

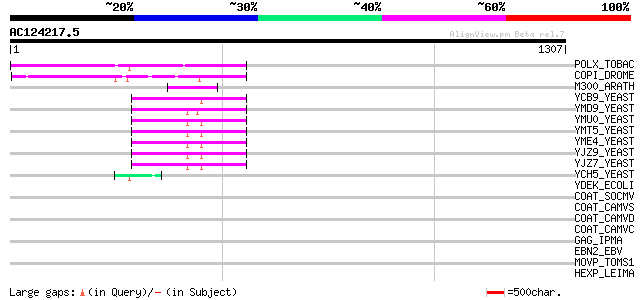
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124217.5 + phase: 0 /pseudo
(1307 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 349 2e-95
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 173 3e-42
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 105 6e-22
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 66 7e-10
YMD9_YEAST (Q03434) Transposon Ty1 protein B 55 1e-06
YMU0_YEAST (Q04670) Transposon Ty1 protein B 54 4e-06
YMT5_YEAST (Q04214) Transposon Ty1 protein B 54 4e-06
YME4_YEAST (Q04711) Transposon Ty1 protein B 54 4e-06
YJZ9_YEAST (P47100) Transposon Ty1 protein B 54 4e-06
YJZ7_YEAST (P47098) Transposon Ty1 protein B 54 4e-06
YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein 46 6e-04
YDEK_ECOLI (P32051) Hypothetical lipoprotein ydeK precursor (ORFT) 37 0.27
COAT_SOCMV (P15627) Coat protein 37 0.35
COAT_CAMVS (P03542) Coat protein 37 0.46
COAT_CAMVD (P03544) Coat protein 37 0.46
COAT_CAMVC (P03543) Coat protein 36 0.60
GAG_IPMA (P11365) Retrovirus-related Gag polyprotein [Contains: ... 36 0.79
EBN2_EBV (P12978) EBNA-2 nuclear protein 35 1.0
MOVP_TOMS1 (Q9YJQ9) Movement protein (Cell-to-cell transport pro... 35 1.3
HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding p... 35 1.3
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 349 bits (896), Expect = 2e-95
Identities = 199/572 (34%), Positives = 321/572 (55%), Gaps = 29/572 (5%)
Query: 3 GSKWDIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAMPATLTQEEKREMIDKAKSA 62
G K+++ KF G N F W+ +M+ +L QQ + L ++ P T+ E+ ++ ++A SA
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 63 IVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQL 122
I L L D V+ ++ E TA + +LESLYM+K+L ++ LK+QLY+ M E + L
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 123 TEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALRT 182
FN ++ LAN+ V E+EDKA+LLL SLP S+++ TIL+GK T L++V +AL
Sbjct: 123 NVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKT-TIELKDVTSALLL 181
Query: 183 KELTKFKDLKVDEGSEGLNVARGRNEHRGKGK-GKSRSKSRSKGFDKSKYK-CFLCHKQG 240
E K + ++G + RGR+ R G+S ++ +SK KS+ + C+ C++ G
Sbjct: 182 NE--KMRKKPENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSKSRVRNCYNCNQPG 239
Query: 241 HFKKDCPDKGGDGSPSVQVAEASNEEGYESTGALVVTSWK----------------SEKS 284
HFK+DCP+ P E S ++ ++T A+V + E
Sbjct: 240 HFKRDCPN------PRKGKGETSGQKNDDNTAAMVQNNDNVVLFINEEEECMHLSGPESE 293
Query: 285 WVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDV 344
WV+D+ S+H P ++ F + G V++GN K+ G+G++ +K G +L+DV
Sbjct: 294 WVVDTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVGCTLVLKDV 353
Query: 345 RFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIGNA 404
R VP+L+ NLIS D GY + + +++ G+L+ KG LY + I G
Sbjct: 354 RHVPDLRMNLISGIALDRDGYESYFANQKWRLTKGSLVIAKGVARGTLYRTNAEICQGEL 413
Query: 405 SVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHRVKFG 464
+ A + +LWH R+GH+SE+GL LAK+ L+ K ++ C++C+ GKQHRV F
Sbjct: 414 NAAQ--DEISVDLWHKRMGHMSEKGLQILAKKSLISYAKGTTVKPCDYCLFGKQHRVSFQ 471
Query: 465 SGMHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKKSDTF*KFKE 524
+ + + V+SD+ GP + + GG YF++ IDD SR++WV++LK K F F++
Sbjct: 472 TSSERKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTKDQVFQVFQK 531
Query: 525 *HTLIENQMGTKLKGLRTDNGLEFVSEQFNDF 556
H L+E + G KLK LR+DNG E+ S +F ++
Sbjct: 532 FHALVERETGRKLKRLRSDNGGEYTSREFEEY 563
