-FTTSTNPIVQQFASGSLDGPIK 281
+ + T +P V F S P+K
Sbjct: 223 DIYDTPNSPFVYGFIGESSSLPVK 246
>CYSA_SYNP7 (P14788) Sulfate/thiosulfate import ATP-binding protein
cysA (EC 3.6.3.25) (Sulfate-transporting ATPase)
Length = 344
Score = 135 bits (341), Expect = 9e-32
Identities = 87/256 (33%), Positives = 134/256 (51%), Gaps = 22/256 (8%)
Query: 17 DSDVLIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKG 76
D V I+ V K FG + + V + G V ++GPSG+GKST+L++IAGL PD G
Sbjct: 4 DKAVGIQVSQVSKQFGSFQAVKDVDLTVETGSLVALLGPSGSGKSTLLRLIAGLEQPDSG 63
Query: 77 EVYIRGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISEL 136
+++ GR +++ + +IG VFQ ALF LTVR+N+ F L E +E++
Sbjct: 64 RIFLTGRD----ATNESVRDRQIGFVFQHYALFKHLTVRKNIAFGL-ELRKHTKEKVRAR 118
Query: 137 VKETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDP 196
V+E L V L G+ +R PS+LSGG ++RVALAR++ +++P+VLL DEP LD
Sbjct: 119 VEELLELVQLTGLGDRYPSQLSGGQRQRVALARAL-------AVQPQVLLLDEPFGALDA 171
Query: 197 IASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGM 256
+ +R +H + + V VTH D+++ ++ GK+ G
Sbjct: 172 KVRKDLRSWLRKLHDEVH---------VTTVFVTHDQEEAMEVADQIVVMNHGKVEQIGS 222
Query: 257 THE-FTTSTNPIVQQF 271
E + P V F
Sbjct: 223 PAEIYDNPATPFVMSF 238
>CYSA_BACHD (Q9K876) Sulfate/thiosulfate import ATP-binding protein
cysA (EC 3.6.3.25) (Sulfate-transporting ATPase)
Length = 357
Score = 135 bits (340), Expect = 1e-31
Identities = 86/254 (33%), Positives = 142/254 (55%), Gaps = 24/254 (9%)
Query: 22 IECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIR 81
I ++V KSFG + L ++ I GE V ++GPSG+GK+++L+IIAGL A D+G++Y
Sbjct: 3 IVIQNVSKSFGSFQALADINLSIETGELVALLGPSGSGKTSLLRIIAGLEAADQGDIYFH 62
Query: 82 GRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENV--GFLLYEHSSMP-EEEISELVK 138
K ++ + ++G VFQ ALF +TV +N+ G + P ++EI+E V+
Sbjct: 63 KDK----VTQTHAASRQVGFVFQHYALFPHMTVADNISYGLRVKPRKERPSKKEIAEKVR 118
Query: 139 ETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIA 198
E LA V L+G+++R P++LSGG ++R+ALAR++ ++EP+VLL DEP LD
Sbjct: 119 ELLALVKLEGMDDRYPAQLSGGQRQRIALARAL-------AVEPKVLLLDEPFGALDAKV 171
Query: 199 STVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTH 258
+ +R +H + + + V VTH DR++ +++GK+ G
Sbjct: 172 RKDLRKWLRKLHNEFQ---------VTSVFVTHDQEEALDVSDRVVVMNQGKIEQVGSPD 222
Query: 259 E-FTTSTNPIVQQF 271
E + +P V F
Sbjct: 223 EVYEQPKSPFVYDF 236
>GLNQ_ECOLI (P10346) Glutamine transport ATP-binding protein glnQ
Length = 240
Score = 135 bits (339), Expect = 1e-31
Identities = 84/235 (35%), Positives = 131/235 (55%), Gaps = 17/235 (7%)
Query: 21 LIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYI 80
+IE ++V K FG ++L+ + I GE V IIGPSG+GKST+L+ I L G++ +
Sbjct: 1 MIEFKNVSKHFGPTQVLHNIDLNIAQGEVVVIIGPSGSGKSTLLRCINKLEEITSGDLIV 60
Query: 81 RGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKET 140
G K + D+ + G+VFQ LF LT ENV F +EE +L +E
Sbjct: 61 DGLKVNDPKVDERLIRQEAGMVFQQFYLFPHLTALENVMFGPLRVRGANKEEAEKLAREL 120
Query: 141 LAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIAST 200