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 173 bits (438), Expect = 3e-42
Identities = 160/581 (27%), Positives = 254/581 (43%), Gaps = 49/581 (8%)
Query: 4 SKWDIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAMPATLTQEEKREMIDKAKSAI 63
+K +I+ F G + +WK +++A+L +Q ++ + G MP + K+ AKS I
Sbjct: 4 AKRNIKPFDGEK-YAIWKFRIRALLAEQDVLKVVDG--LMPNEVDDSWKKAE-RCAKSTI 59
Query: 64 VLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQLT 123
+ L D L + TA + L+++Y KSLA + L+++L S K+ +S+
Sbjct: 60 IEYLSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLLSHFH 119
Query: 124 EFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALRTK 183
F++++ +L E+ DK LL +LP ++ I E TL V+ L +
Sbjct: 120 IFDELISELLAAGAKIEEMDKISHLLITLPSCYDGIITAIETLSEENLTLAFVKNRLLDQ 179
Query: 184 ELTKFKDLKVDEGSEGLNVARGRNEHRGKG----KGKSRSKSRSKGFDKSKYKCFLCHKQ 239
E+ K K+ D + +N N + K ++ K KG K K KC C ++
Sbjct: 180 EI-KIKNDHNDTSKKVMNAIVHNNNNTYKNNLFKNRVTKPKKIFKGNSKYKVKCHHCGRE 238
Query: 240 GHFKKDC-------PDKGGDGSPSVQVAEASNEEGYESTGALVV-----TSWKSEKSWVL 287
GH KKDC +K + VQ A + A +V TS +VL
Sbjct: 239 GHIKKDCFHYKRILNNKNKENEKQVQTATSHGI-------AFMVKEVNNTSVMDNCGFVL 291
Query: 288 DSGCSYHMCPRKEYF----ETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRD 343
DSG S H+ + + E + + V + G +G+ VRL+ + E L D
Sbjct: 292 DSGASDHLINDESLYTDSVEVVPPLKIAVAKQGEFIYATKRGI--VRLR--NDHEITLED 347
Query: 344 VRFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIGN 403
V F E NL+S+ G + IS L+ VK S M L+ VI
Sbjct: 348 VLFCKEAAGNLMSVKRLQEAGMSIEFDKSGVTISKNGLMVVKNSGM-----LNNVPVINF 402
Query: 404 ASVASVVPH-NNSELWHLRLGHVSERGLVELAKQGLLGKDKL-----DKLEFCEHCILGK 457
+ + H NN LWH R GH+S+ L+E+ ++ + L E CE C+ GK
Sbjct: 403 QAYSINAKHKNNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEICEPCLNGK 462
Query: 458 QHRVKFG--SGMHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKK 515
Q R+ F H R VHSD+ GP T +YF+ +D ++ +++K K
Sbjct: 463 QARLPFKQLKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKYK 522
Query: 516 SDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDF 556
SD F F++ E K+ L DNG E++S + F
Sbjct: 523 SDVFSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQF 563
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 105 bits (263), Expect = 6e-22
Identities = 50/117 (42%), Positives = 70/117 (59%), Gaps = 1/117 (0%)
Query: 372 GVCKISHGALITVKGSKMNGLYILDGSIVIGNASVASVVPHNNSELWHLRLGHVSERGLV 431
GV K+ G +KG++ + LYIL GS+ G +++A + + LWH RL H+S+RG+
Sbjct: 27 GVLKVLKGCRTILKGNRHDSLYILQGSVETGESNLAETAK-DETRLWHSRLAHMSQRGME 85
Query: 432 ELAKQGLLGKDKLDKLEFCEHCILGKQHRVKFGSGMHHSSRLFEYVHSDLLGPSKTP 488
L K+G L K+ L+FCE CI GK HRV F +G H + +YVHSDL G P
Sbjct: 86 LLVKKGFLDSSKVSSLKFCEDCIYGKTHRVNFSTGQHTTKNPLDYVHSDLWGAPSVP 142
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 65.9 bits (159), Expect = 7e-10
Identities = 65/292 (22%), Positives = 120/292 (40%), Gaps = 22/292 (7%)
Query: 286 VLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVR 345
++DSG S + Y T + + + +GN+ +G + ++ +
Sbjct: 455 LIDSGASQTLVRSAHYLHHATPNSEINIVDAQKQDIPINAIGNLHFNFQNGTKTSIKALH 514
Query: 346 FVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALIT--VKGSKMNGL---YILDGSI- 399
P + +L+SLS + S G ++ VK L Y++ I
Sbjct: 515 -TPNIAYDLLSLSELANQNITACFTRNTLERSDGTVLAPIVKHGDFYWLSKKYLIPSHIS 573
Query: 400 --VIGNASVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEF-------C 450
I N + + V L H LGH + R + + K+ + K +E+ C
Sbjct: 574 KLTINNVNKSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYLKESDIEWSNASTYQC 633
Query: 451 EHCILGK--QHRVKFGSGMHH--SSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRR 506
C++GK +HR GS + + S F+Y+H+D+ GP SYF+S D+ +R
Sbjct: 634 PDCLIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKSAPSYFISFTDEKTRF 693
Query: 507 VWVFVL--KKKSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDF 556
WV+ L +++ F I+NQ ++ ++ D G E+ ++ + F
Sbjct: 694 QWVYPLHDRREESILNVFTSILAFIKNQFNARVLVIQMDRGSEYTNKTLHKF 745
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 55.5 bits (132), Expect = 1e-06
Identities = 57/294 (19%), Positives = 121/294 (40%), Gaps = 22/294 (7%)
Query: 286 VLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVR 345
+LDSG S + + + + V + + +G+++ D + ++ V
Sbjct: 32 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIK-VL 90
Query: 346 FVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIGNAS 405
P + +L+SL+ + V + S G ++ + ++ ++ N S
Sbjct: 91 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGTVLAPIVKYGDFYWVSKKYLLPSNIS 150
Query: 406 VASVVPHNNSE--------LWHLRLGHVSERGLVELAKQGLL------GKDKLDKLEF-C 450
V ++ + SE H L H + + + K + D+ +++ C
Sbjct: 151 VPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDRSSAIDYQC 210
Query: 451 EHCILGK--QHRVKFGSGMHHSSRL--FEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRR 506
C++GK +HR GS + + + F+Y+H+D+ GP SYF+S D+ ++
Sbjct: 211 PDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKF 270
Query: 507 VWVFVL--KKKSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQ 558
WV+ L +++ F I+NQ + ++ D G E+ + + FL+
Sbjct: 271 RWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLE 324
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 53.5 bits (127), Expect = 4e-06
Identities = 56/294 (19%), Positives = 119/294 (40%), Gaps = 22/294 (7%)
Query: 286 VLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVR 345
+LDSG S + + + + V + + +G+++ D + ++ V
Sbjct: 32 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIK-VL 90
Query: 346 FVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIGNAS 405
P + +L+SL+ + V + S G ++ + ++ ++ N S
Sbjct: 91 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGTVLAPIVKYGDFYWVSKKYLLPSNIS 150
Query: 406 VASVVPHNNSE--------LWHLRLGHVSERGLVELAKQGLLGKDKLDKLEF-------C 450
V ++ + SE H L H + + + K + +++ C
Sbjct: 151 VPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQC 210
Query: 451 EHCILGK--QHRVKFGSGMHHSSRL--FEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRR 506
C++GK +HR GS + + + F+Y+H+D+ GP SYF+S D+ ++
Sbjct: 211 PDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKF 270
Query: 507 VWVFVL--KKKSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQ 558
WV+ L +++ F I+NQ + ++ D G E+ + + FL+
Sbjct: 271 RWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLE 324
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 53.5 bits (127), Expect = 4e-06
Identities = 56/294 (19%), Positives = 119/294 (40%), Gaps = 22/294 (7%)
Query: 286 VLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVR 345
+LDSG S + + + + V + + +G+++ D + ++ V
Sbjct: 32 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIK-VL 90
Query: 346 FVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIGNAS 405
P + +L+SL+ + V + S G ++ + ++ ++ N S
Sbjct: 91 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGTVLAPIVKYGDFYWVSKKYLLPSNIS 150