LA VGL + PSELSGG ++RVA+AR++ +++P+++L+DEPT+ LDP
Sbjct: 121 LAKVGLAERAHHYPSELSGGQQQRVAIARAL-------AVKPKMMLFDEPTSALDP---- 169
Query: 201 VVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEG 255
+R +K L + G + V+VTH+ ++ RL+F+ KG++ +G
Sbjct: 170 ----ELRHEVLKVMQDLAEEG--MTMVIVTHEIGFAEKVASRLIFIDKGRIAEDG 218
>CYSA_SYNY3 (P74548) Sulfate/thiosulfate import ATP-binding protein
cysA (EC 3.6.3.25) (Sulfate-transporting ATPase)
Length = 355
Score = 135 bits (339), Expect = 1e-31
Identities = 85/247 (34%), Positives = 132/247 (53%), Gaps = 22/247 (8%)
Query: 26 DVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIRGRKR 85
+V K FG+ L ++ ++ G+ V ++GPSG+GKST+L+ IAGL PD+G++ I G+
Sbjct: 7 NVSKQFGDFTALKDINLEVPDGKLVALLGPSGSGKSTLLRAIAGLEEPDQGQIIINGQDA 66
Query: 86 DGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKETLAAVG 145
+ +I IG VFQ ALF LT+R+N+ F L E P + E V+E L+ +
Sbjct: 67 THV----DIRKRNIGFVFQHYALFKHLTIRQNIAFGL-EIRKHPPAKTKERVEELLSLIQ 121
Query: 146 LKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIASTVVEDL 205
L+G+ NR PS+LSGG ++RVALAR++ +++P+VLL DEP LD +
Sbjct: 122 LEGLGNRYPSQLSGGQRQRVALARAL-------AVQPQVLLLDEPFGALDAKVRKELRAW 174
Query: 206 IRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHE-FTTST 264
+R +H + + V VTH D ++ + GK+ G E +
Sbjct: 175 LRKLHDEVH---------LTSVFVTHDQEEAMEVADEIVVMSNGKIEQVGTAEEIYEHPA 225
Query: 265 NPIVQQF 271
+P V F
Sbjct: 226 SPFVMGF 232
>CYSA_CUCSA (Q9G4F5) Sulfate/thiosulfate import ATP-binding protein
cysA (EC 3.6.3.25) (Sulfate-transporting ATPase)
Length = 350
Score = 135 bits (339), Expect = 1e-31
Identities = 92/250 (36%), Positives = 134/250 (52%), Gaps = 31/250 (12%)
Query: 22 IECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIR 81
IE R++ K FG+ + LN VS I GE V ++GPSG GK+T+L+IIAGL + D G V
Sbjct: 3 IEVRNLSKRFGQFRALNDVSLHIETGELVALLGPSGCGKTTLLRIIAGLESADNGSVLFA 62
Query: 82 GRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGF-LLYEHSSM--PEEEISELVK 138
G + ++ ++G VFQ ALF +TV ENV F L +H S E++I V
Sbjct: 63 GEDATSV----DVRQRQVGFVFQHYALFKHMTVFENVAFGLRVKHRSQRPSEDQIQRKVH 118
Query: 139 ETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIA 198
+ L V L + +R P++LSGG ++R+ALAR++ ++EP VLL DEP LD
Sbjct: 119 DLLGLVQLDWLADRYPAQLSGGQRQRIALARAL-------AVEPRVLLLDEPFGALDAKV 171
Query: 199 STVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKL------- 251
+ +R +H D L N++S V VTH DR++ ++ G++
Sbjct: 172 RKELRRWLRRLH----DEL----NVAS-VFVTHDQEEALEVADRVVLMNAGRIEQVGTPR 222
Query: 252 -VWEGMTHEF 260
VWEG F
Sbjct: 223 EVWEGPATPF 232
>CYSA_BRAJA (Q89UD2) Sulfate/thiosulfate import ATP-binding protein
cysA (EC 3.6.3.25) (Sulfate-transporting ATPase)
Length = 344
Score = 135 bits (339), Expect = 1e-31
Identities = 95/267 (35%), Positives = 143/267 (52%), Gaps = 30/267 (11%)
Query: 22 IECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIR 81
IE R++ K FG L+GV+ K+ +GE + ++GPSG+GK+T+L+IIAGL PD GEV