Query: 406 VASVVPHNNSE--------LWHLRLGHVSERGLVELAKQGLLGKDKLDKLEF-------C 450
V ++ + SE H L H + + + K + +++ C
Sbjct: 151 VPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQC 210
Query: 451 EHCILGK--QHRVKFGSGMHHSSRL--FEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRR 506
C++GK +HR GS + + + F+Y+H+D+ GP SYF+S D+ ++
Sbjct: 211 PDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKF 270
Query: 507 VWVFVL--KKKSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQ 558
WV+ L +++ F I+NQ + ++ D G E+ + + FL+
Sbjct: 271 RWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLE 324
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 53.5 bits (127), Expect = 4e-06
Identities = 56/294 (19%), Positives = 119/294 (40%), Gaps = 22/294 (7%)
Query: 286 VLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVR 345
+LDSG S + + + + V + + +G+++ D + ++ V
Sbjct: 32 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIK-VL 90
Query: 346 FVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIGNAS 405
P + +L+SL+ + V + S G ++ + ++ ++ N S
Sbjct: 91 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGTVLAPIVKYGDFYWVSKKYLLPSNIS 150
Query: 406 VASVVPHNNSE--------LWHLRLGHVSERGLVELAKQGLLGKDKLDKLEF-------C 450
V ++ + SE H L H + + + K + +++ C
Sbjct: 151 VPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQC 210
Query: 451 EHCILGK--QHRVKFGSGMHHSSRL--FEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRR 506
C++GK +HR GS + + + F+Y+H+D+ GP SYF+S D+ ++
Sbjct: 211 PDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKF 270
Query: 507 VWVFVL--KKKSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQ 558
WV+ L +++ F I+NQ + ++ D G E+ + + FL+
Sbjct: 271 RWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLE 324
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 53.5 bits (127), Expect = 4e-06
Identities = 56/294 (19%), Positives = 119/294 (40%), Gaps = 22/294 (7%)
Query: 286 VLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVR 345
+LDSG S + + + + V + + +G+++ D + ++ V
Sbjct: 459 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIK-VL 517
Query: 346 FVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIGNAS 405
P + +L+SL+ + V + S G ++ + ++ ++ N S
Sbjct: 518 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGTVLAPIVKYGDFYWVSKKYLLPSNIS 577
Query: 406 VASVVPHNNSE--------LWHLRLGHVSERGLVELAKQGLLGKDKLDKLEF-------C 450
V ++ + SE H L H + + + K + +++ C
Sbjct: 578 VPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQC 637
Query: 451 EHCILGK--QHRVKFGSGMHHSSRL--FEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRR 506
C++GK +HR GS + + + F+Y+H+D+ GP SYF+S D+ ++
Sbjct: 638 PDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKF 697
Query: 507 VWVFVL--KKKSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQ 558
WV+ L +++ F I+NQ + ++ D G E+ + + FL+
Sbjct: 698 RWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLE 751
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 53.5 bits (127), Expect = 4e-06
Identities = 56/294 (19%), Positives = 119/294 (40%), Gaps = 22/294 (7%)
Query: 286 VLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVR 345
+LDSG S + + + + V + + +G+++ D + ++ V
Sbjct: 459 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIK-VL 517
Query: 346 FVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIGNAS 405
P + +L+SL+ + V + S G ++ + ++ ++ N S
Sbjct: 518 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGTVLAPIVQYGDFYWVSKRYLLPSNIS 577