Sbjct: 3 IEVRNLVKKFGSFAALDGVNLKVDNGELLALLGPSGSGKTTLLRIIAGLDWPDSGEVSFN 62
Query: 82 GRKRDGLISDDEISGLR---IGLVFQSAALFDSLTVRENVGF-LLYEHSSMPEEE--ISE 135
G D G R +G VFQ ALF +TV ENV F L + ++ +EE I
Sbjct: 63 G-------EDALAQGARERHVGFVFQHYALFRHMTVFENVAFGLRVQPRAVRKEEARIRA 115
Query: 136 LVKETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLD 195
VKE L V L + +R PS+LSGG ++R+ALAR++ +IEP +LL DEP LD
Sbjct: 116 RVKELLDLVQLDWLADRYPSQLSGGQRQRIALARAL-------AIEPRILLLDEPFGALD 168
Query: 196 PIASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEG 255
+ +RS+H N++S + VTH +R++ + KG++ G
Sbjct: 169 AKVRKELRKWLRSLH--------HEINVTS-IFVTHDQEEALEVANRVVVMDKGRIEQIG 219
Query: 256 MTHE-FTTSTNPIVQQFASGSLDGPIK 281
+ + + V F S++ P++
Sbjct: 220 SPEDVYESPATAFVHGFIGESIELPVR 246
>CYSA_PSESM (Q88AS5) Sulfate/thiosulfate import ATP-binding protein
cysA (EC 3.6.3.25) (Sulfate-transporting ATPase)
Length = 326
Score = 134 bits (337), Expect = 2e-31
Identities = 92/258 (35%), Positives = 134/258 (51%), Gaps = 24/258 (9%)
Query: 22 IECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIR 81
IE R+V K+F K LN +S I+ GE V ++GPSG GK+T+L+IIAGL PD G +
Sbjct: 3 IEVRNVSKNFNAFKALNNISLDIQSGELVALLGPSGCGKTTLLRIIAGLETPDDGSIVFH 62
Query: 82 GRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENV--GFLLYEHSSMPEE-EISELVK 138
G +S ++ +G VFQ ALF +TV +NV G + S P E I+E V
Sbjct: 63 GED----VSGHDVRDRNVGFVFQHYALFRHMTVFDNVAFGLRMKPKSERPNETRIAEKVH 118
Query: 139 ETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIA 198
E L V L + +R P +LSGG ++R+ALAR++ ++EP+VLL DEP LD
Sbjct: 119 ELLNMVQLDWLADRYPEQLSGGQRQRIALARAL-------AVEPKVLLLDEPFGALDAKV 171
Query: 199 STVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTH 258
+ + +H + N++S V VTH DR++ ++KG + G
Sbjct: 172 RKELRRWLARLH--------EDINLTS-VFVTHDQEEAMEVADRIVVMNKGVIEQIGSPG 222
Query: 259 E-FTTSTNPIVQQFASGS 275
E + +N V F S
Sbjct: 223 EVYENPSNDFVYHFLGDS 240
>CYS1_AGRT5 (Q8UH62) Sulfate/thiosulfate import ATP-binding protein
cysA 1 (EC 3.6.3.25) (Sulfate-transporting ATPase 1)
Length = 346
Score = 134 bits (336), Expect = 3e-31
Identities = 91/263 (34%), Positives = 133/263 (49%), Gaps = 24/263 (9%)
Query: 22 IECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIR 81
++ + K F LN VS IR GE + ++GPSG+GK+T+L++IAGL P +G+++
Sbjct: 3 VKVSGITKQFDRFPALNDVSLDIRSGELIALLGPSGSGKTTLLRLIAGLEQPTQGQIFF- 61
Query: 82 GRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSM---PEEEISELVK 138
D S + +G VFQ ALF +TV +N+ F L S P+ EI V
Sbjct: 62 ---GDEDASHRTVQERNVGFVFQHYALFRHMTVADNIAFGLKVRPSASRPPKAEIRRRVS 118
Query: 139 ETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIA 198
E L V L G+E R P++LSGG ++RVALAR++ +IEP+VLL DEP LD
Sbjct: 119 ELLDMVHLTGLEKRYPTQLSGGQRQRVALARAV-------AIEPKVLLLDEPFGALDAKV 171
Query: 199 STVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTH 258
+ +R H D G + V VTH DR++ + +GK+ G
Sbjct: 172 RKELRRWLREFH----DRTG-----HTTVFVTHDQEEALELADRVVVMSQGKIEQVGTAD 222
Query: 259 E-FTTSTNPIVQQFASGSLDGPI 280
+ + T +P V F S P+
Sbjct: 223 DVYDTPNSPFVFSFIGESSSLPV 245
>CYSA_VIBCH (Q9KUI0) Sulfate/thiosulfate import ATP-binding protein
cysA (EC 3.6.3.25) (Sulfate-transporting ATPase)
Length = 376
Score = 132 bits (333), Expect = 7e-31
Identities = 86/233 (36%), Positives = 129/233 (54%), Gaps = 23/233 (9%)
Query: 22 IECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIR 81
I ++ K FG+ + L +S KI GE +G++GPSG+GK+T+L+IIAGL D G ++ R
Sbjct: 3 IRLDNISKHFGQFQALAPLSLKIEDGEMIGLLGPSGSGKTTLLRIIAGLEGADSGTIHFR 62
Query: 82 GRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLL--YEHSSMPE-EEISELVK 138
R +++ + R+G VFQ+ ALF +TV +NV F L E + P EI + VK
Sbjct: 63 NRD----VTNVHVRDRRVGFVFQNYALFRHMTVADNVAFGLQVMERAQRPNPAEIQKRVK 118
Query: 139 ETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIA 198
+ L V L + +R P +LSGG K+R+ALAR++ + +PEVLL DEP LD
Sbjct: 119 QLLEIVQLGHLAHRYPEQLSGGQKQRIALARAL-------ATQPEVLLLDEPFGALDAKV 171
Query: 199 STVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKL 251
+ +RS+H D LG + V VTH DR++ + G++
Sbjct: 172 RKELRRWLRSLH----DELG-----FTSVFVTHDQDEALELSDRVVVMSNGRI 215
>NDI1_RHIME (O52618) Nod factor export ATP-binding protein I
(Nodulation ATP-binding protein I)
Length = 355
Score = 132 bits (331), Expect = 1e-30
Identities = 84/239 (35%), Positives = 129/239 (53%), Gaps = 21/239 (8%)
Query: 20 VLIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVY 79
V I+ V KS+G+K ++NG+SF + GE G++GP+G GKSTI ++I G+ P GE+
Sbjct: 55 VAIDVASVTKSYGDKPVINGLSFTVAAGECFGLLGPNGAGKSTITRMILGMTTPGTGEIT 114
Query: 80 IRGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKE 139
+ G + S ++ +RIG+V Q L TVREN+ + + M EI ++
Sbjct: 115 VLGVP---VPSRARLARMRIGVVPQFDNLDLEFTVRENL-LVFGRYFRMSTREIEAVIPS 170
Query: 140 TLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIAS 199
L L+ + S+LSGGMK+R+ LAR++I D P++L+ DEPT GLDP A
Sbjct: 171 LLEFARLENKADARVSDLSGGMKRRLTLARALIND-------PQLLILDEPTTGLDPHAR 223
Query: 200 TVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTH 258
++ + +RS+ +G+ L + TH +R DRL L G + EG H
Sbjct: 224 HLIWERLRSLLARGKTIL----------LTTHIMEEAERLCDRLCVLEAGHKIAEGRPH 272
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.137 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,750,572
Number of Sequences: 164201
Number of extensions: 1396092
Number of successful extensions: 8445
Number of sequences better than 10.0: 997
Number of HSP's better than 10.0 without gapping: 892
Number of HSP's successfully gapped in prelim test: 105
Number of HSP's that attempted gapping in prelim test: 5573
Number of HSP's gapped (non-prelim): 1243
length of query: 281
length of database: 59,974,054
effective HSP length: 109
effective length of query: 172
effective length of database: 42,076,145
effective search space: 7237096940
effective search space used: 7237096940
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 65 (29.6 bits)
Medicago: description of AC124218.7