Query: 406 VASVVPHNNSE--------LWHLRLGHVSERGLVELAKQGLLGKDKLDKLEF-------C 450
V ++ + SE H L H + + + K + +++ C
Sbjct: 578 VPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQC 637
Query: 451 EHCILGK--QHRVKFGSGMHHSSRL--FEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRR 506
C++GK +HR GS + + + F+Y+H+D+ GP SYF+S D+ ++
Sbjct: 638 PDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKF 697
Query: 507 VWVFVL--KKKSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQ 558
WV+ L +++ F I+NQ + ++ D G E+ + + FL+
Sbjct: 698 RWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLE 751
>YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein
Length = 146
Score = 46.2 bits (108), Expect = 6e-04
Identities = 32/121 (26%), Positives = 47/121 (38%), Gaps = 15/121 (12%)
Query: 247 PDKGGDGSPSVQVAEASNEEGYESTGALVVTSW----------KSEKSWVLDSGCSYHMC 296
P+ S S VA E ++ G + SW W+ D+GC+ HMC
Sbjct: 31 PNDKTSRSSSASVAIPDYETQGQTAGQITPKSWLCMLSSTVPATKSSEWIFDTGCTSHMC 90
Query: 297 PRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVRFVPELKRNLIS 356
+ F + T G + + G G V + L DV +VP+L NLIS
Sbjct: 91 HDRSIFSSFTRSSRKDFVRGVGGSIPIMGSGTVNIGTVQ-----LHDVSYVPDLPVNLIS 145
Query: 357 L 357
+
Sbjct: 146 V 146
>YDEK_ECOLI (P32051) Hypothetical lipoprotein ydeK precursor (ORFT)
Length = 1325
Score = 37.4 bits (85), Expect = 0.27
Identities = 30/143 (20%), Positives = 64/143 (43%), Gaps = 11/143 (7%)
Query: 305 LTLKEGGVVRLGNN--KACKVQGMGNVRLKMFDGREFLLRDVRFVPELKRNLISLSMFDG 362
+++ GG+ + N V G+GN+ + DG +F+ +++ F+ + + +L++ D
Sbjct: 434 VSITTGGMWEVNKNVYTTIGVAGVGNLNIS--DGGKFVSQNITFLGDKASGIGTLNLMDA 491
Query: 363 LGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIGNASVASVVPHNNSELWHLRL 422
+ V G + G+ +N + GNAS +V + LW+L+
Sbjct: 492 TSSFDTVGINVGNFGSGIVNVSNGATLNSTGY---GFIGGNASGKGIVNISTDSLWNLKT 548
Query: 423 GHVSERGLVELAKQGLLGKDKLD 445
+ +L + G+LG +L+
Sbjct: 549 SSTN----AQLLQVGVLGTGELN 567
>COAT_SOCMV (P15627) Coat protein
Length = 441
Score = 37.0 bits (84), Expect = 0.35
Identities = 17/51 (33%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Query: 201 NVARGRNEHRGKGKGKSRS--KSRSKGFDKSKYKCFLCHKQGHFKKDCPDK 249
N + N+ + K K R K ++ K K +C+LCH++GH+ +CP K
Sbjct: 349 NFWKWNNQRKKKTFRKKRPFRKQQTCPTGKKKCQCWLCHEEGHYANECPKK 399
>COAT_CAMVS (P03542) Coat protein
Length = 489
Score = 36.6 bits (83), Expect = 0.46
Identities = 21/77 (27%), Positives = 38/77 (49%), Gaps = 5/77 (6%)
Query: 212 KGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDK-GGDGSPSVQVAEASN----EE 266
K K K SK + K +C++C+ +GH+ +CP++ + + +Q AE EE
Sbjct: 393 KPKEKKGSKQKYCPKGKKDCRCWICNIEGHYANECPNRQSSEKAHILQQAEKLGLQPIEE 452
Query: 267 GYESTGALVVTSWKSEK 283
YE + + +K E+
Sbjct: 453 PYEGVQEVFILEYKEEE 469
>COAT_CAMVD (P03544) Coat protein
Length = 490
Score = 36.6 bits (83), Expect = 0.46
Identities = 21/77 (27%), Positives = 38/77 (49%), Gaps = 5/77 (6%)
Query: 212 KGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDK-GGDGSPSVQVAEASN----EE 266
K K K SK + K +C++C+ +GH+ +CP++ + + +Q AE EE
Sbjct: 392 KPKEKKGSKQKYCPKGKKDCRCWICNIEGHYANECPNRQSSEKAHILQQAEKLGLQPIEE 451
Query: 267 GYESTGALVVTSWKSEK 283
YE + + +K E+
Sbjct: 452 PYEGVQEVFILEYKEEE 468
>COAT_CAMVC (P03543) Coat protein
Length = 488
Score = 36.2 bits (82), Expect = 0.60
Identities = 20/78 (25%), Positives = 37/78 (46%), Gaps = 7/78 (8%)
Query: 212 KGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASN------E 265
K K K SK + K +C++C+ +GH+ +CP++ + + +A N E
Sbjct: 391 KPKEKKGSKRKYCPKGKKDCRCWICNIEGHYANECPNRQSSEKAHI-LQQAENLGLQPVE 449
Query: 266 EGYESTGALVVTSWKSEK 283
E YE + + +K E+
Sbjct: 450 EPYEGVQEVFILEYKEEE 467
>GAG_IPMA (P11365) Retrovirus-related Gag polyprotein [Contains:
Protease (EC 3.4.23.-)]
Length = 827
Score = 35.8 bits (81), Expect = 0.79
Identities = 18/38 (47%), Positives = 25/38 (65%), Gaps = 5/38 (13%)
Query: 216 KSRSKSRSKGFDKSKYKCFLCHKQGHFKKDC--PDKGG 251
+S+++S S+ ++ CF C K GHFKKDC PDK G
Sbjct: 446 QSQNRSMSRNDQRT---CFNCGKPGHFKKDCRAPDKQG 480
>EBN2_EBV (P12978) EBNA-2 nuclear protein
Length = 487
Score = 35.4 bits (80), Expect = 1.0
Identities = 17/34 (50%), Positives = 19/34 (55%)
Query: 192 KVDEGSEGLNVARGRNEHRGKGKGKSRSKSRSKG 225
K S G + RGR RG+GKGKSR K R G
Sbjct: 335 KTQGQSRGQSRGRGRGRGRGRGKGKSRDKQRKPG 368
>MOVP_TOMS1 (Q9YJQ9) Movement protein (Cell-to-cell transport
protein) (30 kDa protein)
Length = 264
Score = 35.0 bits (79), Expect = 1.3
Identities = 46/211 (21%), Positives = 83/211 (38%), Gaps = 40/211 (18%)
Query: 32 KCVEALKGEAAMPATLTQEEKREMIDKAKSAIVLCLGDKVLRDVAREATAASM*AKLESL 91
K V+ ++G L + + D + + +CL DK + + A EAT S
Sbjct: 57 KGVKLIEGGYVCLVGLVVSGEWNLPDNCRGGVSVCLVDKRM-ERADEATLGS-------- 107
Query: 92 YMTKSLAHRQLLKQQLYSFKMVESISISEQLTEFN--KILVDLANIEVNTEDEDKALLLL 149
Y T + R + FK+V + I+ + E N ++LV++ N++++
Sbjct: 108 YYTAAAKKR-------FQFKVVPNYGITTKDAEKNIWQVLVNIKNVKMSAG--------- 151
Query: 150 CSLPKSFEHFKDTILYGKEGTTTLEEVQAALRTKELTKFKDLKVDEGSEGLNVA------ 203
P S E I+Y L E ++ + + VDE E + ++
Sbjct: 152 -YCPLSLEFVSVCIVYKNNTKLGLREKVTSVNDGGPMELSEEVVDEFMENVPMSVRLAKF 210
Query: 204 ------RGRNEHRGKGKGKSRSKSRSKGFDK 228
RG + GKG+S + + K FD+
Sbjct: 211 RTKSSKRGPKNNNNLGKGRSGGRPKPKSFDE 241
>HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding
protein)
Length = 271
Score = 35.0 bits (79), Expect = 1.3
Identities = 24/82 (29%), Positives = 38/82 (46%), Gaps = 9/82 (10%)
Query: 197 SEGLNVARGRNEH----RGKGK----GKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPD 248
SE +V R R E R GK + ++ SKG ++S CF C ++GH ++CP+
Sbjct: 2 SETEDVKRPRTESSTSCRNCGKEGHYARECPEADSKGDERST-TCFRCGEEGHMSRECPN 60
Query: 249 KGGDGSPSVQVAEASNEEGYES 270
+ G+ E G+ S
Sbjct: 61 EARSGAAGAMTCFRCGEAGHMS 82
Score = 33.1 bits (74), Expect = 5.1
Identities = 13/39 (33%), Positives = 20/39 (50%)
Query: 213 GKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGG 251
G G+ R +S ++G C+ C GH +DCP+ G
Sbjct: 122 GYGQKRGRSGAQGGYSGDRTCYKCGDAGHISRDCPNGQG 160
Score = 32.3 bits (72), Expect = 8.7
Identities = 14/47 (29%), Positives = 21/47 (43%), Gaps = 5/47 (10%)
Query: 205 GRNEHRGKGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGG 251
G H + S +KGF+ C+ C ++GH +DCP G
Sbjct: 76 GEAGHMSRDCPNSAKPGAAKGFE-----CYKCGQEGHLSRDCPSSQG 117
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.346 0.154 0.537
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 142,286,132
Number of Sequences: 164201
Number of extensions: 5748398
Number of successful extensions: 20618
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 20488
Number of HSP's gapped (non-prelim): 108
length of query: 1307
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1185
effective length of database: 39,941,532
effective search space: 47330715420
effective search space used: 47330715420
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 72 (32.3 bits)
Medicago: description of AC124217.5


